Abstract
Aeromonas hydrophila secretes a number of degradative enzymes and toxins into the external milieu via the type II secretory pathway or secreton. ExeA is an essential component of this system and is necessary for the localization and/or multimerization of the secretin ExeD. ExeA contains two sequence motifs characteristic of the Walker superfamily of ATPases. Previous examination of substitution derivatives altered in these motifs suggested that ATP binding or hydrolysis is required for ExeAB complex formation and subsequent secretion function. To directly examine ExeA function, the N-terminal cytoplasmic domain of ExeA with the addition of a C-terminal hexahistidine tag (cytExeA) was overproduced in Escherichia coli and purified by metal chelate affinity and anion-exchange chromatographic techniques. Purified preparations of cytExeA exhibited ATPase activity in the presence of several divalent cations, Mg2+ being the preferred cation, with an optimum reaction temperature of ∼37 to 42°C and an optimum pH of 7 to 8. cytExeA exhibited an apparent Km for Mg-ATP of 0.22 mM and a Vmax of 0.72 nmol min−1 mg−1 of protein. cytExeA displayed low specificity for nucleoside triphosphate substrates and was significantly inhibited by F-type ATPase inhibitors. Gel filtration analyses of cytExeA, ExeA, and ExeAB indicated that ExeA dimerizes and forms a very large complex with ExeB. These findings support a model whereby ExeAB utilizes energy derived from ATP hydrolysis to facilitate the correct localization and multimerization of the ExeD secretin.
Aeromonads are significant pathogens of aquatic mammals and fish, as well as opportunistic human pathogens (1, 20). Like many other gram-negative organisms, their ability to secrete degradative enzymes and toxins into the external milieux is regarded as a major virulence mechanism. For Aeromonas hydrophila, these extracellular products include the hemolysin aerolysin and the metalloprotease AhpB, which were shown to be important virulence determinants in mouse and rainbow trout toxicity models, respectively (6, 7).
In A. hydrophila, secretion of extracellular proteins such as aerolysin takes place via the type II secretion pathway encoded by the exeC-N operon (17, 21) (or gsp if referring to all type II or general secretory pathway secretion genes), the exeAB operon (19), and the prepilin peptidase tapD (34). This pathway initially involves inner membrane translocation of substrates bearing N-terminal signal sequences in a sec-dependent (10) or tat-dependent (49) manner. It differs from types I and III to V in that type II substrates fold in the periplasm prior to outer membrane translocation. A substrate translocation-competent conformation, which may include modifications such as disulfide bond formation (4), subunit assembly (39), or dimer formation in the case of aerolysin (16), is required for recognition and outer membrane translocation by the type II secretion apparatus or secreton (35). The secreton is proposed to form an envelope-spanning multiprotein complex composed of at least 12 gene products that is dedicated to outer membrane translocation of type II secretion substrates (for a review, see reference 39). Some secreton components, such as ExeE, are very similar to proteins involved in the biogenesis of type IV pili, which function in both adherence and twitching motility (51). In addition, the Klebsiella oxytoca and Pseudomonas aeruginosa secretons have been shown to be capable of assembling a pilus-like structure when overexpressed (11, 43). The type IV pili, however, are not believed to represent (construct) a tubular secretion conduit such as those of the type III and type IV secretion systems (51). Instead, an outer membrane component termed secretin forms a large ring-shaped dodecameric multimer which is thought to function as the translocation channel (29).
ExeE, one of two secreton proteins that contain consensus Walker nucleotide-binding motifs (50), is distantly related to counterparts in type IV extracellular secretion systems as well as the PilB family (36), collectively referred to as the PulE-VirB11 family. Purified EpsE (of the Vibrio cholerae system) was found to exhibit autophosphorylation activity dependent on an intact Walker A box (40) and was recently shown to be a Mg2+-dependent ATPase (5). It is possible that energy generated by ATP hydrolysis is used by EpsE in the extension of a pseudopilus filament. This suggestion is supported by the finding that a distant relative of the GspE proteins, HPO525, found in the type IV extracellular secretion system of Helicobacter pylori, is an ATPase required for the assembly of a conjugation pilus-like structure (24).
ExeA (GspA), a second putative ATPase of the type II secretion pathway, contains two sequence motifs characteristic of the Walker superfamily of ATPases (50). It contains the typical Walker A consensus motif, GX4GKS/T (50-GEVGTGKT-57), known as the phosphate-binding loop or P-loop (42) and the Walker B motif 126-VVLVD-130 (four hydrophobic residues followed by an aspartate). Several kinases have also been found to contain Walker A and B motifs, in this case referred to as kinase-1a and kinase-2 motifs, respectively (48). In addition to the Walker motifs, ExeA is similar to kinases in containing the sequence 228-GGIPR-232, which is highly similar to the kinase-3a motif.
The existence of ExeA and ExeB homologues in type II secretion systems is widespread throughout bacterial species, including V. cholerae, Shewanella putrefaciens, Geobacter sulfurreducens, and Escherichia coli K-12 (13, 14, 38). Although normally silent, the secreton genes of the E. coli gsp system, including gspAB, were shown to be required for secretion of chitinase (13). The importance of GspB proteins in other translocation systems, such as the pul and out systems of Klebsiella and Erwinia, has also been characterized (8, 9).
We have previously shown that ExeA and ExeB form an inner membrane complex that may form a higher-order multimer, as suggested by cross-linking analyses (45). In addition, formation of the multimer and secretion activity could be dependent on the binding and hydrolysis of ATP, as suggested by the examination of a collection of ExeA substitution derivatives altered in the conserved kinase-1a and kinase-3a motifs. The energy derived by ATP hydrolysis could be required to correctly localize and multimerize the ExeD secretin, since ExeAB was recently shown to be required for both transport and assembly of the secretin into the outer membrane (2). In the studies reported here, we directly examined the enzyme activity of ExeA. Although full-length ExeA displayed ATPase activity in extracts fractionated on ion-exchange columns (data not shown), the activity was unstable and the protein could not be purified further. We therefore expressed and purified a His-tagged cytoplasmic fragment of the protein (cytExeA) that contains the putative ATPase domain. This allowed us to investigate the NTPase activity of this enzyme and characterize its inhibitor profile and substrate requirements. The multimerization state of the ExeAB complex in vivo was also examined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The wild-type Aeromonas hydrophila strain Ah65 and Escherichia coli strains used in this study are listed in Table 1. A list of cloning vectors and recombinant plasmids is provided in Table 2. A. hydrophila and E. coli strains containing the appropriate plasmids as indicated were grown at 30°C in brain heart infusion medium and at 37°C in 2× yeast extract-tryptone medium, respectively. E. coli strain BL21(DE3) was made ATPase deficient by cotransduction of Δ(uncB-uncC) with the selectable ilv::Tn10 allele from E. coli strain DK8 (22). P1vir grown in DK8 was used to cotransduce the ilv::Tn10 and Δ(uncB-uncC) alleles into BL21(DE3) by selection on LB containing 10 μg/ml tetracycline and 5 mM sodium citrate. Antibiotics were used at the following final concentrations: ampicillin (Ap), 100 μg ml−1; chloramphenicol (Cm), 2.5 μg ml−1; kanamycin (Km), 50 μg ml−1; nalidixic acid, 5 μg ml−1; streptomycin (St), 100 μg ml−1; and tetracycline (Tc), 10 μg ml−1.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype/phenotype | Source or reference |
|---|---|---|
| Aeromonas hydrophila Ah65 | Wild type | This laboratory |
| Escherichia coli | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| S17-1 | recA thi pro hsdR RP4::2-Tc::Mu::KmTn7 λpir | 55 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15::Tn10] | Stratagene |
| DK8 | bglR thi-1 rel-1 Δ(uncB-uncC) ilv::Tn10 HfrP01 | 22 |
TABLE 2.
Cloning vectors and recombinant plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pBluescript II SK/KS+ | Phagemid; lac promoter; Apr | Stratagene |
| pET30a | Expression vector; T7lac promoter; Kmr | Novagen |
| pRJ31.1 | exeAB 2.5-kb BstXI in SmaI of pMMB207; tac promoter; Cmr | 19 |
| pKRj58.1 | exeAB 2.8-kb DraIII-KpnI in KSII+; lac promoter; Apr | 19 |
| pICS1.1 | cytexeA 0.84-kb NdeI-XhoI PCR fragment in pET30a; T7lac promoter; Kmr | This work |
DNA techniques and plasmid construction.
Plasmid DNA minipreparations (alkaline lysis method), as well as recombinant DNA methods and analyses, were performed as described by Sambrook et al. (37). Agarose purification of DNA fragments was performed after electrophoresis into 0.6% agarose, using a Geneclean II kit (Bio 101 Inc.). Vectors or recombinant plasmids were transformed by electroporation into XL1-Blue electrocompetent E. coli cells as described previously (37). The wide-host-range plasmid pRJ31.1 was transferred to A. hydrophila by conjugation from E. coli S17-1. PCR was used to amplify DNA fragments from recombinant plasmids containing A. hydrophila DNA for subsequent cloning. Plasmid template (10 ng) was used in a 50-μl reaction containing 20 mM Tris, pH 8.4, 50 mM KCl, 1.5 mM MgCl2, deoxynucleoside triphosphates (dNTPs) at a final concentration of 200 μM, a 2 μM concentration of each primer, and 2.5 U Taq DNA polymerase. PCR was performed using 30 cycles at 95°C for 1 min, 50°C for 1 min, and 72°C for 3 min, followed by 73°C for 15 min. Agarose-purified amplicons were cloned into a pBluescript II SK+ vector that was T-tailed at the 3′ ends of an EcoRV cleavage site with Taq DNA polymerase and dTTP (3). Restriction fragments from these recombinant plasmids were then agarose purified and cloned into pET30a as described below and all inserts verified by sequence analysis (41).
Plasmid pICS1.1 contains the cytoplasmic coding region of exeA (residues 1 to 278; Table 2). PCR was used to create NdeI and XhoI sites at the 5′ (upstream) and 3′ (downstream) ends of this segment, respectively, by amplifying exeA in pKRJ58.1 using primers UR 122 (5′-ATCCATATGTACACACAGTTCTTCGGTCTGTCG-3′) and UR 123 (5′-GACCTCGAGGCCGGACTGCCAGGTGCCTTC-3′). The 0.84-kb NdeI-XhoI PCR product was cloned into pET30a yielding pICS1.1, encoding a C-terminal His-tagged cytoplasmic derivative of ExeA, cytExeA.
cytExeA purification.
For the purification of cytExeA, a 500-ml culture of BL21(DE3)(pICS1.1) was grown at 30°C, induced at an optical density of 600 nm (OD600) of 0.6 with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and harvested 2.75 h later for a total of 1,250 OD600 cell units. Cell pellets were resuspended in lysis buffer (20 mM Tris, pH 7.9, 0.25 M NaCl, 10% glycerol, 10 mM β-mercaptoethanol [BME], 1 mM phenylmethylsulfonyl fluoride, 15 mM MgCl2) containing 10 mM imidazole. After the addition of 10 μg RNase ml−1 and 20 μg DNase ml−1, the cells were disrupted by two passes through a French pressure cell (12,000 lb/in2). Lysates were centrifuged at 93,000 × g for 1 h at 4°C, and the supernatant fraction (approximately 18 ml) was applied to a Ni-nitrilotriacetic acid (NTA) column equilibrated in 10 mM imidazole lysis buffer. The column was washed with approximately 18 ml of 35 mM imidazole lysis buffer. cytExeA was eluted with 20 ml of 100 mM imidazole lysis buffer. For use in NTPase characterization and gel filtration, cytExeA elution fractions were dialyzed against dialysis buffer overnight at 4°C, concentrated to 1 mg ml−1 (Amicon YM-10 Centriplus concentrators), and frozen using liquid nitrogen followed by storage at −70°C. Prior to gel filtration, 10 mM MgCl2 and 150 mM NaCl were added to the cytExeA preparation.
NTPase assays.
NTPase assays of cytExeA were performed using a malachite green dye-binding assay as described by Panagiotidis and Shuman (31), which measures the release of inorganic phosphate. Unless otherwise stated, assays were performed at 37°C with approximately 40 μg of dialyzed Ni-NTA-purified protein per reading. Rates were corrected for the nonenzymatic release of Pi from controls without enzyme. Generally, samples were assayed in standard NTPase reaction buffer, consisting of 50 mM HEPES, pH 7.5, 10% glycerol, 10 mM BME, or as otherwise stated, with various cation and nucleotide concentrations. Phosphate content was calculated using a molar extinction coefficient for the malachite green-phosphomolybdate complex (ε660) of 80,000. Nucleotide concentrations were verified using molar extinction coefficients. For the determination of cytExeA substrate specificity, 100 mM stock solutions (Pharmacia Biotech) of nucleotides were used, except for ATP, for which a 50 mM stock solution, pH 7, was used. For the activity data analyses shown in Fig. 4, the concentrations of metal-NTP complexes were determined using stability constants and the COMICS program, as previously described (47).
FIG. 4.
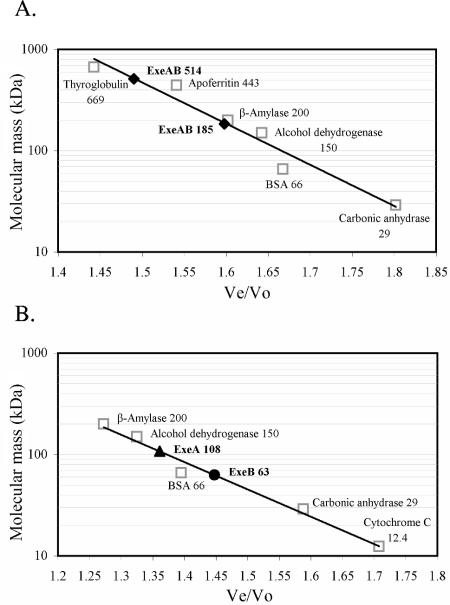
Size determination of native ExeAB, ExeA, and ExeB. Ah65(pRJ31.1) octyl-glucoside membrane extracts were subjected to anion-exchange chromatography prior to gel filtration. (A) An ExeAB anion-exchange coelution peak generated using 0.5% octyl-glucoside was collected and fractionated on a Sephacryl S-400 column. (B) Using 1% octyl-glucoside, separate ExeA and ExeB anion-exchange elution peaks were collected and then fractionated on a Sephacryl S-300 column. Elution of ExeAB, ExeA, and ExeB was compared with the elution of standard proteins of known molecular mass, and from this data the apparent molecular masses of Exe protein complexes were determined. The molecular mass versus the Ve/Vo (elution volume/void volume) for each respective protein standard was plotted. The Ve/Vo values for ExeAB (⧫), ExeA (▴), and ExeB (•) peaks are shown.
Size estimation of ExeA, ExeB, ExeAB, and cytExeA.
In preparation for molecular weight determination by gel filtration, Ah65(pRJ31.1) membranes were prepared from either 14,000 (ExeAB size determination) or 18,000 (ExeA and ExeB size determination) OD600 units of cells. Cell pellets were washed in brain heart infusion and resuspended in 20 mM Tris, pH 7.4, 100 mM NaCl, 20 μg RNase ml−1, 40 μg DNase ml−1, and Complete (Boehringer Mannheim) protease inhibitor cocktail. Cells were disrupted by three passes through a French pressure cell (12,000 lb/in2), after which 1 mM EDTA and 1 mM dithiothreitol (DTT) were added. Membranes were then pelleted by centrifugation at 126,000 × g for 1 h at 4°C. For the preparation of ExeAB samples, membranes were homogenized and then solubilized in 20 mM Tris, pH 7.4, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA containing 0.5% octyl-glucoside for 30 min at 4°C. After centrifugation at 126,000 × g for 1 h at 4°C, the supernatant fractions (solubilized membrane extracts) were applied to a Macro-Q anion-exchange column equilibrated in the 0.5% octyl-glucoside solubilization buffer. Chromatography was performed using a flow rate of 1 ml min−1 with 1.5 column volumes sample application, 1.5 column volumes wash, and 3 column volumes of a 0 to 0.4 M NaCl gradient. An ExeAB coelution peak was found at 0.26 M NaCl, in which fractions between 0.20 M and 0.30 M NaCl were collected and subjected to gel filtration (see below). ExeA and ExeB gel filtration samples were prepared similarly to that of ExeAB, except that solubilization was performed using 1.25% octyl-glucoside and subsequent chromatography was performed using 1% octyl-glucoside. Due to the basic nature of ExeB, this peak sample was collected from pass-through fractions. Under these conditions, ExeA eluted as a single peak at 0.25 M NaCl, in which fractions between 0.19 M and 0.30 M NaCl were collected for gel filtration studies (see below). Prior to gel filtration, 10 mM MgCl2 was added to ExeA and ExeB samples.
Approximately 6.9 mg of purified cytExeA was loaded onto a Sephacryl S-300 (Pharmacia Biotech) gel filtration column equilibrated in 20 mM Tris, pH 7.9, 150 mM NaCl, 10% glycerol, 10 mM BME, and 10 mM MgCl2. For size estimation of ExeA and ExeB, Sephacryl S-300 gel filtration was performed using a buffer consisting of 1% octyl-glucoside, 20 mM Tris, pH 7.9, 150 mM NaCl, 1 mM DTT, and 10 mM MgCl2. For ExeAB size estimation, a Sephacryl S-400 (Pharmacia Biotech) gel filtration column was employed using the buffer 0.5% octyl-glucoside, 20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM DTT, and 1 mM EDTA. Generally, gel filtration was performed using 16- by 700-mm columns at a flow rate of 0.4 ml min−1, with 1.5-ml samples loaded and 2-ml fractions collected. Elution was monitored by measurement of A280, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Coomassie brilliant blue staining, or immunoblotting where necessary. The following proteins were used as molecular mass standards where appropriate (Sigma): thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa).
Protein concentration determination.
Protein concentration was measured spectrophotometrically at 280 nm or by the Bio-Rad protein assay (modified Bradford assay) for cytExeA. Bovine gamma globulin (standard I) was used as a standard for the Bio-Rad assay, which was performed according to the manufacturer's instructions.
SDS-PAGE, Coomassie brilliant blue staining, and immunoblotting.
Samples were electrophoresed on 10% acrylamide SDS-PAGE gels (25). Standard proteins were also loaded on gels where appropriate, and their molecular masses in kDa are indicated. ExeA and ExeB were identified by immunoblot analysis as described previously (18).
RESULTS
In initial attempts to characterize ExeA, octyl-glucoside-solubilized membrane extracts of Ah65(pRJ31.1) cells overexpressing ExeAB were fractionated using Macro-Q anion-exchange chromatography. Eluent fractions were measured for ATPase activity, and one of the four major ATPase peaks observed was found to coincide with the elution profile of ExeA, which eluted at approximately 0.22 M NaCl (data not shown). This peak of ATPase activity was not detected in the elution profile of an identical experiment using C5.84(pRJ11.1) solubilized membrane extracts that do not contain ExeA. Further purification of full-length ExeA in active form was not possible, however.
Overproduction, localization, and purification of cytExeA.
To further examine ExeA enzyme activity, a fragment of the exeA gene was cloned into the expression plasmid pET30a. The plasmid pICS1.1 encodes the cytoplasmic portion of ExeA (cytExeA), consisting of the 278 N-terminal residues of ExeA followed by residues EHHHHHH at the C terminus. The putative ATP binding/hydrolysis motifs kinase-1a, kinase-2, and kinase-3a are found within residues 50 to 232 of ExeA. Only two amino acids belonging to the transmembrane domain that starts at residue 276 of the full-length 547-residue ExeA protein are included in cytExeA. The predicted molecular mass of cytExeA is 31,906 Da, and it has a theoretical pI of 6.42.
cytExeA was overproduced in the E. coli T7 expression strain BL21(DE3)(pICS1.1). Upon IPTG induction, the cells overexpressed an approximately 32-kDa protein that was absent before induction. Immunoblot analysis using ExeA-specific antisera confirmed the identity of this protein as cytExeA. To ascertain the cellular localization of cytExeA, total lysates from BL21(DE3)(pICS1.1) induced cultures were separated into inclusion body, cytoplasmic, and envelope fractions by centrifugation. cytExeA was found mostly in the soluble cytoplasmic fraction, with an appreciable amount associated with inclusion bodies and a minor quantity associated with the envelope.
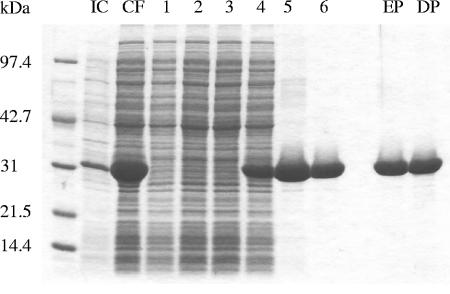
Initial purification of His-tagged cytExeA was accomplished using metal chelate affinity chromatography. Crude lysates were prepared from induced cultures of BL21(DE3)(pICS1.1) by French press and ultracentrifugation. The soluble supernatant was then applied to a Ni-NTA column equilibrated in 10 mM imidazole lysis buffer. No cytExeA could be detected in the flowthrough fractions after application (Fig. 1). Contaminating nonspecifically bound proteins as well as some of the cytExeA were eluted with a lysis buffer containing 35 mM imidazole. cytExeA was then eluted in a peak of high purity and yield with lysis buffer containing 100 mM imidazole (Fig. 2). The protein was further purified using anion-exchange chromatography. cytExeA eluted from a Macro-Q column as a single peak at approximately 0.11 M NaCl (data not shown).
FIG. 1.
Purification of cytExeA by Ni-NTA column chromatography. Lane IC, induced culture sample of BL21(DE3)(pICS1.1) 2.75 h after addition of 0.1 mM IPTG; lane CF, soluble cytoplasmic fraction following French pressure cell lysis and high-speed ultracentrifugation (applied to Ni-NTA column); lanes 1 to 3, flowthrough generated upon application of sample to Ni-NTA column; lanes 4 to 6, 35 mM imidazole wash; lane EP, elution pool of cytExeA samples corresponding to lanes 7 to 13 (Fig. 3A); lane DP, elution pool after dialysis.
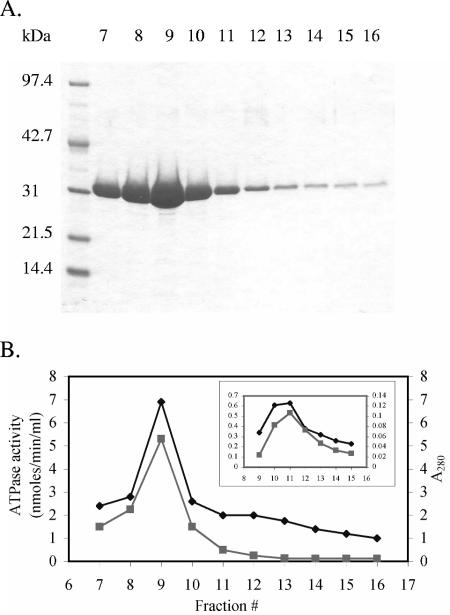
FIG. 2.
Ni-NTA purification and ATPase activity of cytExeA. (A) Purified cytExeA collected in fractions 7 to 16 by elution with 100 mM imidazole. (B) A280 and ATPase activity of the cytExeA elution fractions from panel A were measured. ATPase activity was measured at 37°C with 5 mM ATP as substrate using the malachite green dye-binding assay, for which each data point represents the average of two independent readings. Rates were corrected for the nonenzymatic release of Pi from a control containing only 100 mM imidazole lysis buffer. ATPase activity, ⧫; A280, ▪. The inset in panel B shows the elution profile and ATPase activity of cytExeA purified from BL21(DE3)Δ(uncB-uncC)(pICS1.1) cells.
cytExeA is an ATPase.
To assess ATPase activity of cytExeA, the malachite green dye-binding assay, a colorimetric method for the detection of inorganic phosphate, was employed (31). Elution fractions from the Ni-NTA purification of cytExeA were analyzed for ATPase activity (Fig. 2). The ATPase activity profile of the Ni-NTA fractions correlated well with the elution profile of cytExeA, suggesting that cytExeA has ATPase activity.
To eliminate the possibility that the activity observed may be due to low-level quantities of contaminating F1Fo ATP synthase (cf. Table 3) that went undetected by Coomassie brilliant blue staining, cytExeA was also purified in a similar manner from strain BL21(DE3)Δ(uncB-uncC)(pICS1.1), which contains a deletion of the entire unc operon. This strain grew poorly when induced for cytExeA synthesis, resulting in much lower yields; however, ATPase activity again closely followed the elution profile of cytExeA (Fig. 2, inset). The ATPase activity also continued to correlate with the elution of cytExeA from the Macro-Q column, eliminating the possibility that the observed activity was due to a contaminating ATPase other than F1 (data not shown).
TABLE 3.
Effect of ATPase inhibitors on cytExeA ATPase activity
| Compound | % cytExeA ATPase activitya | Specific forb |
|---|---|---|
| None | 100 | |
| 0.4 mM vanadate | 97 | P, I |
| 10 mM KNO3 | 74 | V |
| 10 mM NaN3 | 10 | F, A |
| 10 μM DCCDc | 76 | F, P, V |
| 5 mM DCCDc | 8 | F, P, V |
| 1 mM AMP-PNPd | 12 | F, V |
ATPase activity was measured using the malachite green method under standard conditions with 2.5 mM MgCl2 and 4 mM ATP. cytExeA was preincubated with the compounds for 10 minutes prior to the addition of ATP.
P, phosphorylation-type ATPase; V, vacuolar-type ATPase; F, Fo F1-type ATPase; A, SecA component of the Sec translocase; I, ATPase of the bacterial type I extracellular secretory pathway (or ABC ATPase).
DCCD, 1,3-dicyclohexylcarbodiimide.
AMP-PNP, 5′-adenylylimidodiphosphate.
Characterization of cytExeA NTPase activity.
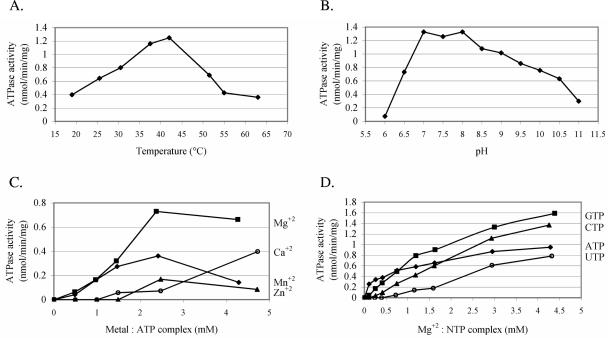
ATP hydrolysis was found to be proportional to cytExeA concentration and linear with time (up to 90 min, 37°C) (data not shown). Further characterization of cytExeA NTPase activity involved determination of the optimal reaction temperature and pH, as well as divalent cation and nucleotide specificity. In addition, the effects of several known ATPase inhibitors on cytExeA activity were examined. cytExeA ATPase activity was measured over the range of 19 to 63°C in dialysis buffer containing 5 mM MgCl2 and 5 mM ATP (Fig. 3A). ATPase activity appeared to rise linearly up to an optimum temperature of approximately 42°C, after which activity dropped sharply, coincident with cytExeA precipitation. The near-optimal and nondenaturing temperature of 37°C was then used as a standard assay condition. When similar ATPase assays were carried out under various pH conditions using PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6 to 7), HEPES (pH 7.5 to 8), AMPSO [(N-(1,1-dimethyl-2-hydroxyethyl)-3-amino-2-hydroxypropanesulfonic acid] (pH 8.5 to 9.5), and CAPS (3-cyclohexylamino-1-propanesulfonic acid) (pH 10 to 11) buffers, optimal activity was observed within a pH range of 7 to 8 (Fig. 3B). Activity declined sharply with acidic pH, compared to the gradual reduction observed with pH greater than 8. Therefore, 50 mM HEPES, pH 7.5, was used as the standard assay buffer for further analyses. The optimal concentration of Mg2+ was found to be 2.5 mM, with higher concentrations resulting in a slight loss of activity (Fig. 3C). Although less effective, Mn2+ and Zn2+ provided similar trends, with maximal activities approximately 50% and 25%, respectively, of that observed with Mg2+. Interestingly, cytExeA was able to use Ca2+ effectively, such that at 10 mM Ca2+, activity approached that observed with 2.5 mM MgCl2. The activity profile of cytExeA varied considerably depending upon the identity and concentration of NTP used in the assay (Fig. 3D). At low Mg-NTP concentrations, ATP was utilized most effectively; however, at higher Mg-NTP concentrations, greater activities were observed with GTP and CTP. The least utilized substrate was UTP, with activities approaching those of ATP only near 5 mM concentrations. As derived from a Lineweaver-Burk plot (data not shown), the apparent Km for Mg-ATP was 0.22 mM with a Vmax of 0.72 nmol min−1 mg−1 of protein. Estimated Km values for GTP, CTP, and UTP were significantly higher than that for ATP, indicating lower affinity for these substrates (data not shown). Finally, sensitivities of cytExeA to known inhibitors of the ATPase families F, P, and V (generally ion-translocating ATPases), SecA of the Sec translocase, as well as ATPases involved in type I extracellular secretion (such as HlyB) were examined (Table 3). Only compounds known to affect F-type ATPases were found to significantly inhibit cytExeA. An inhibitor of V-type ATPases, KNO3, only slightly affected the activity, with an approximate 25% reduction observed at 10 mM KNO3. In addition, ATPase activity was relatively unaltered by the addition of 0.4 mM vanadate, an inhibitor of P- and HlyB-type ATPases.
FIG. 3.
Enzymatic characterization of cytExeA. NTPase assays were performed using the malachite green method at 37°C with approximately 40 μg of purified cytExeA per reading. Rates were corrected for the nonenzymatic release of Pi from controls without cytExeA. Each data point represents the average of two independent readings. (A) Temperature optimum was quantified using 5 mM ATP and 5 mM MgCl2 at pH 7.9. (B) pH optimum was assessed with 5 mM ATP and 5 mM MgCl2 in 50 mM buffers of various pHs (6 to 7, 7.5 to 8, 8.5 to 9.5, and 10 to 11) composed of PIPES, HEPES, AMPSO, and CAPS, respectively. (C) Divalent cation specificity was determined by addition of 5 mM ATP at pH 7.5 in the presence of Ca2+ (○), Mg2+ (▪), Mn2+ (⧫), or Zn2+ (▴). (D) Nucleotide specificity of cytExeA was investigated by the addition of 5 mM MgCl2 and 5 mM GTP (▪), CTP (▴), ATP (⧫), or UTP (○) at pH 7.5. In panels C and D, the metal-NTP complex concentrations are plotted on the x axis.
ExeA and ExeB are part of a large complex.
Previous characterization of the ExeAB complex via cross-linking analyses suggested that ExeA may form a dimeric structure in addition to forming a complex with ExeB (45). It thus appeared that the 85-kDa ExeAB complex may be part of a higher-order multimer. To obtain an estimate of native ExeAB size, gel filtration chromatography was performed using a Sephacryl S-400 matrix. Material fractionated in this analysis was prepared by solubilization of Ah65(pRJ31.1) membranes with 0.5% octyl-glucoside followed by anion-exchange chromatography of these soluble membrane extracts. Octyl-glucoside (0.5%) solubilization and chromatography conditions maintained the ExeAB complex largely in a nondissociated state, as evidenced by the coelution of ExeA and ExeB during anion-exchange chromatography. This Macro-Q coelution peak, containing intact ExeAB, was collected and subjected to gel filtration analysis with 0.5% octyl-glucoside (Fig. 4). ExeA and ExeB, localized by immunoblotting, were found to cofractionate in two separate peaks. Comparison of the elution volumes of these two peaks with those of known molecular mass markers indicated that ExeAB was present in solution as approximately 185-kDa and 500-kDa complexes. Assuming these complexes contain only ExeA and ExeB and that they are comprised of stoichiometric amounts of the 85-kDa ExeAB complex identified by cross-linking (45), these complexes would consist of approximately two and six ExeAB subunits, respectively. It should be noted, however, that the ExeAB peak eluting from the Macro-Q column and applied to the gel filtration column is not pure, meaning that the ExeAB complexes identified could contain other proteins, and in addition an unknown quantity of the detergent would also be bound to the solubilized complex.
Octyl-glucoside concentrations of 1% or higher resulted in dissociation of the ExeAB complex during membrane solubilization. Therefore, to further elucidate the native ExeAB structure and to assess ExeA and ExeB oligomerization, analyses similar to those described above were performed except that the membranes were solubilized using 1.25% octyl-glucoside to dissociate ExeAB complexes, with subsequent chromatography carried out using 1% detergent. Separate anion-exchange elution peaks containing either ExeA or ExeB were collected and fractionated using the gel filtration matrix Sephacryl S-300. Figure 4B shows the results from two independent experiments in which both ExeA and ExeB eluted in a single peak. With reference to the elution profile of known molecular mass markers, the results indicate that ExeA and ExeB fractionated with apparent molecular masses of approximately 108 kDa and 63 kDa, respectively. These estimates are 1.8 and 2.2 times larger, respectively, than the values derived from in vitro expression and SDS-PAGE analysis (19). If these complexes contained only ExeA or ExeB, the results suggest that both proteins form stable dimers; again, however, bound detergent and possibly other unidentified proteins would be present in the ExeA- and ExeB-containing complexes.
In summary, the ExeAB, ExeA, and ExeB gel filtration analyses suggest the existence of a large native ExeAB complex which may be comprised of three dimers of each protein, although other proteins could be part of these large complexes. Furthermore, the observation that purified cytExeA is present in solution as a monomer as determined by gel filtration (data not shown) suggests that the soluble N-terminal domain is not an ExeA multimerization determinant.
DISCUSSION
ExeA and ExeB are components of the type II secretion pathway of A. hydrophila. The multicomponent secreton is required for extracellular secretion of several exotoxins, such as aerolysin. Previous studies have suggested that ExeA and ExeB form a complex that provides energy required by the secreton to transport exotoxins (45). In the studies reported here, we sought to characterize the putative ATPase activity of ExeA and further examine the oligomeric structure of the ExeAB complex.
Due to the difficulties encountered during native ExeA purification attempts, we expressed and purified the N-terminal cytoplasmic domain of ExeA (cytExeA) that contains the putative ATPase domain fused with a C-terminal six-histidine tag (Fig. 1). One possible explanation for the observed long-term instability of the ExeA activity site in detergent-solubilized extracts is that ExeA contains six Cys residues, four located in the cytoplasmic compartment and two adjacent residues located in the periplasm. Since the cytoplasm is a reducing environment, whereas the periplasm is an oxidizing one, it is possible that proper ExeA conformation requires variable reduction states of its Cys residues. In contrast to ExeA, cytExeA contains only the four cytoplasmic Cys residues and as such, under reducing conditions, is likely to mimic the native state conformation.
This study directly demonstrated that cytExeA and therefore native ExeA is an NTPase. Based on the assay used, which measures the release of inorganic phosphate from nucleotides, it is unlikely that this enzyme acts as a kinase (phosphorylating specific residues within proteins) or a phosphatase (selectively removing phosphate groups from amino acids). Purified cytExeA displayed Mg2+-dependent ATPase activity with a Vmax of 0.72 nmol min−1 mg−1 of protein and a Michaelis constant (Km) of 0.22 mM at pH 7.5, as determined from a linear Lineweaver-Burk plot. However, since we measured the ATPase activity of the truncated cytExeA derivative, which may not contain the appropriate structural context for optimal activity, we cannot rule out the possibility that native ExeA activities are significantly higher. Furthermore, since previous cross-linking analysis (45) and our gel filtration data (Fig. 4) suggest that native ExeA exists at least as a dimer whereas cytExeA is monomeric, it is possible that ExeA requires multimer formation for optimal activity.
Mg2+ was the preferred divalent cation (Fig. 3C), and cytExeA was able to utilize both purine and pyrimidine nucleotides as substrates (Fig. 3D). Interestingly, the maximal hydrolytic rates for GTP and CTP were higher than for ATP, although at low NTP concentrations ATP was hydrolyzed with the greatest efficiency. Due to the relatively low abundance of the other nucleotides in vivo and the observed lower Km value for ATP, ATP is likely the substrate utilized intracellularly. ExeA likely does not use GTP as the main substrate because GTPases are very specific for GTP, with Km values in the μM range, and are also strongly inhibited by GDP (27). Since ExeA exhibited a high Km for GTP and low substrate specificity, this suggests that the observed GTPase activity is likely due to the catalytic activity of an ATPase. In addition, the NTPase activities more closely exhibited hyperbolic substrate kinetics when reactions contained equimolar amounts of nucleotides and MgCl2, a characteristic observed for other ATPases (23). This finding may suggest that binding of the cation prior to that of the nucleotide may inhibit catalytic efficiency, and indeed reactions carried out using various NTP concentrations and a fixed MgCl2 concentration of 2.5 mM exhibited sigmoidal substrate kinetics (data not shown).
ExeA belongs to the Walker superfamily of A/GTPases (50) based on two conserved sequence motifs typical of these proteins, the Walker A and B boxes. Several other enzyme categories have been established, subclassifying ATPases based on sequence (Walker types and others), structure, function, and activity characteristics. These include the AAA (ATPases associated with various cellular activities), ABC (ATP-binding-cassette), F-type (F1Fo), A-type (A1A0, archaeal), V-type (V1V0, vacuolar), P-type (phosphorylated), PulE-VirB11, and SecA families. The AAA members are characterized by the presence of a highly conserved 230-amino-acid domain containing the Walker boxes, as well as by exhibiting sensitivity to N-ethyl-maleimide (mM concentrations) and insensitivity to other ATPase inhibitors (52). Like the AAA proteins, ABC ATPases are also characterized by a highly conserved ∼200-amino-acid motif containing the Walker boxes, as well as a signature sequence motif of up to 15 residues in length commencing with LSGGQ. Most ABC ATPases are inhibited by vanadate, with a small proportion sensitive to azide (44). The inhibitors used in Table 3 are typically used to distinguish between P-type, V-type, and F-type ATPases (33). The A-type ATPases exhibit an inhibitor profile which appears to be a hybrid of both V- and F-type profiles (12). P-type ATPases form an acylphosphate intermediate during their catalytic cycle, which is a hallmark of these enzymes and the reason they are inhibited by vanadate, a transition state analog of phosphate (26). The H+- or Na+-translocating ATPases F, V, and A are multisubunit complexes with at least three dissimilar subunits embedded as a complex in the membrane comprising Fo, with at least five others forming the F1 subunit attached to Fo (32). V-type ATPases are distinguished based on their vacuolar location, whereas A-type ATPases are found in archaea. The recently classified PulE-VirB11 family of ATPases is characterized by a highly conserved region of approximately 100 amino acids containing the Walker boxes, and they appear to be relatively unaffected by common ATPase inhibitors (36). However, it should be noted that ATPase activity has been demonstrated with only a few members, and information on the activity characteristics of this protein class is limited. Finally, SecA, which energizes the Sec translocase, is inhibited by azide (30).
Apart from the Walker A and B motifs, ExeA lacks significant sequence similarity with any of the families described above. To determine whether there are any mechanistic similarities and to further define cytExeA, the effects of various known ATPase inhibitors on cytExeA activity were examined (Table 3). Vanadate had no effect on cytExeA ATPase activity at a concentration of 0.4 mM. As well, cytExeA activity was almost insensitive to NO3−, with a reduction in activity of only approximately 25% with 10 mM KNO3. Surprisingly, inhibitors of F-type ATPases greatly reduced cytExeA activity, including 10 mM NaN3, 5 mM 1,3-dicyclohexylcarbodiimide, and 1 mM 5′-adenylylimidodiphosphate. The divalent cation specificity of cytExeA (Fig. 3C) is also similar to that of F1 ATPase, in that Ca2+ effectively supports activity (46). However, this may not be an important distinguishing factor, since Ca2+-supported ATPase activity has also been observed among members of several other ATPase families (24). Although the inhibitor profile of cytExeA suggests that ExeA belongs to the F-type ATPase family, an interesting discrepancy is that bacterial F1 ATPase is unable to utilize CTP (28), whereas cytExeA exhibits greater maximal activities with this substrate than with ATP (Fig. 3D). To summarize, characteristics of ExeA, including its inhibitor profile, the association with ExeB, and the function of ExeAB in type II extracellular secretion, suggest that ExeA represents a novel ATPase family.
Although both ExeA and ExeB are required for the multimerization of ExeD (2), the role of each of these proteins remains to be determined. One possible role of ExeB may involve a direct interaction with ExeD, as OutB and OutD can be cross-linked with formaldehyde in E. coli cells (9). To date, A. hydrophila cross-linking studies have not identified such a complex. Since gel filtration studies suggest that native ExeAB may exist as a very large complex (Fig. 4), it is tempting to speculate that this complex may directly associate with the putative ExeD homododecamer either during or after transport/assembly. In addition, it has been proposed that ExeAB may fulfill the role in A. hydrophila that is performed by GspS in K. oxytoca and Erwinia (2) in assisting in the stability and outer membrane localization of GspD (15). It is interesting that PulS and PulD have recently been found to form an outer membrane complex composed of 12 subunits of each protein (29). However, the Out system of Erwinia chrysanthemi requires both OutB and OutS, and it is unclear whether they perform overlapping or separate functions. Surprisingly, there have been no GspA homologues identified in Klebsiella and Erwinia. Perhaps the GspA, GspB, and GspS proteins carry out similar or overlapping functions in the localization and assembly of GspD, with the different systems requiring various combinations of these factors for the successful assembly of the secretin.
In conclusion, this study has confirmed that ExeA and ExeB form a large oligomeric inner membrane complex that is capable of providing energy for type II secretion. The novel NTPase activity demonstrated here for ExeA is presumably involved in the recently demonstrated role of this oligomer in mediating the transport and assembly of the ExeD secretin into the outer membrane.
Acknowledgments
We thank Jamie Kalanack for technical assistance and Hughes Goldie for E. coli strain DK8 and helpful discussions.
This research was supported by a research operating grant (MT10470) from the Canadian Institutes of Health Research.
REFERENCES
- 1.Altwegg, M., and H. K. Giess. 1989. Aeromonas as a human pathogen. Crit. Rev. Microbiol. 16:253-286. [DOI] [PubMed] [Google Scholar]
- 2.Ast, V. M., I. C. Schoenhofen, G. R. Langen, C. W. Stratilo, M. D. Chamberlain, and S. P. Howard. 2002. Expression of the ExeAB complex of Aeromonas hydrophila is required for the localization and assembly of the ExeD secretion port multimer. Mol. Microbiol. 44:217-231. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons Ltd., Chichester, England.
- 4.Bortoli-German, I., E. Brun, B. Py, M. Chippaux, and F. Barras. 1994. Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol. Microbiol. 11:545-553. [DOI] [PubMed] [Google Scholar]
- 5.Camberg, J. L., and M. Sandkvist. 2005. Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J. Bacteriol. 187:249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascon, A., J. Yugueros, A. Temprano, M. Sanchez, C. Hernanz, J. M. Luengo, and G. Naharro. 2000. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect. Immun. 68:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty, T., B. Huhle, H. Hof, H. Bergbauer, and W. Goebel. 1987. Marker exchange mutagenesis of the aerolysin determinant in Aeromonas hydrophila demonstrates the role of aerolysin in A. hydrophila-associated systemic infections. Infect. Immun. 55:2274-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condemine, G., C. Dorel, N. Hugouvieux-Cotte-Pattat, and J. Robert-Baudouy. 1992. Some of the out genes involved in the secretion of pectate lyases in Erwinia chrysanthemi are regulated by kdgR. Mol. Microbiol. 6:3199-3211. [DOI] [PubMed] [Google Scholar]
- 9.Condemine, G., and V. E. Shevchik. 2000. Overproduction of the secretin OutD suppresses the secretion defect of an Erwinia chrysanthemi outB mutant. Microbiology 146:639-647. [DOI] [PubMed] [Google Scholar]
- 10.Duong, F., J. Eichler, A. Price, M. Leonard, and W. Wickner. 1997. Biogenesis of the Gram-negative bacterial envelope. Cell 91:567-573. [DOI] [PubMed] [Google Scholar]
- 11.Durand, É., A. Bernadac, G. Ball, A. Lazdunski, J. N. Sturgis, and A. Filloux. 2003. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185:2749-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eddy, M. L., and P. E. Jablonski. 2000. Purification and characterization of a membrane-associated ATPase from Natronococcus occultus, a haloalkaliphilic archaeon. FEMS Microbiol. Lett. 189:211-214. [DOI] [PubMed] [Google Scholar]
- 13.Francetic, O., D. Belin, C. Badaut, and A. P. Pugsley. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francetic, O., and A. P. Pugsley. 1996. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J. Bacteriol. 178:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie, K. R., A. Schulze, M. W. Parker, and J. T. Buckley. 1995. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulphide bond formation. Mol. Microbiol. 17:1035-1044. [DOI] [PubMed] [Google Scholar]
- 17.Howard, S. P., J. Critch, and A. Bedi. 1993. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J. Bacteriol. 175:6695-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard, S. P., H. G. Meiklejohn, D. Shivak, and R. Jahagirdar. 1996. A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol. Microbiol. 22:595-604. [DOI] [PubMed] [Google Scholar]
- 19.Jahagirdar, R., and S. P. Howard. 1994. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J. Bacteriol. 176:6819-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda, J. M., L. S. Guthertz, R. P. Kokka, and T. Shimada. 1994. Aeromonas species in septicemia: laboratory characteristics and clinical observations. Clin. Infect. Dis. 19:77-83. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, B., and S. P. Howard. 1992. The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol. Microbiol. 6:1351-1361. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky, D. J., W. S. A. Brusilow, and R. D. Simoni. 1984. In vivo evidence for the role of the epsilon subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli. J. Bacteriol. 160:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koronakis, V., C. Hughes, and E. Koronakis. 1993. ATPase activity and ATP/ADP-induced conformational change in the soluble domain of the bacterial protein translocator HlyB. Mol. Microbiol. 8:1163-1175. [DOI] [PubMed] [Google Scholar]
- 24.Krause, S., W. Pansegrau, R. Lurz, F. de la Cruz, and E. Lanka. 2000. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182:2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Macara, I. G. 1980. Vanadium—an element in search for a role. Trends Biochem. Sci. 5:92-94. [Google Scholar]
- 27.Moll, R., S. Schmidtke, and G. Schafer. 1999. Domain structure, GTP-hydrolyzing activity and 7S RNA binding of Acidianus ambivalens ffh-homologous protein suggest an SRP-like complex in archaea. Eur. J. Biochem. 259:441-448. [DOI] [PubMed] [Google Scholar]
- 28.Noji, H., D. Bald, R. Yasuda, H. Itoh, M. Yoshida, and K. Kinosita. 2001. Purine but not pyrimidine nucleotides support rotation of F(1)-ATPase. J. Biol. Chem. 276:25480-25486. [DOI] [PubMed] [Google Scholar]
- 29.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and W. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver, D. B., R. J. Cabelli, K. M. Dolan, and G. P. Jarosik. 1990. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc. Natl. Acad. Sci. USA 87:8227-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panagiotidis, C. H., and H. A. Shuman. 1998. Maltose transport in Escherichia coli: mutations that uncouple ATP hydrolysis from transport. Methods Enzymol. 292:30-39. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen, P. L., and L. M. Amzel. 1992. F-type ATPases. Introduction. J. Bioenerg. Biomembr. 24:427-428. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen, P. L., and E. Carafoli. 1987. Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem. Sci. 12:146-150. [Google Scholar]
- 34.Pepe, C. M., M. W. Eklund, and M. S. Strom. 1996. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol. Microbiol. 19:857-869. [DOI] [PubMed] [Google Scholar]
- 35.Possot, O. M., G. Vignon, N. Bomchil, F. Ebel, and A. P. Pugsley. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 182:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivas, S., S. Bolland, E. Cabezon, F. M. Goni, and F. de la Cruz. 1997. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 272:25583-25590. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 40.Sandkvist, M., M. Bagdasarian, S. P. Howard, and V. J. DiRita. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14:1664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15:430-434. [DOI] [PubMed] [Google Scholar]
- 43.Sauvonnet, N., G. Vignon, A. P. Pugsley, and P. Gounon. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 45.Schoenhofen, I. C., C. Stratilo, and S. P. Howard. 1998. An ExeAB complex in the type II secretion pathway of Aeromonas hydrophila: effect of ATP-binding cassette mutations on complex formation and function. Mol. Microbiol. 29:1237-1247. [DOI] [PubMed] [Google Scholar]
- 46.Senior, A. E. 1990. The proton-translocating ATPase of Escherichia coli. Annu. Rev. Biophys. Biophys. Chem. 19:7-41. [DOI] [PubMed] [Google Scholar]
- 47.Sudom, A., R. Walters, L. Pastushok, D. Goldie, L. Prasad, L. T. J. Delbaere, and H. Goldie. 2003. Mechanisms of activation of phosphoenolpyruvate carboxykinase from Escherichia coli by Ca2+ and of desensitization by trypsin. J. Bacteriol. 185:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traut, T. W. 1994. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur. J. Biochem. 222:9-19. [DOI] [PubMed] [Google Scholar]
- 49.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 52.Wolf, S., I. Nagy, A. Lupas, G. Pfeifer, Z. Cejka, S. A. Muller, A. Engel, R. De Mot, and W. Baumeister. 1998. Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J. Mol. Biol. 277:13-25. [DOI] [PubMed] [Google Scholar]