Abstract
Although the finding of severe acute respiratory syndrome coronavirus (SARS-CoV) in caged palm civets from live animal markets in China has provided evidence for interspecies transmission in the genesis of the SARS epidemic, subsequent studies suggested that the civet may have served only as an amplification host for SARS-CoV. In a surveillance study for CoV in noncaged animals from the wild areas of the Hong Kong Special Administration Region, we identified a CoV closely related to SARS-CoV (bat-SARS-CoV) from 23 (39%) of 59 anal swabs of wild Chinese horseshoe bats (Rhinolophus sinicus) by using RT-PCR. Sequencing and analysis of three bat-SARS-CoV genomes from samples collected at different dates showed that bat-SARS-CoV is closely related to SARS-CoV from humans and civets. Phylogenetic analysis showed that bat-SARS-CoV formed a distinct cluster with SARS-CoV as group 2b CoV, distantly related to known group 2 CoV. Most differences between the bat-SARS-CoV and SARS-CoV genomes were observed in the spike genes, ORF 3 and ORF 8, which are the regions where most variations also were observed between human and civet SARS-CoV genomes. In addition, the presence of a 29-bp insertion in ORF 8 of bat-SARS-CoV genome, not in most human SARS-CoV genomes, suggests that it has a common ancestor with civet SARS-CoV. Antibody against recombinant bat-SARS-CoV nucleocapsid protein was detected in 84% of Chinese horseshoe bats by using an enzyme immunoassay. Neutralizing antibody to human SARS-CoV also was detected in bats with lower viral loads. Precautions should be exercised in the handling of these animals.
Coronaviruses (CoV) are found in a wide variety of animals in which they can cause respiratory, enteric, hepatic, and neurological diseases of varying severity. As a result of the unique mechanism of viral replication, CoV have a high frequency of recombination (1). Their tendency for recombination and high mutation rates may allow them to adapt to new hosts and ecological niches. Among CoV that infect humans, including human CoV 229E (HCoV-229E), human CoV OC43 (HCoV-OC43), severe acute respiratory syndrome CoV (SARS-CoV), human CoV NL63 (HCoV-NL63), and CoV HKU1 (CoV-HKU1), SARS-CoV causes the most severe disease, with >700 fatalities reported since the SARS epidemic in 2003 (2-8).
The isolation of SARS-CoV from caged animals, including Himalayan palm civets and a raccoon dog, from wild live markets in mainland China suggested that these animals are the reservoir for the origin of the SARS epidemic (9). However, subsequent studies suggested that the civet may have served only as an amplification host for SARS-CoV and provided the environment for major genetic variations permitting efficient animal-to-human and human-to-human transmissions (10-13). Because civets are often mixed with different species in overcrowded conditions at markets, we conducted a surveillance study for CoV in noncaged animals from the wild areas of the Hong Kong Special Administrative Region (HKSAR). In this report, we describe the identification and molecular characterization of a SARS-CoV-related virus from Chinese horseshoe bats in Hong Kong and propose that this virus be named bat SARS CoV (bat-SARS-CoV).
Methods
Wild Animal Surveillance and Sample Collection. The study was approved by the HKSAR's Department of Agriculture, Fisheries, and Conservation and the University of Hong Kong Committee on the Use of Live Animals in Teaching and Research. From summer 2004 to spring 2005, 127 bats, 60 rodents, and 20 monkeys from 11 locations in the HKSAR were captured (Fig. 1a and Table 2, which is published as supporting information on the PNAS web site). Nasopharyngeal and anal swabs and blood samples were collected by a veterinary surgeon. Swabs were taken with sterile swabs and kept in viral transport medium at 4°C before processing (14). Where possible, blood was collected for serological studies.
Fig. 1.
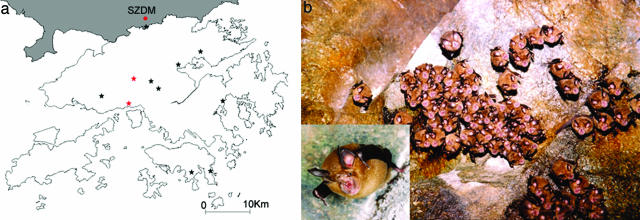
Surveillance for bat-SARS-CoV in the wild areas of the HKSAR. (a) Map of HKSAR showing locations of wild animal surveillance indicated by stars. Areas belonging to Shenzhen of mainland China are shaded. Red stars represent the two locations with bats positive for bat-SARS-CoV, both being water tunnels. Other sampling sites include abandoned mines, sea caves and forested areas. SZDM, Shenzhen Dongmen market, the nearest wildlife market with SARS-CoV-infected civets. (b) A group of Chinese horseshoe bats roosting in a water tunnel in the HKSAR. The common name describes the horseshoe-shaped area of thick skin surrounding the nostrils that covers the upper lip (close-up view)
RNA Extraction, RT-PCR, and DNA Sequencing. Nasopharyngeal and anal swabs were tested for CoV RNA by RT-PCR. Viral RNA extraction was performed by using the QIAamp viral RNA mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA was generated by using random hexamers and the SuperScript II kit (Invitrogen) as described in ref. 7.
CoV screening was performed by using conserved primers (5′-GGTTGGGACTATCCTAAGTGTGA-3′ and 5′-CCATCATCAGATAGAATCATCATA-3′) targeted to a 440-bp fragment of the pol gene as described in ref. 7. Standard precautions were taken to avoid PCR contamination, and no false-positive was observed in negative controls. The sequences of the PCR products were compared with known sequences of the pol genes of CoV in GenBank.
Viral Cultures. Attempts to isolate bat-SARS-CoV were made by inoculating RT-PCR-positive specimens to FRhK-4, HRT-18G, Huh-7, Vero E6, C6/36 and Caco-2 cells, and chicken embryonated eggs. Viral replication was detected by observation for cytopathic effects and quantitative RT-PCR described below.
Complete Genome Sequencing and Genome Analysis. The complete genome of bat-SARS-CoV was sequenced by using RNA extracted from three anal swabs from three bats (B24, B41, and B43) as template. RNA was converted to cDNA by a combined random-priming and oligo(dT)-priming strategy described in ref. 7. A total of 63 sets of primers, available on request, were used for PCR. The 5′ end of the viral genome was confirmed by rapid amplification of cDNA ends by using the 5′/3′ RACE kit (Roche Diagnostics). Sequences were assembled and manually edited to produce complete sequences of the three viral genomes, which have been deposited into GenBank (Table 3, which is published as supporting information on the PNAS web site). The nucleotide and deduced amino acid sequences were compared with those of other CoV in GenBank (Table 3) by multiple sequence alignment using clustalw software (www.ebi.ac.uk/clustalw). Phylogenetic tree construction was performed by using the neighbor-joining method with growtree software (Genetics Computer Group, Madison, WI) using Jukes-Cantor correction. Prediction of signal peptides and cleavage sites was performed by using signalp software, transmembrane domains by using tmpred and tmhmm software, potential N-glycosylation sites by using scanprosite software, and protein family analysis by using pfam and interproscan software (15-21).
Sequencing of Complete Spike (S) Genes of bat-SARS-CoV. The complete S genes of bat-SARS-CoV from 14 positive samples, with adequate amount of RNA available, were sequenced by using primers targeted to S. The nucleotide and deduced amino acid sequences were compared with those of SARS-CoV by multiple alignment.
Western Blot Analysis Using Recombinant Nucleocapsid (N) Protein of bat-SARS-CoV. Cloning and purification of (His)6-tagged recombinant N protein of bat-SARS-CoV were performed as described in ref. 22. Primers (5′-CGCGGATCCGATGTCTGATAATGGACCC-3′ and 5′-CGGAATTCTTATGCCTGAGTAGAATCA-3′) were used to amplify the N gene of bat-SARS-CoV by RT-PCR. Western blot analysis, using 900 ng of purified bat-SARS-CoV (His)6-tagged N protein and sera at 1:1,000 dilution, was performed as described in ref. 22. Antigen-antibody interaction was detected with 1:4,000 horseradish peroxidase-conjugated protein G (Zymed) and an enhanced chemiluminescence fluorescence system (Amersham Pharmacia).
Enzyme Immunoassay (EIA) Using Recombinant N Protein of bat-SARS-CoV. Sera from eight bats of four different species, six rodents, and two monkeys negative for bat-SARS-CoV antibody by Western blot analysis were used to set up the baseline for the EIA performed as described in ref. 22. Nunc immunoplates coated with 20 ng of purified (His)6-tagged recombinant bat-SARS-CoV N protein per well were used. Detection was performed by using 1:2,000 horseradish peroxidase-conjugated protein G and 3,3′,5,5′-tetramethylbenzidine, both from Zymed. Each sample was tested in duplicate, and the mean absorbance for each serum was calculated.
Specificity of Recombinant bat-SARS-CoV N Protein-Based Western Blot Analysis and EIA. To evaluate the specificity of the recombinant N protein-based Western blot assay and EIA, convalescent human serum samples from patients with recent infections by HCoV-OC43 (n = 13), HCoV-229E (n = 9), HCoV-NL63 (n = 5), and CoV-HKU1 (n = 11), positive for specific antibodies against the respective CoV, were subject to Western blot assay and EIA against recombinant N protein of bat-SARS-CoV.
Neutralization Assays. Because attempts to passage bat-SARS-CoV in cell cultures were not successful, neutralization assays for human SARS-CoV were carried out as described in ref. 9. Animal sera serially diluted from 1:20 to 1:640 were mixed with 100 tissue culture 50% infective dose of SARS-CoV isolate HKU-39849. Human sera from SARS patients with neutralizing antibody titer of 1:160 were included as a positive control. After incubation for 1 h at 37°C, the mixture was inoculated in triplicate onto 96-well plates of FRhK-4 cell cultures. Results were recorded after 3 days of incubation at 37°C.
Quantitative RT-PCR. Quantitaive RT-PCR was performed on anal swabs of bats positive for bat-SARS-CoV by RT-PCR or antibody by EIA. cDNA was amplified in SYBR Green I fluorescence reactions (Roche) using specific primers (5′-TGTGACAGAGCCATGCCTAA-3′ and 5′-ATCTTATTACCATCAGTTGAAAGA-3′) as described in ref. 7. A plasmid with the target sequence was used for generating the standard curve. At the end of the assay, PCR products (280-bp fragment of pol) were subjected to melting curve analysis (65-95°C, 0.1°C/s) to confirm the specificity of the assay.
Results
Wild Animal Surveillance, Identification of bat-SARS-CoV, and Viral Cultures. A total of 414 nasopharyngeal and anal swabs from 127 bats, 60 rodents, and 20 monkeys were obtained from rural areas in the HKSAR (Fig. 1a and Table 2). RT-PCR for a 440-bp fragment of pol gene of CoV was positive in anal swabs from 29 bats. Sequencing results suggested the presence of three different CoV (Fig. 4, which is published as supporting information on the PNAS web site). The sequences of three samples from lesser bent-winged bat (Miniopterus pusillus) possessed >97% nucleotide identities to a group 1 CoV recently identified from bats in the HKSAR (14). The sequences of three samples from Chinese horseshoe bat (Rhinolophus sinicus) had only 79% nucleotide identities to HCoV-229E, suggesting a previously uncharacterized group 1 CoV (bat-CoV-HKU2). Detailed analysis of these two CoV is not included in this study. The sequences of the remaining 23 positive samples, also from Chinese horseshoe bats (Fig. 1b), possessed 88% nucleotide identities to SARS-CoV (Table 2). Attempts to stably passage this virus in cell lines were unsuccessful.
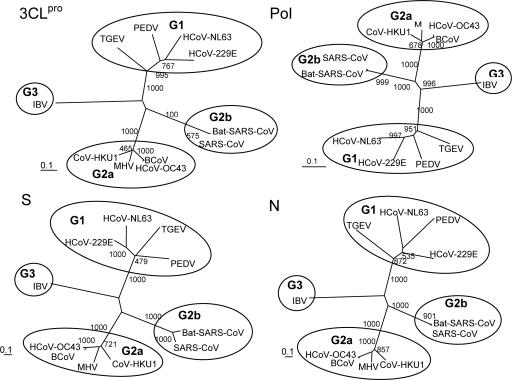
Complete Genome Analysis of bat-SARS-CoV. Complete genome sequencing was performed directly from three samples collected at different dates (Table 3). Their genome sizes were all 29,709 bp (G + C content 41.15%). Their genome organization and coding potential, similar to those of SARS-CoV (23-25) except for the regions in ORF 3 and ORF 8, is summarized in Table 4, which is published as supporting information on the PNAS web site. Two genomes (B24 and B43, from the same location) were identical, whereas the other (B41, from a different location) differed by 19 nucleotides, of which eight (42%) were nonsynonymous mutations (Table 5, which is published as supporting information on the PNAS web site). Phylogenetic analysis of the chymotrypsin-like protease, polymerase, S, and N sequences showed similar topologies, clustering with SARS-CoV (Fig. 2). Together with SARS-CoV, the virus belongs to a group distantly related to known group 2 CoV. It has been illustrated that SARS-CoV represents an early split-off from group 2 CoV and has been classified as a group 2b CoV (25, 26). Our results support the presence of a SARS-CoV-related CoV in bats, proposed to be named bat-SARS-CoV.
Fig. 2.
Phylogenetic analysis of chymotrypsin-like protease (3CLpro), RNA-dependent RNA polymerase (Pol), spike (S), and nucleocapsid (N) of bat-SARS-CoV. Bootstrap values were calculated from 1,000 trees. Included for analysis were 306, 932, 1242, and 421 amino acid positions in 3CLpro, Pol, S, and N, respectively. The scale bar indicates the estimated number of substitutions per 10 amino acids. PEDV, porcine epidemic diarrhea virus; TGEV, porcine transmissible gastroenteritis virus; MHV, murine hepatitis virus; BCoV, bovine CoV; IBV, infectious bronchitis virus
The three genomes had 88% nucleotide and 93% amino acid identities to 10 human and civet SARS-CoV isolated from different locations and at differing times (Table 3). Most differences between the bat-SARS-CoV genomes and human and civet SARS-CoV genomes were observed in the S gene, ORF 3, and ORF 8 (Table 4), which also were the regions where most variations were observed in human SARS-CoV and civet SARS-CoV genomes (10). Compared with SARS-CoV from humans and civets, there were 11 insertions and 15 deletions in the bat-SARS-CoV genome, located in the 5′ noncoding region, nsp3 of ORF 1ab, S gene, ORF 3, and ORF 8. However, the 29-bp region, deleted in most human SARS-CoV, is present, as in civet SARS-CoV (Fig. 5, which is published as supporting information on the PNAS web site).
ORF 3 (274 aa) had 81% identities to ORF 3a of SARS-CoV. ORF 3b of SARS-CoV is prematurely terminated in bat-SARS-CoV because of nucleotide substitution at position 25748 (TCA → TAA) (Fig. 6, which is published as supporting information on the PNAS web site). Because the transcription regulatory sequence is not found upstream of ORF 3b, this ORF of SARS-CoV may not be functional.
Among all ORFs, ORF 8 (121 aa) had the lowest identity to the corresponding protein of SARS-CoV. Similar to civet SARS-CoV, a 29-bp insertion, absent in most human SARS-CoV, was found, resulting in a longer ORF than ORF 8a of human SAR-CoV with the absence of ORF 8b (Fig. 7, which is published as supporting information on the PNAS web site). As for the 29-bp insertion (5′-CCAATACATTACTATTCGGACTGGTTTAT-3′), there were 12 nucleotide substitutions, compared with its counterpart in civet SARS-CoV (5′-CCTACTGGTTACCAACCTGAATGGAATAT-3′). The ORF 8 displayed only 33% amino acid identities to that of civet SARS-CoV, which suggests that the encoded proteins may function differently in these viruses.
Because the S protein had only 79-80% amino acid identities with that of SARS-CoV and encodes the receptor-binding site, the complete S genes were sequenced from 11 additional samples. When these 14 sequences were compared with human and civet SARS-CoV, 958 nucleotide polymorphisms (59% nonsynonymous) were noted (Tables 6 and 7, which are published as supporting information on the PNAS web site). A total of 890 (58% nonsynonymous) of the 958 nucleotide polymorphisms appeared to distinguish between bat-SARS-CoV and SARS-CoV, suggesting that the S of bat-SARS-CoV is distinct from that of human and civet SARS-CoV. Whereas 35 of the 43 nucleotide polymorphisms among the 10 human and civet SARS-CoV sequences were nonsynonymous changes, only 2 of the 8 nucleotide polymorphisms among the 14 sequences of bat-SARS-CoV were nonsynonymous (Table 8, which is published as supporting information on the PNAS web site). Similar to SARS-CoV, the basic amino acid cleavage site found in S protein of group 2 and 3 CoV is absent. Nevertheless, a recent study detected a cleaved S2 fragment in the lysate of cells infected with SARS-CoV, suggesting proteolytic processing of S protein of SARS-CoV in host cells. The proposed cleavage site in SARS-CoV (RS667-668) also is found in bat-SARS-CoV (RS653-654) (27). Further studies are required to determine whether the S protein of bat-SARS-CoV also is cleaved into S1 and S2 subunits. For SARS-CoV, both angiotensin-converting enzyme 2 (ACE2) and CD209L (L-SIGN) were shown to be receptors (28, 29). Although the important amino acid residues for binding to CD209L are yet to be defined, a 193-aa fragment in the S1 domain of SARS-CoV (residues 318-510) has been found essential for binding to ACE2 (30). A shorter homologous fragment of 176 aa with 73% identities was identified in bat-SARS-CoV (Fig. 8, which is published as supporting information on the PNAS web site). Within this fragment, the two residues that contribute most substantially to ACE2 association (E452 and D454) (30) and five of the seven cysteines in SARS-CoV also are present. However, the other two cysteines (C467 and C474) in SARS-CoV, alterations of which are shown to prevent efficient precipitation of ACE2 (30), are absent in bat-SARS-CoV.
The rest of the bat-SARS-CoV genome shared similar features with SARS-CoV. A putative transcription regulatory sequence motif, 5′-ACGAAC-3′, identical to that of SARS-CoV (31), was found at the 3′ end of the putative 5′ leader sequence and precedes each ORF except ORF 7b.
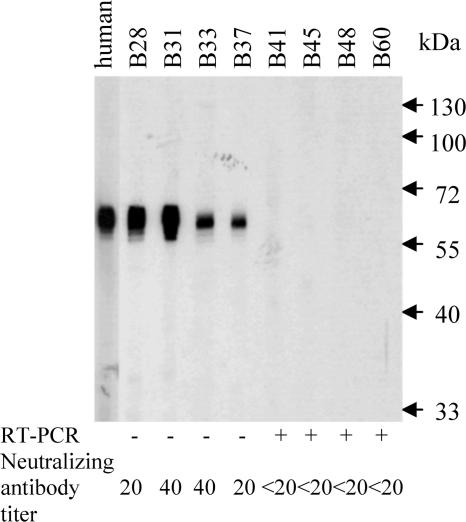
Serological Studies. Only 1 of the 13 human serum samples from patients with recent infections by HCoV-OC43 was positive in the recombinant bat-SARS-CoV N protein-based EIA (with titer of 1:400) and Western blot analysis (with a very faint band). None of the other 12 sera from patients with recent infections by HCoV-OC43, the 9 sera from patients with recent infections by HCoV-229E, the 5 sera from patients with recent infections by HCoV-NL63, and the 11 sera from patients with recent infections by CoV-HKU1, tested positive by the recombinant bat-SARS-CoV N protein-based EIA or Western blot analysis. Antibody detection in animal serum samples with sufficient quantities was performed by bat-SARS-CoV N protein-based Western blot analysis and EIA, and neutralization tests against human SARS-CoV. Among tested sera from Chinese horseshoe bats, 12 (67%) of 18 were positive for bat-SARS-CoV antibody by Western blot analysis (Fig. 3) and 31 (84%) of 37 by EIA with titer ≥1:400, compared with only 8 (42%) of 19 for human SARS-CoV-neutralizing antibody with titer ≥1:20 (Table 1). Interestingly, bats with neutralizing antibody had a lower viral load in their anal swabs (P = 0.016; Student's t test). Quantitative RT-PCR also identified the presence of bat-SARS-CoV in five additional bats negative by RT-PCR. Antibodies were not detected in samples from other bat species or animals from the present surveillance study.
Fig. 3.
Western blot analysis with animal sera against the purified (His)6-tagged recombinant bat-SARS-CoV N protein antigen. Prominent immunoreactive protein bands of about 50 kDa, consistent with the expected size of 49.5 kDa of the recombinant protein, were detected with a convalescent serum sample from a patient with SARS as positive control and four of the eight bat serum samples shown, indicating antigen-antibody interactions between the recombinant bat-SARS-CoV N protein and serum antibodies. Results of neutralization tests for SARS-CoV and RT-PCR of anal swabs for bat-SARS-CoV also are shown
Table 1. Viral load and antibody test results in Chinese horseshoe bats.
| Antibody response
|
|||||
|---|---|---|---|---|---|
| Animal no. | RT-PCR for bat-SARS-CoV | Viral load in anal swabs by quantitative RT-PCR, copies per ml | Western blot analysis using bat-SARS-CoV N protein | EIA titer using bat-SARS-CoV N protein | Neutralizing antibody titer to human SARS-CoV |
| B21 | + | 4.82 × 105 | + | 800 | <20 |
| B22 | + | 7 × 104 | + | 1,600 | 40 |
| B23 | + | 6.21 × 104 | + | >3,200 | 40 |
| B24 | + | 1.35 × 107 | + | 1,600 | <20 |
| B26 | + | 7.69 × 105 | + | 800 | <20 |
| B39 | + | 1.07 × 103 | QI | QI | QI |
| B40 | + | 1.34 × 106 | QI | <400 | QI |
| B41 | + | 4.21 × 106 | – | <400 | <20 |
| B43 | + | 2.21 × 107 | QI | QI | QI |
| B45 | + | 4.71 × 106 | – | 400 | <20 |
| B46 | + | 8.25 × 105 | QI | >3,200 | QI |
| B47 | + | 2.18 × 106 | QI | QI | QI |
| B48 | + | 8.64 × 106 | – | <400 | <20 |
| B50 | + | 1.55 × 104 | QI | 3,200 | QI |
| B51 | + | 1.66 × 106 | + | 800 | <20 |
| B52 | + | 5.46 × 102 | – | 400 | QI |
| B53 | + | 3.33 × 105 | QI | 400 | QI |
| B54 | + | 6.54 × 104 | QI | 3,200 | QI |
| B55 | + | 2.61 × 106 | QI | >3,200 | <20 |
| B56 | + | 2.56 × 106 | + | >3,200 | 20 |
| B59 | + | 9.79 × 106 | QI | >3,200 | QI |
| B60 | + | 1.13 × 107 | – | <400 | <20 |
| B61 | + | 2.98 × 106 | QI | 1,600 | QI |
| B5 | – | 0 | – | <400 | <20 |
| B6 | – | 0 | QI | 1,600 | QI |
| B18 | – | 0 | QI | >3,200 | QI |
| B25 | – | 0 | + | 400 | 20 |
| B27 | – | 1.29 × 103 | QI | 1,600 | QI |
| B28 | – | 0 | + | 3,200 | 20 |
| B29 | – | 0 | QI | 3,200 | QI |
| B30 | – | 0 | QI | 3,200 | QI |
| B31 | – | 0 | + | >3,200 | 40 |
| B33 | – | 0 | + | 800 | 40 |
| B37 | – | 0 | + | 800 | 20 |
| B39 | – | ND | QI | <400 | QI |
| B42 | – | 3.89 × 102 | QI | >3,200 | QI |
| B44 | – | 4.25 × 102 | QI | 1,600 | <20 |
| B49 | – | 0 | QI | 400 | QI |
| B57 | – | 7.07 × 103 | QI | 1,600 | QI |
| B58 | – | 8.68 × 102 | QI | >3,200 | QI |
QI, quantity insufficient; ND, not done.
Discussion
The isolation of SARS-CoV from Himalayan palm civets from wild live markets in mainland China has led to the belief that wild animals are the reservoir for the origin of the SARS epidemic (9). However, several lines of evidence suggested that the civets may have served only as an amplification host for SARS-CoV and provided the environment for efficient interspecies transmissions. First, the Ka/Ks ratios (rate of nonsynonymous mutation/rate of synonymous mutation) of the S, orf3a, and nsp3 genes of the civet strains in both the 2003 and the minor 2004 outbreaks were high, suggesting a rapidly evolving process of gene adaptation in the animals (10). Second, although the S proteins of SARS-CoV isolated from humans during the 2003 epidemic were shown to bind to and use both human and palm-civet ACE2 efficiently, isolates from the 2004 minor outbreaks and those from palm civets used human ACE2 markedly less efficiently (11). Furthermore, substantial functional changes have been detected in the S proteins from a patient in late 2003 from Guangdong Province and from two palm civets, which depended less on the human ACE2 receptor and were markedly resistant to antibody inhibition (12). Third, although as many as 80% of the animals from markets in Guangzhou had significant levels of antibody to SARS-CoV, civets on farms were largely free from SARS-CoV infection (13). These findings provide strong evidence for the presence of yet unknown primary or introductory hosts of the SARS-CoV or related viruses. Because civets are often mixed with different species under the conditions of overcrowding in the markets, wild animals in the area would be the target for searching for the origin of SARS-CoV. Therefore, we carried out a surveillance study and screened for CoV in wild animals in HKSAR.
A CoV closely related to SARS-CoV, bat-SARS-CoV, was identified from 23 of 59 (39%) anal swabs from the species R. sinicus in this study. Serological studies showed that 67% and 84% of tested sera from Chinese horseshoe bats were positive for antibodies against recombinant bat-SARS-CoV N protein by Western blot analysis and EIA, respectively. The presence of cross-reacting neutralizing antibodies to human SARS-CoV is not unexpected, because there is high degree of homology between the predicted proteins of bat-SARS-CoV and SARS-CoV. Despite this cross-reaction, using sera of patients infected with group 1 (HCoV-229E and HCoV-NL63) and group 2 (HCoV-OC43 and CoV-HKU1) CoV, we showed that the Western blot assay and EIA were likely to be specific for SARS-CoV-like viruses. Such results are in line with previous findings that cross-reactions of HCoV-229E and HCoV-OC43 to human SARS-CoV N protein were uncommon (32, 33). Although cross-reactions between bat-SARS-CoV and yet unidentified, closely related CoV cannot be excluded, it is unlikely that significant proportions of positive reactions by the present Western blot analysis and EIA were due to cross-reacting antibodies against the group 1 CoV identified in bats. Moreover, only bat-SARS-CoV genomes but not human or civet SARS-CoV were detected in bats positive for antibodies against bat-SARS-CoV N protein in the present study.
The phylogenetic distance from SARS-CoV and the presence of the 29-bp insertion in ORF 8 of bat-SARS-CoV genomes suggest that bat-SARS-CoV is unlikely to be a result of transmission of SARS-CoV from humans to bats. Instead, bat-SARS-CoV and civet SARS-CoV are likely to have a common ancestor. Because these positive samples were collected within a short period, it is not possible to estimate the evolutionary rate of bat-SARS-CoV and the time of possible interspecies transmission. BCoV is suspected to have jumped to humans as HCoV-OC43 in 1890, according to estimation from molecular clock analysis (34). The phylogenetic distance between BCoV and HCoV-OC43 is comparable to that between bat-SARS-CoV and SARS-CoV (Fig. 2). Although the HKSAR is an urbanized, subtropical city, it has extensive natural areas with 52 terrestrial mammals, including 22 bat species. Chinese horseshoe bat (R. sinicus), belonging to the family Rhinolophidae of the order Chiroptera under Microchiroptera (microbats), is an insectivorous species widely distributed in forested areas throughout the HKSAR and China (www.hkbiodiversity.net). R. sinicus was previously called R. rouxii subspecies sinicus, but recent karyotyping study has elevated its status to a separate species (35). Although no local data on its migration patterns are available, members of Rhinolophus may migrate up to 30 km for hibernation in winter (36, 37). Interestingly, the nearest wildlife market previously found to have animals with SARS-CoV in Shenzhen is only 17 km away from the locations with bats harboring bat-SARS-CoV in the HKSAR (Fig. 1a). All samples that were positive for bat-SARS-CoV were collected during winter and spring. Nevertheless, the present data do not allow direct inference on the direction of interspecies transmission of SARS-CoV-like viruses or their ancestral relationships.
The pathogenicity and host range of bat-SARS-CoV remain to be determined. In this study, the virus could be detected only in anal swabs, suggesting that it may have enteric tropism, in line with findings in SARS-CoV (38). All bats with bat-SARS-CoV did not display obvious signs of disease. Although analysis of the S gene revealed the presence of most amino acid residues in SARS-CoV crucial for binding ACE2, more extensive surveillance studies and receptor-binding experiments are necessary to determine the host range and potential of interspecies transmission of bat-SARS-CoV.
Bats, the only flying mammals, account for 20% of the 4,800 mammalian species recorded in the world. They are an important reservoir of emerging zoonotic viruses, including rabies virus, lyssavirus, Hendra and Nipah viruses, St. Louis encephalitis virus, and fungi such as Histoplasma (39, 40). The feces of bats (excrementum vespertilionis 
 ) are used in traditional Chinese medicine (www.aompress.com/pdf/singleherbs.pdf). The Chinese and Manadonese populations of Malaysia and Indonesia consider bat meat a delicacy. Many Chinese also believe that eating bat meat can cure asthma, kidney ailments, and general malaise (41). In this study, three different CoV were identified from wild bats in the HKSAR, suggesting that these animals are an important reservoir for CoV. Continuous surveillance for CoV in these flying mammals with roosting behavior is indicated to assess their potential threats to human health.
) are used in traditional Chinese medicine (www.aompress.com/pdf/singleherbs.pdf). The Chinese and Manadonese populations of Malaysia and Indonesia consider bat meat a delicacy. Many Chinese also believe that eating bat meat can cure asthma, kidney ailments, and general malaise (41). In this study, three different CoV were identified from wild bats in the HKSAR, suggesting that these animals are an important reservoir for CoV. Continuous surveillance for CoV in these flying mammals with roosting behavior is indicated to assess their potential threats to human health.
Supplementary Material
Acknowledgments
We thank Prof. York Y. N. Chow (Secretary for Health, Welfare, and Food, HKSAR); Thomas C. Y. Chan, Chik-Chuen Lay, and Ping-Man So [HKSAR Department of Agriculture, Fisheries, and Conservation (AFCD)]; Dr. Che-Hung Leong; and the Hong Kong Police Force for facilitation and support; Prof. Lap-Chee Tsui (University of Hong Kong) for critical comments on the manuscript; Chung-Tong Shek and Cynthia S. M. Chan from AFCD for their excellent technical assistance; Dr. King-Shun Lo (Laboratory Animal Unit) and Dr. Cassius Chan for the collection of animal specimens; and the AFCD Mammal Working Group for their help in taking the photographs of Chinese horseshoe bats. This work was partly supported by the Henry Fok Foundation; the Research Grant Council; the University Development Fund, University of Hong Kong; the Tung Wah Group of Hospitals' Fund for Research in Infectious Diseases; and the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare, and Food Bureau.
Author contributions: S.K.P.L., P.C.Y.W., and K.-Y.Y. designed research; S.K.P.L., P.C.Y.W., K.S.M.L., Y.H., H.-W.T., B.H.L.W., S.S.Y.W., S.-Y.L., K.-H.C., and K.-Y.Y. performed research and analyzed data; and S.K.P.L., P.C.Y.W., and K.-Y.Y. wrote the paper.
Abbreviations: CoV, coronavirus; HCoV, human CoV; BCoV, bovine CoV; SARS, severe acute respiratory syndrome; HKSAR, Hong Kong Special Administrative Region; S, spike; N, nucleocapsid; EIA, enzyme immunoassay; ACE2, angiotensin-converting enzyme 2.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ022305, DQ084199, and DQ084200).
References
- 1.Lai, M. M. & Cavanagh, D. (1997) Adv. Virus Res. 48, 1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris, J. S., Lai, S. T., Poon, L. L., Guan, Y., Yam, L. Y., Lim, W., Nicholls, J., Yee, W. K., Yan, W. W., Cheung, M. T., et al. (2003) Lancet 361, 1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten, C., Gunther, S., Preiser, W., van der Werf, S., Brodt, H. R., Becker, S., Rabenau, H., Panning, M., Kolesnikova, L., Fouchier, R. A., et al. (2003) N. Engl. J. Med. 348, 1967-1976. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J. A., Lim, W., et al. (2003) N. Engl. J. Med. 348, 1953-1966. [DOI] [PubMed] [Google Scholar]
- 5.van der Hoek, L., Pyrc, K., Jebbink, M. F., Vermeulen-Oost, W., Berkhout, R. J., Wolthers, K. C., Wertheim-van Dillen, P. M., Kaandorp, J., Spaargaren, J. & Berkhout, B. (2004) Nat. Med. 10, 368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouchier, R. A., Hartwig, N. G., Bestebroer, T. M., Niemeyer, B., de Jong, J. C., Simon, J. H. & Osterhaus, A. D. (2004) Proc. Natl. Acad. Sci. USA 101, 6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo, P. C., Lau, S. K., Chu, C. M., Chan, K. H., Tsoi, H. W., Huang, Y., Wong, B. H., Poon, R. W., Cai, J. J., Luk, W. K., et al. (2005) J. Virol. 79, 884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo, P. C., Lau, S. K., Tsoi, H. W., Poon, R. W., Chu, C. M., Lee, R. A., Luk, W. K., Wong, G. K., Wong, B. H., Cheng, V. C., et al. (2005) J. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 9.Guan, Y., Zheng, B. J., He, Y. Q., Liu, X. L., Zhuang, Z. X., Cheung, C. L., Luo, S. W., Li, P. H., Zhang, L. J., Guan, Y. J., et al. (2003) Science 302, 276-278. [DOI] [PubMed] [Google Scholar]
- 10.Song, H. D., Tu, C. C., Zhang, G. W., Wang, S. Y., Zheng, K., Lei, L. C., Chen, Q. X., Gao, Y. W., Zhou, H. Q., Xiang, H., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 2430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, W., Zhang, C., Sui, J., Kuhn, J. H., Moore, M. J., Luo, S., Wong, S. K., Huang, I. C., Xu, K., Vasilieva, N., et al. (2005) EMBO J. 24, 1634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, Z. Y., Werner, H. C., Kong, W. P., Leung, K., Traggiai, E., Lanzavecchia, A. & Nabel, G. J. (2005) Proc. Natl. Acad. Sci. USA 102, 797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu, C., Crameri, G., Kong, X., Chen, J., Sun, Y., Yu, M., Xiang, H., Xia, X., Liu, S., Ren, T., et al. (2004) Emerg. Infect. Dis. 10, 2244-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon, L. L., Chu, D. K., Chan, K. H., Wong, O. K., Ellis, T. M., Leung, Y. H., Lau, S. K., Woo, P. C., Suen, K. Y., Yuen, K. Y., et al. (2005) J. Virol. 79, 2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M. D., et al. (2001) Nucleic Acids Res. 29, 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gattiker, A., Gasteiger, E. & Bairoch, A. (2002) Appl. Bioinformatics 1, 107-108. [PubMed] [Google Scholar]
- 18.Combet, C., Blanchet, C., Geourjon, C. & Deléage, G. (2000) Trends Biochem. Sci. 25, 147-150. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann, K. & Stoffel, W. (1993) Biol. Chem. Hoppe-Seyler 374, 166. [DOI] [PubMed] [Google Scholar]
- 20.Sonnhammer, E. L., von Heijne, G. & Krogh, A. (1998) Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175-182. [PubMed] [Google Scholar]
- 21.Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 22.Woo, P. C., Lau, S. K., Tsoi, H. W., Chan, K. H., Wong, B. H., Che, X. Y., Tam, V. K., Tam, S. C., Cheng, V. C., Hung, I. F., et al. (2004) Lancet 363, 841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., et al. (2003) Science 300, 1394-1399. [DOI] [PubMed] [Google Scholar]
- 24.Marra, M. A., Jones, S. J., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., et al. (2003) Science 300, 1399-1404. [DOI] [PubMed] [Google Scholar]
- 25.Snijder, E. J., Bredenbeek, P. J., Dobbe, J. C., Thiel, V., Ziebuhr, J., Poon, L. L., Guan, Y., Rozanov, M., Spaan, W. J. & Gorbalenya, A. E. (2003) J. Mol. Biol. 331, 991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorbalenya, A. E., Snijder, E. J. & Spaan, W. J. (2004) J. Virol. 78, 7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, X. D., Shang, B., Yang, R. F., Yu, H., Ma, Z. H., Shen, X., Ji, Y. Y., Lin, Y., Wu, Y. D., Lin, G. M., et al. (2004) Cell Res. 14, 400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., et al. (2003) Nature 426, 450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffers, S. A., Tusell, S. M., Gillim-Ross, L., Hemmila, E. M., Achenbach, J. E., Babcock, G. J., Thomas, W. D., Jr., Thackray, L. B., Young, M. D., Mason, R. J., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong, S. K., Li, W., Moore, M. J., Choe, H. & Farzan, M. (2004) J. Biol. Chem. 279, 3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain, S., Pan, J., Chen, Y., Yang, Y., Xu, J., Peng, Y., Wu, Y., Li, Z., Zhu, Y., Tien, P., et al. (2005) J. Virol. 79, 5288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo, P. C., Lau, S. K., Wong, B. H., Chan, K. H., Hui, W. T., Kwan, G. S., Peiris, J. S., Couch, R. B. & Yuen, K. Y. (2004) J. Clin. Microbiol. 42, 5885-5888. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Che, X. Y., Qiu, L. W., Liao, Z. Y., Wang, Y. D., Wen, K., Pan, Y. X., Hao, W., Mei, Y. B., Cheng, V. C. & Yuen, K. Y. (2005) J. Infect. Dis. 191, 2033-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijgen, L., Keyaerts, E., Moes, E., Thoelen, I., Wollants, E., Lemey, P., Vandamme, A. M. & Van Ranst, M. (2005) J. Virol. 79, 1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, Y., Harada, M. & Li, Y. (2004) Acta Theriol. Sin. 24, 30-35. [Google Scholar]
- 36.Neuweiler, G. (2000) The Biology of Bats, trans. Covey, E. (Oxford Univ. Press, New York), p. 266.
- 37.Nowak, R. M. & Paradiso, J. L. (1983) Walker's Mammals of the World (Johns Hopkins Univ. Press, Baltimore), p. 230.
- 38.Hung, I. F., Cheng, V. C., Wu, A. K., Tang, B. S., Chan, K. H., Chu, C. M., Wong, M. M., Hui, W. T., Poon, L. L., Tse, D. M., et al. (2004) Emerg. Infect. Dis. 10, 1550-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyon, G. M., Bravo, A. V., Espino, A., Lindsley, M. D., Gutierrez, R. E., Rodriguez, I., Corella, A., Carrillo, F., McNeil, M. M., Warnock, D. W., et al. (2004) Am. J. Trop. Med. Hyg. 70, 438-442. [PubMed] [Google Scholar]
- 40.Mackenzie, J. S. & Field, H. E. (2004) Arch. Virol. Suppl. 18, 97-111. [DOI] [PubMed] [Google Scholar]
- 41.Fujita, M. (1988) Bats 6, 4-9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.