Abstract
We used panhandle PCR to clone the der(11) genomic breakpoint junction in three leukemias with t(4;11) and devised reverse-panhandle PCR to clone the breakpoint junction of the other derivative chromosome. This work contributes two elements to knowledge on MLL translocations. First is reverse-panhandle PCR for cloning breakpoint junctions of the other derivative chromosomes, sequences of which are germane to understanding the MLL translocation process. The technique revealed duplicated sequences in one case of infant acute lymphoblastic leukemia (ALL) and small deletions in a case of treatment-related ALL. The second element is discovery of a three-way rearrangement of MLL, AF-4, and CDK6 in another case of infant ALL. Cytogenetic analysis was unsuccessful at diagnosis, but suggested t(4;11) and del(7)(q21q31) at relapse. Panhandle PCR analysis of the diagnostic marrow identified a breakpoint junction of MLL intron 8 and AF-4 intron 3. Reverse-panhandle PCR identified a breakpoint junction of CDK6 from band 7q21-q22 and MLL intron 9. CDK6 encodes a critical cell cycle regulator and is the first gene of this type disrupted by MLL translocation. Cdk6 is overexpressed or disrupted by translocation in many cancers. The in-frame CDK6-MLL transcript is provocative with respect to a potential contribution of the predicted Cdk6-MLL fusion protein in the genesis of the ALL, which also contains an in-frame MLL-AF4 transcript. The sequences in these three cases show additional MLL genomic breakpoint heterogeneity. Each breakpoint junction suggests nonhomologous end joining and is consistent with DNA damage and repair. CDK6-MLL is a new fusion of both genes.
The MLL gene was cloned 10 years ago as a common target of translocations in human acute leukemias (1–3), especially in infants. The translocations fuse the breakpoint cluster region (bcr) that spans exons 5–11 of MLL with one of many partner genes, 31 of which have been cloned so far [J. L. Huret, (2001) http://www.infobiogen.fr/services/chromcancer/Anomalies/11q23ID1030.html]. The genomic breakpoint junction sequences provide clues to the translocation mechanism and suggest DNA damage and repair (4–7). Backtracking nonconstitutional MLL translocations to the prenatal period (8–10) indicates that the damage occurs in utero, but the agent(s) is unknown. Because similar translocations occur in leukemias related to chemotherapy with DNA topoisomerase II inhibitors, DNA topoisomerase II may be implicated in the damage (reviewed in ref. 11). Maternal prenatal consumption of dietary DNA topoisomerase II inhibitors may increase the risk of infant acute myeloid leukemia (12). An inactivating NAD(P)H:quinone oxidoreductase polymorphism is associated with infant leukemias with MLL translocations (13), and the NQO1 substrate benzoquinone interferes with DNA topoisomerase II (14). A model for the translocation process involves DNA topoisomerase II-mediated chromosomal breakage and formation of the translocations when the breakage is repaired.
Nonetheless, the genomic breakpoint junction sequences of both derivative chromosomes have been examined in few de novo and treatment-related leukemias that represent the spectrum of partner genes of MLL. The large number of potential partner genes can impede genomic cloning. We have used panhandle PCR approaches to clone the der(11) genomic breakpoint junctions (7, 15–17). Amplification of the genomic breakpoint junctions of the other derivative chromosomes with primers based on der(11) sequences may be unsuccessful if there are large duplications or deletions or if the rearrangements are complex. Here, we developed reverse-panhandle PCR to clone the breakpoint junctions of the other derivative chromosomes and identified a new CDK6-MLL fusion in a cryptic, complex translocation.
Methods
IRBs at the Children's Hospital of Philadelphia and Memorial Sloan–Kettering Cancer Center approved this research.
Case Histories.
Patient 45 was diagnosed with French–American–British (FAB) L1 acute lymphoblastic leukemia (ALL) at age 3 weeks. She presented with hepatosplenomegaly and a WBC count of 86 × 109/liter, but no evidence of central nervous system disease. The bone marrow karyotype in five metaphases was 46,XX,t(4;11). The immunophenotype was Tdt+, CD19+, CD10−, CD20−; no myeloid antigens were expressed. At age 5 months, a progressive seizure disorder with loss of milestones developed. Head MRI and CT scans were normal. By age 10 months, myeloblasts in the cerebrospinal fluid suggested CNS relapse with lineage shift. She suffered rapid neurologic deterioration and died.
Patient t-120 was diagnosed with stage IV neuroblastoma at age 2 years. His primary posterior mediastinal tumor was metastatic to the bone and marrow. Memorial Sloan–Kettering N7 treatment included four cycles of cyclophosphamide, doxorubicin, and vincristine, three cycles of cisplatin and etoposide (PVP), surgical resection, local radiation, radiolabeled anti-GD2 mAb (3F8), and autologous marrow rescue with cells harvested after chemotherapy cycle 5 (PVP) and purged ex vivo with 3F8. Eleven months after starting treatment and 2 weeks after transplant, the WBC count was 46 × 109/liter and FAB L2 ALL was diagnosed. The karyotype in 17 metaphases was 46,XY,t(4;11)(q21;q23).
The presentation of patient 38 at infant ALL diagnosis was as described (15). The 3-month-old girl presented with hepatosplenomegaly and a WBC count of 399 × 109/liter. The marrow was replaced with FAB L1, Tdt+, CD19+, CD10−, CD20−, CD34+ blasts. Cytogenetic analysis of the diagnostic marrow was unsuccessful (15). She received CCG 1883-like chemotherapy (18) but relapsed in the marrow at age 4 years, 25 months from completion of this treatment, when the marrow karyotype in three metaphases was 47,XX,t(4;11)(q21;q23),del(7)(q21q31),+8. She died from Pseudomonas sepsis during reinduction.
Detection of MLL Gene Rearrangements.
Rearrangements were examined by Southern blot analysis of BamHI-digested DNA with the B859 fragment of ALL-1 cDNA (1).
Cloning of der(11) Genomic Breakpoint Junctions.
For the leukemia of patient 38, panhandle PCR analysis of the MLL-AF-4 genomic breakpoint junction was described (GenBank accession no. AF031403) (15). The der(11) genomic breakpoint junctions in the leukemias of patients 45 and t-120 were amplified by panhandle PCR as described (15), except that primers 3 and 4 were those used for cDNA panhandle PCR (7). Panhandle PCR products were subcloned by recombination PCR (7); subclones were screened by PCR and sequenced. der(11) breakpoint junctions were confirmed by amplification of genomic DNAs with MLL- and AF-4-specific primers and direct sequencing.
Cloning of Genomic Breakpoint Junctions of Other Derivative Chromosomes.
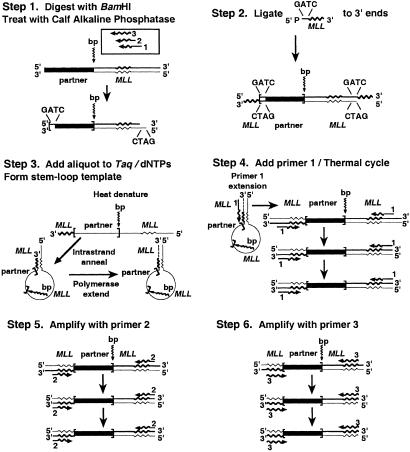
Reverse-panhandle PCR was accomplished by ligation of a phosphorylated oligonucleotide containing known sense sequence from MLL intron 10/exon 11 to the 3′ ends of BamHI-digested DNA and formation of a stem-loop template from the antisense strand. The template contained unknown partner sequence, the breakpoint junction of the other derivative chromosome, and MLL sequence in the loop. MLL sequence and its complement at either end of the “handle” enabled amplification of the breakpoint junction in three sequential, single-primer, two-sided PCRs with primers all antisense with respect to MLL exon 11 or intron 10/exon 11 sequences (Fig. 1).
Figure 1.
Reverse-panhandle PCR analysis of genomic breakpoint junction of other derivative chromosomes of MLL translocation.
In Step 1, 2.5 μg of genomic DNA was digested with 20 units of BamHI (New England Biolabs). The DNA was treated with 0.025 units of calf intestinal alkaline phosphatase (Boehringer Mannheim) at 37°C for 30 min and purified with a Geneclean III kit (Bio 101). In Step 2, a single-stranded, 5′ phosphorylated oligonucleotide (5′-GATCTCTAGATCTGTACCAAGTGTGTTCGCTGTAAGAGC-3′) was ligated to the 3′ ends. The 4-base 5′ end of the oligonucleotide was complementary to the 5′ overhang of the BamHI-digested DNA; its 3′ end corresponded to sense positions 8299–8333 from MLL intron 10/exon 11 (GenBank accession no. U04737). Each 50-μl ligation reaction mixture contained 2.5 μg of DNA, a 50-fold molar excess of 5′ phosphorylated oligonucleotide, 1 Weiss unit of T4 DNA ligase, and 1× ligase buffer (Boehringer Mannheim). Ligations were performed at 4°C. The DNA was purified with a Geneclean III kit (Bio 101). The stem-loop template was formed from the antisense strand in Step 3. After heating the other components to 80°C for 5 min, 20 ng of digested, ligated DNA was added to 1.75 units of Taq/Pwo DNA polymerase mix, 368 μM each dNTP, and 1.05× PCR buffer (Expand Long Template PCR System; Boehringer Mannheim) in a 47.5-μl reaction mixture. The DNA was made single-stranded by heating the reaction mixture at 94° for 1 min. The template was generated by a 2-min ramp to 72°C and incubation at 72°C for 30 s to promote intrastrand annealing of the ligated oligonucleotide to the complementary sequence in the antisense strand and polymerase extension of the recessed 3′ end (15). In Step 4, primer 1 extension made the template double-stranded, allowing exponential amplification with primer 1, which anneals to both ends. After 2.5 μl of a 5-pmol/μl solution of primer 1 corresponding to MLL exon 11 antisense positions 8342–8315 (5′-GGATCCACAGCTCTTACAGCGAACACAC-3′) was added, each final, 50-μl PCR contained 12.5 pmol of primer 1, 350 μM each dNTP, and 1× PCR buffer. After denaturation at 94°C for 1 min, 10 cycles at 94°C for 10 s and 68°C for 7 min and 20 cycles at 94°C for 10 s and 68°C for 7 min (increment, 20 s/cycle) were used, followed by final elongation at 68°C for 7 min. Steps 5 and 6 were sequential, two-sided, single-primer-nested PCRs with primers 2 and 3, respectively, which are antisense with respect to MLL exon 11 positions 8336–8305 (5′-ACAGCTCTTACAGCGAACACACTTGGTACAGA-3′) and MLL exon 11/intron 10 positions 8333–8299 (5′-GCTCTTACAGCGAACACACTTGGTACAGATCTAGA-3′). Nested PCR conditions were the same as in the initial PCR; 1-μl aliquots of the products of respective preceding PCRs were used as templates.
Reverse-panhandle PCR products were subcloned by recombination PCR (7). pUC19 was linearized by HindIII digestion. MLL ends complementary to the ends of the products of the last nested PCR in reverse-panhandle PCR were added to the linearized vector by amplification with primers 5′- ACCAAGTGTGTTCGCTGTAAGAGCTGGCGTAATCATGGTCATAGC-3′ and 5′-ACCAAGTGTGTTCGCTGTAAGAGCCATGCCTGCAGGTCGACTCTAGAG-3′.
The breakpoint junctions of the other derivative chromosomes were validated by PCR amplification of genomic DNA with gene-specific primers and direct sequencing.
PCR was performed on genomic DNA from the diagnostic marrow of patient 38 with the sense primer 5′-GAAATGGGTGCAGTGTTCCA-3′ from AF-4 intron 3 and the antisense primer 5′-TGGATTACGGGATAGGGACA-3′ from CDK6 intron 2 to determine whether a reciprocal AF-4-CDK6 rearrangement had occurred.
Analysis of Fusion Transcripts.
Reverse transcriptase–PCR analysis of the MLL-AF-4 transcript in the marrow cells of patient 38 at diagnosis has been described (GenBank accession no. AF031404) (15). The same method was used to generate randomly primed, first-strand cDNA and characterize the der(11) transcript in the ALL cells of patient 45 (15). The der(11) transcript in the ALL cells of patient t-120 was identified by cDNA panhandle PCR (7) and confirmed by amplification of the same first-strand cDNA with MLL- and AF-4-specific primers.
der(4) transcripts in the leukemia cells of patients 45 and t-120 were identified by amplification of the above-generated first-strand cDNAs with the AF-4 exon 3 sense primer 5′-CTCCCCTCAAAAAGTGTTGC-3′ (GenBank accession no. L13773) and the MLL exon 9 antisense primer 5′-CAATTTTCCAGCTGGTCCTC-3′ (GenBank accession no. L04284).
The CDK6-MLL transcript was identified in the ALL cells of patient 38 with the sense primer 5′-CGTGGTCAGGTTGTTTGATG-3′ from CDK6 exons 1–2 (GenBank accession no. NM_001259) and the MLL exon 13 antisense primer 5′-GCCGCTCAGTACAGTTCACA-3′. PCR was performed with the same first-strand cDNA and the AF-4 exon 3 sense primer 5′-CTCCCCTCAAAAAGTGTTGC-3′ and the CDK6 exon 4 antisense primer 5′-GACTTCGGGTGCTCTGTACC-3′.
Results
Characterization of der(11) and der(4) Genomic Breakpoint Junctions and Fusion Transcripts in Infant ALL Cells.
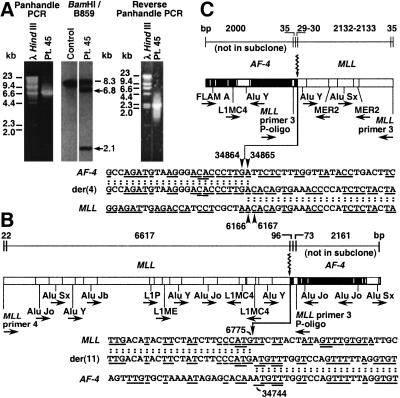
Southern blot analysis revealed 6.8- and 2.1-kb MLL bcr rearrangements in the infant ALL of patient 45 (Fig. 2A). Panhandle PCR amplified the der(11) genomic breakpoint junction (Fig. 2 A and B). The 6,808-bp product suggested that the 6.8- and 2.1-kb rearrangements were from the der(11) and der(4) chromosomes, respectively. The MLL der(11) breakpoint was 3′ in the bcr at position 6775 in intron 8 (GenBank accession no. U04737) (Fig. 2B). The der(11) breakpoint in the partner gene was position 34744 in AF-4 intron 3 (GenBank accession no. AJ238093).
Figure 2.
(A) MLL bcr rearrangements in ALL of patient 45 (Center) and panhandle PCR (Left) and reverse-panhandle PCR (Right) products. The 8.3-kb fragment on Southern blot is from unrearranged MLL allele (Center, dash); arrows show rearrangements. (B) Summary of der(11) genomic breakpoint junction in recombination PCR-generated subclones from panhandle PCR. One subclone was sequenced in its entirety; the breakpoint junction was verified in three more subclones. The 5′ 6,639 bp include MLL primer 4 and MLL bcr sequence. Ninety-six base pairs of 3′ sequence are AF-4 DNA. Seventy-three base pairs of 3′ sequence extend from ligated oligonucleotide (P-Oligo) through MLL primer 3 (Top). Arrow shows MLL and AF-4 breakpoint positions (Bottom). Underlines show short homologies (bottom). Repetitive sequences are shown (Middle). (C) Summary of der(4) genomic breakpoint junction in recombination PCR-generated subclone from reverse-panhandle PCR. In reverse-panhandle PCR, nested primer 3 from MLL exon 11/intron 10 anneals to both ends of the template. Thirty-five base pairs of 5′ sequence extend from MLL primer 3 through ligated oligonucleotide (P-Oligo). Twenty-nine to 30 bp of 5′ sequence are AF-4. The 3′ 2167–2168 bp are MLL bcr sequence through MLL primer 3 (Top). Arrowheads show AF-4 and MLL breakpoint positions; “A” residue in both genes precluded precise assignments (Bottom). Short homologies are underlined (Bottom). Repetitive sequences are shown (Middle). One subclone was sequenced in its entirety; three PCRs with gene-specific primers confirmed der(4) breakpoint junction.
AF-4 and MLL primers were designed to amplify the der(4) genomic breakpoint junction predicted by the der(11) sequence. The expected product size was 494 bp but a ≈1.2-kb product was obtained (data not shown). Reverse-panhandle PCR was tested in this case with a known der(4) breakpoint junction sequence. The 2,232-bp product was consistent with the 2.1-kb MLL bcr rearrangement on the Southern blot (Fig. 2A). The sequence was the same as in the PCR product obtained with gene-specific primers, validating reverse-panhandle PCR as a cloning strategy for the breakpoint junction of the other derivative chromosome of an MLL translocation. The AF-4 der(4) breakpoint was position 34864 or 34865 in intron 3; the MLL der(4) breakpoint was position 6166 or 6167 in intron 8. “A” residues at the breakpoints in both genes precluded more precise breakpoint assignments (Fig. 2C). Depending on the exact breakpoint positions, 609–610 bases from AF-4 and 121–122 bases from MLL were present in both derivative chromosomes, suggesting duplication (Fig. 2 B and C). Other identical 1- to 4-base sequences in MLL and AF-4 were present near the der(11) and der(4) breakpoint junctions. There were AluJo repeats in MLL and AF-4, within ≈1,681 bp and ≈259 bp, respectively, of the der(11) breakpoint junction (Fig. 2B). The MLL der(4) breakpoint was within an AluY, and there was an AluY in AF-4 intron 3 ≈988 bp from the der(4) breakpoint junction (Fig. 2C).
der(11) and der(4) transcripts were produced. The der(11) transcript fused MLL exon 8 in-frame to AF-4 exon 4. The der(4) transcript fused AF-4 exon 3 in-frame to MLL exon 9.
Characterization of der(11) and der(4) Genomic Breakpoint Junctions and Fusion Transcripts in Treatment-Related ALL Cells.
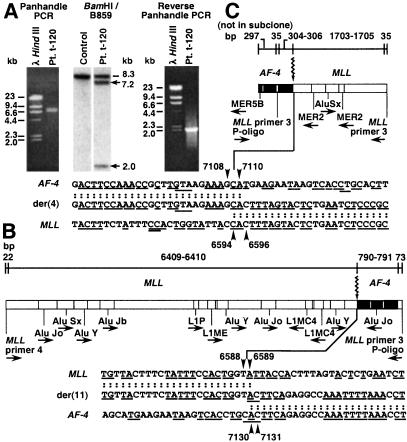
Southern blot analysis revealed 7.2- and 2.0-kb MLL bcr rearrangements in the treatment-related ALL of patient t-120 (Fig. 3A). Panhandle PCR products (7,295 bp) suggested that the 7.2- and 2.0-kb rearrangements were from the der(11) and der(4) chromosomes, respectively (Fig. 3A). The MLL der(11) breakpoint was position 6588 or 6589 in intron 8, also 3′ in the bcr; the der(11) breakpoint in the partner gene was AF-4 intron 3 position 7130 or 7131 (Fig. 3B). These breakpoints could not be localized precisely because both genes contain an “A” residue at the breakpoint junction. Other 1- to 4-base homologies were present near the breakpoints in both genes (Fig. 3B). The MLL and AF-4 der(11) breakpoints were near AluJo and other repetitive sequence elements.
Figure 3.
(A) MLL bcr rearrangements in ALL of patient t-120 (Center) and panhandle PCR (Left) and reverse-panhandle PCR (Right) products. The 8.3-kb fragment on Southern blot (Center, dash) and larger, 8.3-kb panhandle PCR product (Left) are from the unrearranged MLL allele; arrows show rearrangements (Center). (B) Summary of der(11) genomic breakpoint junction in recombination PCR-generated subclones from panhandle PCR. One subclone was sequenced in its entirety; the breakpoint junction was verified in another subclone. The 5′ 6431–6432 bp include MLL primer 4 and MLL bcr sequence. 790–791 bp of 3′ sequence are AF-4. Seventy-three base pairs of 3′ sequence extend from ligated oligonucleotide (P-Oligo) through MLL primer 3 (Top). Arrowheads show AF-4 and MLL breakpoint positions; “A” residue in both genes precluded precise assignments (Bottom). Underlines indicate short homologies (Bottom). Repetitive sequences are shown (Middle). (C) Summary of der(4) genomic breakpoint junction in recombination PCR-generated subclones from reverse-panhandle PCR. One subclone was sequenced in its entirety; the breakpoint junction was verified in three more subclones. Thirty-five base pairs of 5′ sequence extend from MLL primer 3 through ligated oligonucleotide (P-Oligo). 304–306 bp of 5′ sequence are AF-4. The 3′ 1738–1740 bp include MLL bcr sequence through MLL primer 3. Arrowheads show AF-4 and MLL breakpoint positions; “CA” in both genes precluded precise assignments (Bottom). Short homologies are underlined (Bottom). Repetitive sequences are shown (Middle).
The 2,079-bp reverse-panhandle PCR product was consistent with the 2.0-kb rearrangement on the Southern blot (Fig. 3A). The AF-4 der(4) breakpoint was position 7108, 7109, or 7110 in intron 3; the MLL der(4) breakpoint was position 6594, 6595, or 6596 in intron 8 (Fig. 3C). In addition to the 5′-CA-3′ immediately at the breakpoints in both genes that precluded more precise assignments, other short, homologous sequences flanked the der(4) breakpoint junction (Fig. 3C). The closest repetitive sequences to the der(4) breakpoints in both genes were MERs (Fig. 3C). Depending on the exact breakpoint positions, 4–7 bp from MLL and 19–22 bp from AF-4 were lost during the translocation.
cDNA panhandle PCR identified an in-frame der(11) transcript joining MLL exon 8 to AF-4 exon 4. A der(4) transcript fusing AF-4 exon 3 in-frame to MLL exon 9 also was produced.
Reverse-Panhandle PCR Identifies Complex Translocation of MLL, AF-4, and CDK6.
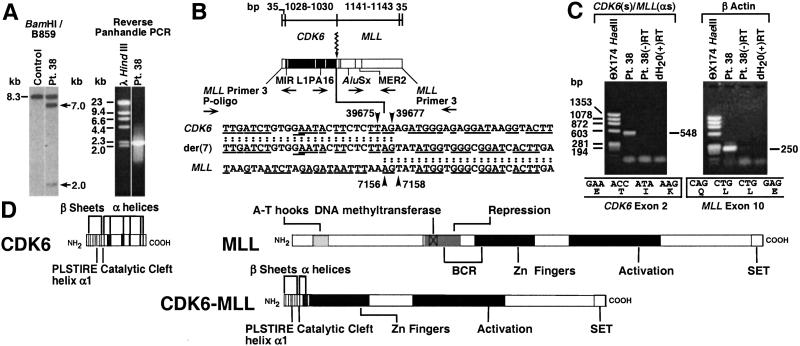
Southern blot analysis of the diagnostic marrow of patient 38 revealed 7.0- and 2.0-kb MLL bcr rearrangements (Fig. 4A) (15). Although cytogenetic analysis was unsuccessful, panhandle PCR identified an MLL intron 8-AF-4 intron 3 genomic breakpoint junction of a putative der(11) chromosome. The MLL breakpoint was position 3802 in intron 8; the AF-4 breakpoint was position 16039 in intron 3 (GenBank accession no. AF031403) (15). Because PCR with AF-4- and MLL-specific primers designed from this sequence did not identify the predicted der(4) breakpoint junction (data not shown), reverse-panhandle PCR was used to identify the genomic breakpoint junction of the other derivative chromosome, the presence of which was suggested by the two MLL bcr rearrangements on the Southern blot. The panhandle PCR product size suggested that the 7.0-kb rearrangement was from the putative der(11) chromosome (15). The Southern blot and panhandle PCR product size predicted a reverse-panhandle PCR product of ≈2.0 kb. A 2,241-bp reverse-panhandle PCR product was obtained (Fig. 4A); the sequence showed that the 3′ portion of the MLL bcr had not fused with AF-4 but with the CDK6 (cyclin-dependent kinase 6) gene from chromosome band 7q21-q22 (Fig. 4B). The CDK6 intron 2 breakpoint corresponded to position 39675–39677 of the genomic clone AC004128. The MLL breakpoint was position 7156–7158 in intron 9. A total of 3355–3357 were lost from MLL in the complex rearrangement. Homologous 5′-AG-3′ sequences in CDK6 and MLL precluded more precise breakpoint assignments. Other short, homologous sequences in CDK6 and MLL flanked the breakpoint junction (Fig. 4B). An MLL exon 8-AF-4 exon 4 fusion transcript was produced as described previously (GenBank accession no. AF031404) (15). The CDK6-MLL rearrangement produced an in-frame fusion transcript of CDK6 exon 2 with MLL exon 10 (Fig. 4C). Although there were no mitoses on cytogenetic analysis of the diagnostic bone marrow (15), the bone marrow karyotype at relapse demonstrated del(7)(q21q31) in addition to the t(4;11) (Fig. 5). No material was available for fluorescence in situ hybridization analysis, but the molecular analyses of the diagnostic marrow are consistent with a three-way translocation. Genomic DNA from the diagnostic marrow was analyzed with AF-4- and CDK6-specific primers, but no reciprocal AF-4-CDK6 product was obtained. In addition, no AF-4-CDK6 fusion transcript was detected (data not shown).
Figure 4.
(A) MLL bcr rearrangements in ALL of patient 38 (15) (arrows, Left) and reverse-panhandle PCR product (Right) consistent with 2.0-kb rearrangement. The 8.3-kb fragment (Left) was from unrearranged MLL allele; 7.0-kb fragment was from MLL-AF-4 rearrangement (15). (B) Sequence of genomic breakpoint junction of other derivative chromosome in recombination PCR-generated subclone from reverse-panhandle PCR. Thirty-five base pairs of 5′ sequence are from MLL primer 3 through ligated oligonucleotide (P-Oligo). 1028–1030 bp of 5′ sequence are CDK6. The 3′ 1176–1178 bp include MLL bcr sequence from intron 9 through MLL primer 3. Arrowheads show CDK6 and MLL breakpoint positions; “AG” in both genes precluded precise assignments (Bottom). Short homologies are underlined (Bottom). Repetitive sequences are shown (Middle). (C) Detection of CDK6-MLL fusion transcript. Reverse transcriptase–PCRs with CDK6 exons 1–2 and MLL exon 13 primers and randomly primed first-strand cDNA gave a 548-bp product (Top). Reaction with β-actin primers and RNA-negative reagent control reaction (dH2O) were performed (Top). Sequencing showed in-frame fusion of CDK6 exon 2 at position 486 of the 1,249-bp CDK6 cDNA (GenBank accession no. NM_001259) to MLL exon 10 (Bottom). (D) Cdk6 and MLL proteins and predicted Cdk6-MLL fusion protein.
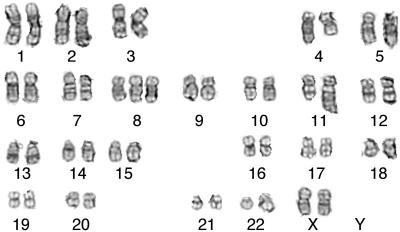
Figure 5.
Representative G-banded karyotype of relapse marrow of patient 38. The karyotype was described as 47,XX,t(4;11)(q21;q23),del(7)(q21q31),+8.
Discussion
Examination of the genomic breakpoint junctions of both derivative chromosomes is essential to understanding the MLL translocation process. It is customary to attempt isolation of the genomic breakpoint junction of the other derivative chromosome by PCR with partner gene- and MLL-derived primers designed based on the der(11) sequence (5, 7) or, in the case of the t(4;11), based on karyotypic evidence of potential involvement of a known partner gene of MLL (4). This approach fails when there are large duplications, deletions, inversions, or complex rearrangements or when the der(11) sequence or the partner gene is unknown. Because MLL has many partner genes, we previously implemented panhandle PCR and panhandle variant PCR for der(11) genomic breakpoint junctions and cDNA panhandle PCR for der(11) transcripts in which all primers are from MLL (7, 9, 15, 17, 19, 20). For the present study, we devised a reverse-panhandle PCR approach to clone the genomic breakpoint junctions of the other derivative chromosomes that has features similar to panhandle PCR and panhandle variant PCR (7, 9, 15, 17). Stem-loop templates are created in all three genomic methods by BamHI digestion, which creates a fragment size amenable to PCR, and ligation of known MLL bcr sequence to the unknown partner sequence in the fragment (7, 9, 15–17). All four panhandle PCR approaches lead readily to known and unknown partner sequences.
Although both the der(11) and der(4) genomic breakpoint junctions have been amplified with gene-specific primers in many de novo leukemias with t(4;11) (4, 5), both genomic breakpoint junctions have been characterized in few de novo leukemias with other MLL translocations (6) and few leukemias after chemotherapy with DNA topoisomerase II inhibitors (7, 21–23). As in the ALL of patient 45, the sequences in de novo cases have shown regions up to several hundred bases from MLL and/or the partner gene on both derivative chromosomes, suggesting duplication (4–6). Deletions of several hundred bases also have been observed (4, 6). Except in the ML-1 cell line (22), in treatment-related leukemias with MLL translocations including the ALL of patient t-120, the sequences suggest more precise recombinations with deletions or duplications of relatively few bases (7, 21, 23).
The association of DNA topoisomerase II-targeted chemotherapy with leukemias with MLL translocations has suggested that repair of DNA topoisomerase II-mediated chromosomal breakage may cause the translocations (reviewed in ref. 11). Sequence information on both genomic breakpoint junctions may suggest different types of DNA topoisomerase II cleavage in de novo and treatment-related cases. DNA topoisomerase II creates 4-base, staggered, double-stranded breaks in DNA, but DNA topoisomerase II also introduces single-stranded nicks as kinetic intermediates of double-stranded breaks (24). The more precise recombination in treatment-related cases may be more consistent with the processing of 4-base, staggered, double-stranded breaks (23). Duplicated sequences in the de novo cases may arise from staggered, single-stranded nicks in both DNA strands and template-directed polymerization of the intervening sequence (4, 5). The large, deleted regions in some de novo cases may arise from multiple breaks or extensive processing (4, 6). Sequence differences between treatment-related and de novo cases may suggest that different types of breakage are induced by different agents. Microhomologies at the breakpoint junctions in the leukemias studied here, as those at other MLL genomic breakpoint junctions (4–7, 9, 15), may suggest that nonhomologous end-joining is involved in the repair (4, 23).
The MLL-AF-4 genomic breakpoint junction in the ALL of patient 38 predicted a reciprocal AF-4-MLL genomic breakpoint junction, but reverse-panhandle PCR led instead to the discovery of the CDK6-MLL junction in a cryptic, complex, three-way rearrangement. The ≈226-kb CDK6 gene located at chromosome 7q21-q22 contains 7 exons (25), which encode a 325-aa, 40-kDa protein (26, 27). Cdk6 is a D cyclin-dependent kinase signaling at the G1/S cell cycle transition and the major Cdk in human lymphoid cells (27–29). Upon D cyclin activation, Cdk6 phosphorylates and inactivates Rb, inhibiting its growth-suppressive function, activating E2F transcription factors, and enabling entry into S phase (30). An in-frame CDK6-MLL fusion transcript was identified. The corresponding full-length fusion transcript would include the first 123 codons of CDK6 and the last 2,476 codons of MLL. The PLSTIRE helix α1, β sheets, catalytic cleft, and 22 residues from the carboxyl-terminal α-helices of Cdk6 (26) and the zinc fingers, transactivation, and SET domains of MLL would be preserved in the predicted fusion protein (Fig. 4D). Although the term “partner gene” generally refers to the gene whose 3′ sequence is fused to the 5′ sequence of MLL, CDK6 is the first gene of this type identified in an MLL translocation.
MLL translocations are thought to be leukemogenic by producing chimeric oncoproteins from the der(11) chromosome in which the amino terminus of MLL is joined to the carboxyl terminus of the partner protein (31). Murine models have used 5′-MLL-partner-3′ constructs to establish that MLL translocations are leukemogenic (31). Latency to leukemia in these models suggests that additional alterations also may be important. At least one such construct (5′-MLL-FBP17-3′) shows minimal transformation in serial replating assays (32). However, experiments on the potential functional contribution of partner-MLL fusion proteins have not been performed. Although an MLL-AF-4 transcript was produced in the leukemia cells of patient 38 (15) and the der(11) gene product is considered critical in leukemogenesis (31), it is possible that the Cdk6-MLL fusion protein predicted by the maintenance of a productive ORF in the fusion transcript may have contributed as well.
Cdk6 overexpression occurs in T cell lymphoblastic lymphoma, T cell ALL, natural killer/T cell nasal lymphoma, and glioblastoma multiforme (27, 33, 34). B cell splenic lymphomas with villous lymphocytes are characterized by t(2;7)(p12;q21) juxtaposing CDK6 to the Ig κ gene (35). A t(7;21) disrupting CDK6 was observed in a splenic marginal zone lymphoma (35). The central role of Cdk6 in cell cycle progression and its recurrent alteration in human cancer suggest that the CDK6-MLL juxtaposition may have been a cooperating mutation in leukemogenesis in patient 38. G-banded and spectral karyotype analyses identified t(7;11)(q22;q23) in infant ALL, MDS, and non-Hodgkin lymphoma [F. Mitelman, B. Johansson, and F. Mertens (2000) http://cgap.nci.nih.gov/Chromosomes/Mitelman], possibly suggesting that CDK6-MLL junctions will be found in other cases.
Genomic breakpoint junction sequences and the fusion transcripts resulting from three-way rearrangements provide insights into the translocation process and molecular alterations leading to leukemias with MLL translocations. Other three-way MLL translocations have been identified (32, 36–39). Reverse-panhandle PCR is a significant advance for cloning breakpoint junctions of the other derivative chromosomes and is well suited to complex, three-way rearrangements. The duplicated sequences in the ALL of patient 45, small deletions in the treatment-related ALL of patient t-120, and complex translocation in the ALL of patient 38 indicate heterogeneity in MLL genomic breakpoint junctions and are consistent with DNA damage and repair.
Acknowledgments
We thank Nancy Spinner for cytogenetic analysis of relapse marrow of patient 38. C.A.F. was supported by National Institutes of Health Grants CA66140, CA80175, CA77683, and CA85469, a Leukemia and Lymphoma Society Translational Research Award, and the Joshua Kahan Foundation.
Abbreviations
- bcr
breakpoint cluster region
- ALL
acute lymphoblastic leukemia
Footnotes
References
- 1.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 2.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 3.Ziemin-van der Poel S, McCabe N R, Gill H J, Espinosa R, III, Patel Y, Harden A, Rubinelli P, Smith S D, LeBeau M M, Rowley J D, et al. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillert E, Leis T, Repp R, Reichel M, Hosch A, Breitenlohner I, Angermuller S, Borkhardt A, Harbott J, Lampert F, et al. Oncogene. 1999;18:4663–4671. doi: 10.1038/sj.onc.1202842. [DOI] [PubMed] [Google Scholar]
- 5.Felix C A, Hosler M R, Slater D J, Megonigal M D, Lovett B D, Williams T M, Nowell P C, Spinner N B, Owens N L, Hoxie J, et al. Mol Diagnosis. 1999;4:269–283. doi: 10.1016/s1084-8592(99)80002-2. [DOI] [PubMed] [Google Scholar]
- 6.Super H G, Strissel P L, Sobulo O M, Burian D, Reshmi S C, Roe B, Zeleznik-Le N J, Diaz M O, Rowley J D. Genes Chromosomes Cancer. 1997;20:185–195. [PubMed] [Google Scholar]
- 7.Megonigal M D, Cheung N-K V, Rappaport E F, Nowell P C, Wilson R B, Jones D H, Addya K, Leonard D G B, Kushner B H, Williams T M, et al. Proc Natl Acad Sci USA. 2000;97:2814–2819. doi: 10.1073/pnas.050397097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford A M, Ridge S A, Cabrera M E, Mahmoud H, Steel C M, Chan L C, Greaves M. Nature (London) 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 9.Megonigal M D, Rappaport E F, Jones D H, Williams T M, Lovett B D, Kelly K M, Lerou P H, Moulton T, Budarf M L, Felix C A. Proc Natl Acad Sci USA. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gale K, Ford A, Repp R, Borkhardt A, Keller C, Eden O, Greaves M. Proc Natl Acad Sci USA. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felix C A. Med Pediatr Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 12.Ross J A. Int. J. Cancer. 1998. , Suppl 11, 26–28. [Google Scholar]
- 13.Wiemels J L, Pagnamenta A, Taylor G M, Eden O B, Alexander F E, Greaves M F. Cancer Res. 1999;59:4095–4099. [PubMed] [Google Scholar]
- 14.Chen H, Eastmond D A. Carcinogenesis. 1995;16:2301–2307. doi: 10.1093/carcin/16.10.2301. [DOI] [PubMed] [Google Scholar]
- 15.Felix C, Kim C, Megonigal M, Slater D, Jones D, Spinner N, Stump T, Hosler M, Nowell P, Lange B, et al. Blood. 1997;90:4679–4686. [PubMed] [Google Scholar]
- 16.Felix C A, Jones D H. Leukemia. 1998;12:976–981. doi: 10.1038/sj.leu.2401026. [DOI] [PubMed] [Google Scholar]
- 17.Megonigal M D, Rappaport E F, Jones D H, Kim C S, Nowell P C, Lange B J, Felix C A. Proc Natl Acad Sci USA. 1997;94:11583–11588. doi: 10.1073/pnas.94.21.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reaman G, Sposto R, Sensel M, Lange B, Feusner J, Heerema N, Leonard M, Holmes E, Sather H, Pendergrass T, et al. J Clin Oncol. 1999;17:445–455. doi: 10.1200/JCO.1999.17.2.445. [DOI] [PubMed] [Google Scholar]
- 19.Megonigal M D, Rappaport E F, Wilson R B, Jones D H, Lange B J, Whitlock J A, Ortega J A, Slater D J, Nowell P C, Felix C A. Proc Natl Acad Sci USA. 2000;97:9597–9602. doi: 10.1073/pnas.150241797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pegram L D, Megonigal M D, Lange B J, Nowell P C, Rowley J D, Rappaport E F, Felix C A. Blood. 2000;96:4360–4362. [PubMed] [Google Scholar]
- 21.Domer P H, Head D R, Renganathan N, Raimondi S C, Yang E, Atlas M. Leukemia. 1995;9:1305–1312. [PubMed] [Google Scholar]
- 22.Strout M P, Mrozek K, Heinonen K, Sait S N J, Shows T B, Aplan P D. Genes Chromosomes Cancer. 1996;16:204–210. doi: 10.1002/(SICI)1098-2264(199607)16:3<204::AID-GCC8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Lovett B D, Lo Nigro L, Rappaport E F, Blair I A, Osheroff N, Zheng N, Megonigal M D, Williams W R, Nowell P C, Felix C A. Proc Natl Acad Sci USA. 2001;98:9802–9807. doi: 10.1073/pnas.171309898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zechiedrich E L, Christiansen K, Andersen A H, Westergaard O, Osheroff N. Biochemistry. 1989;28:6229–6236. doi: 10.1021/bi00441a014. [DOI] [PubMed] [Google Scholar]
- 25.Thomas J W, Lee-Lin S Q, Green E D. Mamm Genome. 1999;10:764–767. doi: 10.1007/s003359901088. [DOI] [PubMed] [Google Scholar]
- 26.Brotherton D H, Dhanaraj V, Wick S, Brizuela L, Domaille P J, Volyanik E, Xu X, Parisini E, Smith B O, Archer S J, et al. Nature (London) 1998;395:244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- 27.Chilosi M, Doglioni C, Yan Z, Lestani M, Menestrina F, Sorio C, Benedetti A, Vinante F, Pizzolo G, Inghirami G. Am J Pathol. 1998;152:209–217. [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas J J, Szepesi A, Modiano J F, Domenico J, Gelfand E W. J Immunol. 1995;154:6275–6284. [PubMed] [Google Scholar]
- 29.Wagner E F, Hleb M, Hanna N, Sharma S. J Immunol. 1998;161:1123–1131. [PubMed] [Google Scholar]
- 30.Sherr C J. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 31.Ayton P M, Cleary M L. In: Transcription Factors: Normal and Malignant Development of Blood Cells. Ravid K, Licht J D, editors. New York: Wiley; 2001. [Google Scholar]
- 32.Fuchs U, Rehkamp G, Haas O A, Slany R, Konig M, Bojesen S, Bohle R M, Damm-Welk C, Ludwig W D, Harbott J, et al. Proc Natl Acad Sci USA. 2001;98:8756–8761. doi: 10.1073/pnas.121433898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lien H C, Lin C W, Huang P H, Chang M L, Hsu S M. Lab Invest. 2000;80:893–900. doi: 10.1038/labinvest.3780093. [DOI] [PubMed] [Google Scholar]
- 34.Costello J F, Plass C, Arap W, Chapman V M, Held W A, Berger M S, Su Huang H J, Cavenee W K. Cancer Res. 1997;57:1250–1254. [PubMed] [Google Scholar]
- 35.Corcoran M M, Mould S J, Orchard J A, Ibbotson R E, Chapman R M, Boright A P, Platt C, Tsui L C, Scherer S W, Oscier D G. Oncogene. 1999;18:6271–6277. doi: 10.1038/sj.onc.1203033. [DOI] [PubMed] [Google Scholar]
- 36.Taki T, Hayashi Y, Taniwaki M, Seto M, Ueda R, Hanada R, Suzukawa K, Yokota J, Morishita K. Oncogene. 1996;13:2121–2130. [PubMed] [Google Scholar]
- 37.So C W, Ma S K, Wan T S, Chan G C, Ha S Y, Chan L C. Cancer Genet Cytogenet. 2000;117:24–27. doi: 10.1016/s0165-4608(99)00136-3. [DOI] [PubMed] [Google Scholar]
- 38.Bernasconi P, Cavigliano P M, Boni M, Malcovati L, Astori C, Castagnola C, Pagnucco G, Vanelli L, Calatroni S, Caresana M, et al. Cancer Genet Cytogenet. 2000;116:111–118. doi: 10.1016/s0165-4608(99)00117-x. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi N, Miura I, Ohshima A, Nimura T, Hashimoto K, Hatano Y, Utsumi S, Kume M, Saito K, Kobayashi Y, et al. Cancer Genet Cytogenet. 1996;88:26–29. doi: 10.1016/0165-4608(95)00297-9. [DOI] [PubMed] [Google Scholar]