Abstract
The angiotensinogen M235T polymorphism in humans is linked to differential expression of the human angiotensinogen gene (AGT) gene and hypertension, but the homeostatic responses resulting from this polymorphism are not known. We therefore investigated how mice respond to five genetically determined levels of mouse angiotensinogen gene (Agt) expression covering the range associated with the M235T variants. By using high-throughput molecular phenotyping, tissue RNAs were assayed for expression of 10 genes important in hypertension. Significant positive and negative responses occurred in both sexes as Agt expression increased twofold, including a three-fold increase in aldosterone synthase expression in adrenal gland, and a two-fold decrease in renin expression in kidney. In males, cardiac expression of the precursor of atrial natriuretic peptide B and of adrenomedullin also increased approximately twofold. The relative expression of all genes studied except Agt differed significantly in the two sexes, and several unexpected relationships were encountered. A highly significant correlation between renal expression of the angiotensin type 1a receptor and kallikrein, independent of Agt genotype, is present in females (P < 0.0001) but not males (P = 0.4). The correlation between blood pressure (BP) and liver Agt expression within the five Agt genotypes is significant in females (P = 0.0005) but not in males (P = 0.2), whereas correlation of BP with differences between the genotypes is less in females (P = 0.06) than in males (P = 0.001). The marked gender differences in gene expression in wild-type mice and the changes induced by moderate alterations in Agt expression and BP emphasize the need to look for similar differences in humans.
Essential hypertension is a deleterious condition with a multifactorial etiology that includes both genetic and environmental determinants. However, despite decades of intense research into the physiological mechanisms that regulate blood pressure (BP), the primary genetic determinants of essential hypertension remain elusive. A substantial source of the difficulties associated with attempts to understand the regulation of BP is the existence of sophisticated homeostatic systems that permit desirable physiological changes in biological variables in vivo but prevent the changes from extending into pathological ranges. These homeostatic changes, combined with environmental effects and the difficulties of obtaining measurements of BP sufficiently precise to detect small differences, make dissection of the hypertensive phenotype difficult.
Because humans are genetically very heterogenous, polymorphic forms of genes are common. Consequently, among the polymorphic forms, it is reasonable to expect that different alleles will exist that dictate expression at different intrinsic levels. Such genetic variations may cause BP changes if the recruiting homeostatic adjustments are incomplete. The M235 and T235 alleles of the human angiotensinogen gene (AGT), described by Jeunemaitre et al. (1), affect BP. Thus, it is important to understand in detail the homeostatic adjustments that occur in response to this type of genetic alteration and to identify genes that are called into action during homeostasis. The responding genes themselves then become candidates for having effects on BP. Obviously, however, a precise analysis of this type is difficult in humans because of their heterogeneous genetic background.
For these reasons, we have recently been carrying out experiments in mice with the aim of identifying genes whose quantitative expression affects BP or induces homeostatic compensations. To do this, we first use targeted gene disruption and gene duplication to generate mice having different numbers of the target gene (2) and then use this “gene titration” series of mice to uncover changes that directly influence BP and those that represent homeostatic compensations. Our first application of this strategy has centered on the renin-angiotensin system (RAS), which plays a central role in salt and water homeostasis and the maintenance of vascular tone. Angiotensinogen (AGT) is the sole source of angiotensin II (Ang II), the most active peptide of the RAS, which is produced from AGT by the serial actions of renin and the angiotensin-converting enzyme. Of all of the components in the RAS, AGT is the one most definitively identified as genetically important in the causation of essential hypertension in humans (1, 3) and in animals (4–6).
The mice we have studied here carry one, two, three, or four functional copies of the murine wild-type mouse angiotensinogen gene (Agt) at its normal chromosomal location (2, 6). Plasma AGT levels increase progressively from 35% in the one-copy mice to 145% of normal in the four-copy animals, and their BPs show significant and almost linear increases of ≈8 mmHg (1 mmHg = 133 Pa) per gene copy, even though their normal homeostatic systems are intact. Thus, the ranges of AGT alteration and BP in the mice include those observed in humans with variant AGT alleles. We have reported on the nature of the homeostatic compensations that occur in the one-copy mice (7). In the present article we extend this work to include increases as well as decreases in the expression of AGT, and we move from studying expression at the level of proteins and peptides to the study of expression at the level of mRNA. We do so for several reasons. First, the quantitation of the proteins and peptides requires a unique method for each component. Second, distinguishing between related components is often difficult. Third, sensitivities and precision are not always adequate. Fourth, high-throughput determinations are only available for some components. These difficulties can all be circumvented in experimental animals when expression is studied at the mRNA level and quantitative reverse transcription (RT)-PCR is used in conjunction with automated RNA preparation. Our application of this molecular phenotyping system to animals in the Agt gene titration series illustrates its power for recognizing subtle phenotypic changes in animals with minimal genetic differences and reveals several unexpected findings.
Experimental Procedures
Animals.
The experimental mice used in this investigation have different numbers of the functional Agt gene (6). Their genotypes are Agt 0/1 (with one wild-type copy of the gene), Agt 0/2 (with a tandemly duplicated copy of the gene), Agt 1/1 (wild type with two singleton copies of the gene), Agt1/2 (one wild-type and one duplicated copy), and Agt 2/2 (two duplicated copies). The original gene targeting was in embryonic stem cells derived from strain 129 Ola. All mice used in this work were, however, from a seventh-generation back cross to strain C57BL/6J (B6). The mice were fed regular chow and were handled following the National Institutes of Health guidelines for the care and use of experimental animals. Most were 3–4 months in age.
High-Throughput Tissue Processing and RNA Isolation.
Livers, kidneys, adrenal glands, and whole hearts were stored with 5× (vol/wt) volumes of “RNA later” solution (Ambion, Austin, TX). Small pieces of tissue (50–100 mg) were added to 2-ml special tubes containing 1 ml of lysis buffer made by diluting 2× RNA lysis buffer (PE Biosystems, Foster City, CA) with Ca2+- and Mg2+-free PBS. The tissues were macerated with a 1/4-inch cylindrical bead at speed 4 for 30 sec in a Fast Prep 120 mixer (QBIOgene, Vista, CA). Tissue homogenates were transferred into Eppendorf tubes, heated to 50°C for 2 min, then centrifuged at 25°C for 1 min at 10,000 × g to remove tissue debris. Tissue solutions were stored at −20°C for up to 16 h before use for RNA isolations in an RNA purification tray, using the ABI Prism 6700 automated nucleic acid workstation (PE Biosystems) and following the manufacturer's protocol. Total RNA samples were stored at −20°C.
Primers and Probes.
The nucleotide sequences of the PCR primers and their fluorogenic probes for the target genes (provided as supporting information, which is published on the PNAS web site, www.pnas.org) were designed by using the computer program primer express (PE Biosystems). Each fluorescent probe has a reporter dye (FAM for the target RNA and TET for the β-actin control) covalently attached at its 5′ end and a quencher dye (TAMRA) attached at its 3′ end. Before use, the probes were purified in the PolyPak II cartridge (Glen Research, Sterling, VA) following the manufacturer's instructions.
RT-PCR Amplifications.
Real-time RT-PCR amplifications (8) were performed in a 96-well plate in the ABI Prism 7700 sequence detector (PE Biosystems) in a total volume of 30 μl, which included 10 μl of RNA sample from the ABI Prism 6700 plus 20 μl of a reaction mixture made with minor modifications of the manufacturer's instructions. Each RT-PCR amplification was performed in duplicate: 30 min at 48°C for the RT reaction, then 10 min at 94°C, followed by a total of 40 temperature cycles (15 sec at 94°C and 1 min at 60°C). During the amplification, the fluorescence of FAM (or TET), TAMRA, and ROX (a passive reference dye) was measured by the 7700 sequence detector in each well of the 96-well plate. The numbers of copies of the PCR template in the starting sample were calculated by using the sequence detector software incorporated in the ABI Prism 7700 Sequence Detector System.
Relative Quantification of Gene Expression.
Standard plasmids containing a DNA fragment for each of the genes of interest were used as external controls, and amplification of the β-actin gene was used as an endogenous control. Sense RNAs were synthesized from the standard plasmids by the manufacturer's protocols, using a MAXIscript transcription kit (Ambion). The concentrations of purified sense RNAs were determined as micrograms per optical density unit, and serial dilutions of the sense RNA, using bacterial tRNA as a carrier, were used to generate standard curves. When quantification was relative to an endogenous control, standard curves were prepared for both the target and the endogenous control. For each experimental sample, the amounts of the target and of the endogenous control were determined from the appropriate standard curves. We assume that β-actin mRNA is present in all tested and control samples of tissue RNA at a constant proportion and normalize the amount of total RNA in our test samples by comparing their β-actin fluorescent signal after PCR with that from mouse embryonic stem cell RNA freed from DNA by DNase treatment.
Statistics.
Unless otherwise indicated, ANOVA and multivariate regression analyses were used for statistical evaluations with jmp software (SAS Institute, Cary, NC).
Results
Expression of 10 Genes Relevant to the Control of BP.
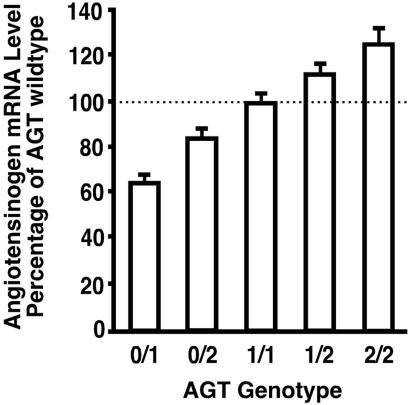
The levels of Agt mRNA in the livers of mice (five females and five males) with each of five different Agt genotypes are presented in Fig. 1 as percentages of wild type. Their genotypes and expression are as follows: Agt 0/1, 65 ± 3% (0/1 mice have one copy of the wild-type Agt gene and one inactivated copy); Agt 0/2, 84 ± 4% (0/2 mice have one tandemly duplicated pair of Agt genes); Agt 1/1, 100 ± 3% (1/1 mice have two copies of the wild-type gene); Agt 2/1, 112 ± 3% (2/1 mice have one duplicated pair and one wild-type gene); Agt 2/2, 125 ± 6% (2/2 mice have two duplicated pairs). There were no differences in the liver Agt mRNA levels in males and females (P = 0.20, Table 1), thus the data from both sexes were combined in Fig. 1. The mRNA levels in the Agt 0/2 mice were less than those in the Agt 1/1mice, although both have two copies of the gene, indicating that the two genes in the tandemly duplicated Agt gene pair are expressed at approximately 80% of two wild-type genes. Together, the gradation in the liver Agt expression in mice of the five Agt genotypes is gently progressive. Despite this gentle progression (which averages only 15% among each of the genotypes), the molecular phenotyping procedure allows the five genotypes to be distinguished from each other (P < 0.05) solely on the basis of Agt mRNA measurements.
Figure 1.
AGT mRNA levels in mice with different Agt genotypes. The levels of mRNAs in the livers are presented as percentages of wild type (1/1). Five males and five females were used for each genotype. The bars show means ± SEM.
Table 1.
mRNA levels for 10 genes vs. Agt genotype and gender
| Gene | Gender |
Agt genotype
|
P (ANOVA)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 0/1 | 0/2 | 1/1 | 1/2 | 2/2 | Genotype | Gender | Interaction | ||
| Agt | F | 94 ± 6 | 122 ± 10 | 154 ± 9 | 170 ± 9 | 192 ± 17 | <0.0001 | 0.2 | 0.8 |
| M | 94 ± 5 | 119 ± 6 | 141 ± 5 | 161 ± 5 | 177 ± 10 | ||||
| Ren | F | 89 ± 11 | 72 ± 10 | 55 ± 5 | 58 ± 8 | 45 ± 3 | <0.0001 | <0.0001 | 0.4 |
| M | 47 ± 7 | 30 ± 3 | 30 ± 2 | 20 ± 3 | 23 ± 3 | ||||
| Ace | F | 172 ± 25 | 151 ± 18 | 153 ± 22 | 143 ± 18 | 150 ± 15 | 0.8 | 0.5 | 0.9 |
| M | 161 ± 11 | 156 ± 16 | 161 ± 11 | 148 ± 15 | 180 ± 19 | ||||
| Atr1a | F | 12.7 ± 0.3 | 12.2 ± 0.5 | 10.9 ± 0.4 | 12.9 ± 1.0 | 14.5 ± 8.9 | 0.001 | 0.002 | 0.007 |
| M | 9.9 ± 0.6 | 11.0 ± 0.7 | 10.4 ± 0.5 | 12.2 ± 0.6 | 12.8 ± 0.6 | ||||
| AS | F | 397 ± 52 | 704 ± 89 | 691 ± 80 | 992 ± 76 | 1149 ± 82 | <0.0001 | <0.0001 | 0.02 |
| M | 594 ± 16 | 941 ± 56 | 957 ± 94 | 1249 ± 76 | 1885 ± 176 | ||||
| Klk1 | F | 6.5 ± 0.1 | 7.5 ± 0.5 | 6.3 ± 0.3 | 7.6 ± 0.7 | 8.3 ± 0.6 | 0.09 | 0.0003 | 0.06 |
| M | 6.1 ± 0.6 | 6.2 ± 0.2 | 6.5 ± 0.3 | 5.6 ± 0.1 | 6.4 ± 0.4 | ||||
| Nos3 | F | 8.9 ± 0.4 | 8.7 ± 0.5 | 10.2 ± 0.4 | 9.0 ± 0.4 | 9.1 ± 0.9 | 0.2 | <0.0001 | 0.06 |
| M | 8.1 ± 0.9 | 7.9 ± 0.8 | 6.6 ± 0.4 | 5.2 ± 0.6 | 6.7 ± 0.8 | ||||
| Nppa | F | 1304 ± 70 | 915 ± 89 | 1341 ± 144 | 1024 ± 45 | 1232 ± 90 | 0.09 | <0.0001 | 0.01 |
| M | 413 ± 69 | 495 ± 98 | 503 ± 25 | 539 ± 56 | 466 ± 41 | ||||
| Nppb | F | 101 ± 6 | 88 ± 12 | 106 ± 13 | 89 ± 4 | 87 ± 12 | <0.0001 | 0.9 | <0.0001 |
| M | 58 ± 8 | 50 ± 4 | 67 ± 3 | 148 ± 19 | 141 ± 18 | ||||
| Adm | F | 68 ± 8 | 66 ± 9 | 82 ± 10 | 74 ± 9 | 73 ± 9 | 0.04 | 0.02 | 0.1 |
| M | 42 ± 7 | 53 ± 8 | 54 ± 3 | 78 ± 7 | 76 ± 6 | ||||
Values are pg/μg of total RNA ± SEM; n = 5 for each genotype and gender.
We next assessed the effects of the genetically induced changes in Agt expression on the expression of nine additional genes: renin (Ren), Ang II receptor 1a (Atr1a), kallikrein 1 (Klk1), and endothelial nitric oxide synthase (Nos3) in the kidney; angiotensin-converting enzyme (Ace) in the lung; aldosterone synthase (Cyp11b2, hereafter referred to AS) in the adrenal gland; and the genes coding for the precursors of atrial natriuretic peptides A and B (Nppa and Nppb) and for adrenomedullin (Adm) in the heart. Data were first analyzed by ANOVA with Agt genotype and gender as two variables (Table 1). With the exception of Ace mRNA levels in the lung, strong Agt genotype-dependent differences were found in the expression of the genes in the RAS pathway. Thus, a gradual decrease in the mRNA levels of Ren as Agt expression increases twofold in the five genotypes resulted in a two-fold difference between the Agt2/2 and Agt0/1 mice (P < 0.0001). This decrease of Ren expression is the major homeostatic response that we observe consequent to the increase in BP caused by the genetic increase in AGT. In contrast, in response to the increased AGT, mRNA levels of AS progressively increase over a three-fold range in animals of the five genotypes. Activation of aldosterone synthesis by Ang II (9) is well known, and we presume that this increase of AS mRNA indicates the direct effects on AS expression of an increase in intra-adrenal Ang II. Our data indicate that any negative feedback between the elevated BP and aldosterone synthesis, if it exists, is ineffective. We observe modest but significant genotype- and gender-dependent differences in Atr1a expression, although there is no clear trend in these differences. In contrast to the genes in the RAS, expression of the Klk1 and Nos3 genes in the kidney, and of Nppa and Adm in the heart, do not differ significantly among the five genotypes. There is, however, a significant increase in the cardiac expression of Nppb in 1/2 and 2/2 males compared with males in the other groups (P < 0.001, Tukey–Kramer). This change was not observed in females (see below).
Gender-Specific Effects.
As illustrated in Table 1, the expressions of 8 of 10 of the genes we studied, with the exception of Agt and Ace, are significantly affected by gender, and in some cases in opposite directions, with probabilities ranging from <0.0001 for Ren, AS, Nppa, and Nos3, to <0.02 for Adm, and with male/female ratios ranging from 2.05 for Ren to 0.42 for cardiac Nppa. The expression of Agt and Ace were not affected by gender. Furthermore, the effects of Agt genotype on the expression of the eight gender-sensitive genes were frequently different in the two sexes. For example, the cardiac expressions of Nppb and Adm in females were not influenced by Agt genotype (P = 0.45 and 0.7, respectively) but were highly influenced by Agt genotype in males (P < 0.0001 and <0.002). Because of these strong gender effects, the data for the male and female mice are presented and analyzed separately.
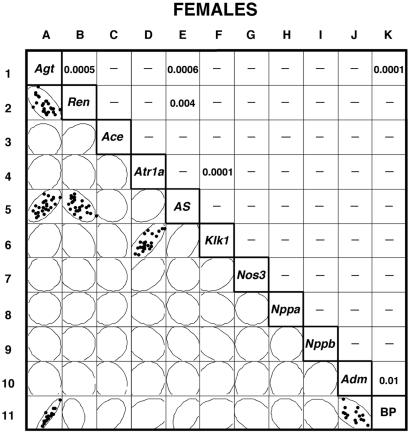
The left lower half of Fig. 2 summarizes the data from the females in the form of a scatterplot matrix of the individual measurements for the 25 females studied (5 per genotype) for each of the 10 mRNAs assayed with a 95% bivariate normal density ellipse superimposed on each scatterplot. (The BP measurements included in the figure are discussed below.) To simplify the figure, individual data points are shown only when the corresponding correlation is significant. The right upper half of the figure tabulates the probability (P) that the observed correlations occur by chance based on analysis of the Pearson product moment correlations. Again, for simplicity, only significant probabilities are shown. The degree of asymmetry of the 95% bivariate ellipses provides visual representations of the significance of each correlation. For example, the ellipse in square A2 is markedly asymmetric, indicating a strong inverse correlation between kidney Ren mRNA and liver Agt expression; square B1 shows the corresponding P value, 0.0005. One other strong direct correlation is detected in the mRNA data from females, namely that between liver Agt expression and adrenal gland AS mRNA (squares A5 and E1, P = 0.0006).
Figure 2.
Comparisons of gene expression and BP in individual female mice. The left lower half of the figure presents the data in the form of a bivariate scatterplot matrix of the individual measurements and the corresponding 95% confidence ellipses. In those pairwise comparisons that have a significant correlation, the dots in the squares represent individual values; the right upper half of the figure tabulates the corresponding probabilities. In the pairwise comparisons without a significant correlation, only the 95% confidence ellipses are shown. More detailed versions of Figs. 2 and 3 are provided as supporting information.
Pairwise correlations outside the primary experimental design (which was to investigate how genetic variations in Agt expression affect other molecular parameters) can reveal previously unexpected relationships, but estimating their significance requires caution. A secondary correlation between the expression of Ren in the kidney and of AS in the adrenal gland (Fig. 2, squares B5 and E2, P = 0.004) is the result of the corresponding two components each being correlated with Agt mRNA expression in the primary analysis as described in the preceding paragraph. Assessing the significance of other secondary correlations must take into account the number of pairwise comparisons tested. A total of 28 comparisons involving the expression of pairs of genes other than those with Agt are included in the tabulation. If Bonferroni's correction is applied to these comparisons, a P of <0.002 (0.05/28) would conservatively indicate a significant correlation. One pairwise correlation reaches well beyond this level—that between kidney expression of Atr1a mRNA and kidney expression of Klk1 (squares D6 and F4, P < 0.0001)–although neither is significantly influenced by changes in Agt expression (squares A4 and A6).
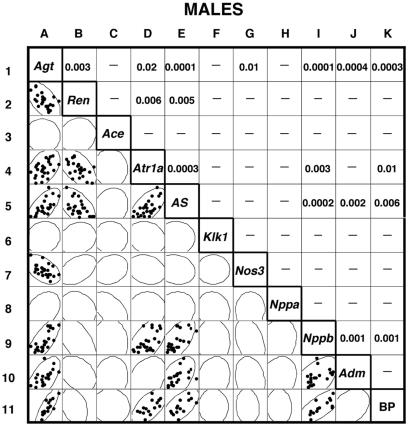
Fig. 3 presents the equivalent comparisons for male mice. They are considerably more complicated. The primary correlations with Agt expression seen in females also occur in males—kidney Ren expression (squares A2 and B1, P = 0.003) and adrenal gland AS expression (squares A5 and E1, P = 0.0001). But in male mice, there are two other strong primary correlations: liver Agt expression with cardiac Nppb expression (squares A9 and I1, P < 0.0001) and with cardiac Adm (squares A10 and J1, P = 0.0004). The secondary correlations between these primaries all have P < 0.01, except for kidney Ren with cardiac Adm and Nppb. Two weaker primary correlations are seen between liver Agt and kidney Atr1a (squares A4 and D1, P = 0.02) and between liver Agt and kidney Nos3 (squares A7 and G1, P = 0.01).
Figure 3.
Comparisons of gene expression and BP in individual male mice.
Our overall conclusion from these comparisons is that genetic changes in Agt gene expression markedly affect kidney renin and adrenal gland aldosterone synthase expression in both females and males. In females, but not in males, the expression of kidney Atr1a and Klk1 is strongly correlated with the other but not with Agt expression. In males, but not in females, changes in Agt expression are associated with marked changes in the cardiac expression of Nppb and Adm, and with less marked changes in kidney Atr1a and Nos3.
BP and Gene Expression.
To obtain an overview of the relevance of the molecular phenotyping data to increases in BP and hypertension, we measured the tail-cuff BP of the mice with 2, 3, or 4 copies of Agt gene. Row 11 in Fig. 2 presents the relevant data for the 15 females. In agreement with previous reports (1, 6), we find that BP is positively correlated with variation in Agt expression (squares A11 and K1). However, the degree to which Agt expression and BP are correlated (P < 0.0001) is very remarkable. One other correlation is clear in females, namely an inverse relationship between BP and cardiac Adm expression (squares J11 and K10, P = 0.01). Adrenal AS expression in females is not significantly correlated with BP (squares E11 and K5), indicating a lack of negative feedback of BP on aldosterone synthesis.
Row 11 in Fig. 3 presents comparable BP data for the 15 males with 2, 3, or 4 copies of the Agt gene. As in females, the correlation between liver Agt expression and BP is very strong (squares A11 and K1, P = 0.0003). However, unlike females, cardiac Adm expression in males is not correlated with BP (squares J11 and K10), although cardiac Nppb expression is correlated with BP (squares I11 and K9, P = 0.001). Adrenal gland expression of AS is likewise correlated with BP in males (squares E11 and K5, P = 0.006), as is renal expression of the Atr1a (squares D11 and K4, P = 0.01).
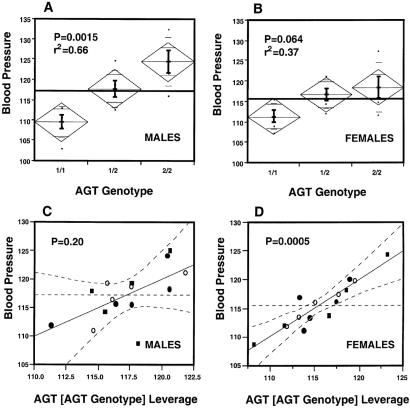
Fig. 4 A and B presents the results of an analysis by ANOVA of BP vs. Agt genotype in the males (A) and females (B), and the plots of BP against the leverage of Agt expression nested within the Agt genotypes in males (C) and in females (D). When the data are averaged by Agt genotype, ANOVA shows that BPs in males are significantly affected by their genotypes [P = 0.0015, r2 = 0.66 (Fig. 4A)] but this effect is borderline in females [P = 0.06, r2 = 0.37 (Fig. 4B)]. However, when the data are analyzed by individual animals, as presented in Fig. 2, a highly significant correlation is seen between Agt mRNA levels and BP in both sexes (P = 0.0003, r2 = 0.65 in males and P < 0.0001, r2 = 0.85 in females). This remarkably strong correlation between BP and liver Agt expression in the individual female mice is considerably greater than that observed when the BP and Agt expression are averaged by genotype. Yet in individual males, r2 for BP vs. liver Agt expression is high (0.68), although not appreciably more so than the r2 for BP vs. Agt expression averaged for each Agt genotype (0.66). These similarities and differences suggest that in females some factor in addition to the Agt genotype affects Agt expression and/or BP.
Figure 4.
Comparison of correlation between BP and Agt expression. The data in A (males) and B (females) are from an analysis by ANOVA of BPs and Agt genotypes. C (males) and D (females) present leverage plots of BP against hepatic Agt mRNA expression nested within the Agt genotypes. The Agt genotypes in C and D are 1/1 (○), 1/2 (●), and 2/2 (■).
To explore this possibility further, we used ANOVA to test whether the BPs of individual animals were correlated with differences in liver expression of Agt within the Agt genotypes in addition to their correlation with expression differences between the genotypes. The results of this analysis are illustrated by leverage plots for males (Fig. 4C) and females (Fig. 4D) of BP vs. Agt levels nested within the genotypes. The data are compelling. The effects of Agt expression within the genotype are highly significant in females (P = 0.0005) but not in males (P = 0.20).
Taken together, our data demonstrate that the homeostatic adjustments in the RAS and elsewhere in the body are not sufficient to normalize the BPs caused by modest alterations in Agt expression. Furthermore, the data show that, although Agt levels, because of our genetic manipulations, primarily account for BP differences in individual males, in individual females other factors are so strong that the effects of the genetically induced changes in the Agt gene expression are partially masked.
Discussion
Single nucleotide polymorphisms (SNPs) represent a substantial portion of human genetic variation, and considerable effort is being expended in attempts to correlate their occurrence with the incidence of disease. A corollary of this endeavor is the need to be able to determine in experimental animals whether subtle genetic differences have any phenotypic consequences. Our present study explores the use of “molecular phenotyping” by automated quantitative RT-PCR on RNA isolated from liver, heart, kidney, and adrenals of experimental animals to uncover phenotypic effects caused by subtle differences in the expression of the gene (Agt) coding for AGT. We selected this gene because an SNP in its promoter has been reported to have a quantitative effect on its expression (10), and because this polymorphism has been implicated with the incidence of hypertension (11). Our first tests established that the molecular phenotyping procedure can detect Agt expression differences between animals of different genotypes of the order of 15%. Because the sensitivity of the system can be adjusted readily by changing the concentrations of reagents, the phenotyping procedure is not limited to abundantly expressed genes or to larger organs. Accordingly, we chose to examine the expression of 10 genes that are relevant to the control of BP or to the homeostatic changes induced when BP deviates from normal.
Our finding that expression of the Ren gene in the kidney decreases systematically over an approximately two-fold range as the levels of hepatic Agt gene expression increase over a comparable two-fold range confirms and extends our earlier comparisons between wild-type mice and mice with only one functional copy of the Agt gene (7). These homeostatic changes in renal Ren gene expression are the largest compensatory adjustments that we have observed consequent to the genetically determined differences in Agt expression.
This adjustment of renin production by the RAS is, however, insufficient to normalize BP as demonstrated by the systematic increase of BP as the expression of Agt is genetically increased. Part of this insufficiency is probably the result of an approximately three-fold increase in the expression of the AS gene in the adrenal glands as Agt expression increases twofold, which we find in both sexes (P < 0.001), presumably because of increased intra-adrenal Ang II levels directly acting on the promoter of the AS gene (12) coupled with the absence of effective negative feedback regulation between BP and aldosterone synthesis. A support of this interpretation is our preliminary observation that the plasma levels of Ang II and of aldosterone are both increased in the Agt 2/2 animals to about 1.4 times their levels in wild-type animals. Human patients with glucocorticoid-remediable aldosteronism carry an additional chimeric gene that fuses the regulatory sequences of steroid 11β hydroxylase to the coding sequences of AS; these patients have 1.5 times plasma aldosterone levels and are hypertensive (13, 14).
Gender-related differences in gene expressions are to be expected, but we were surprised at both the generality of their occurrence and the magnitude of the differences. More than half of the 10 genes studied showed significant differences between the two sexes in either their mean expression in the wild-type animals or the changes of the mean expression across the five Agt genotypes. The degree of difference was also remarkable, ranging from more than twofold higher in males (for cardiac expression of Nppa) to more than twofold less in males (for kidney Ren). But of greater interest in relation to developing an understanding of gender effects on BP was the sometimes opposite responses of genes to the genetic changes in Agt expression and BP in animals of the two sexes. Changes in expression of the precursors of BNP and Adm illustrate this situation. Plasma levels of BNP have recently been used in humans as an indicator of cardiac hypertrophy (15), and an increase in cardiac expression of ANP has long been associated in experimental animals with cardiac hypertrophy. Both BNP and ANP have antihypertrophic effects by means of their binding to the natriuretic peptide receptor A (NPRA). Yet the cardiac expression of these natriuretic peptides changed in relation to Agt expression only in males. Similarly, we find that in male mice the cardiac expression of the Adm gene, which codes for the BP-lowering peptide adrenomedullin, is markedly increased (P = 0.0004) by increases in Agt expression and BP, but in females there is no correlation between these factors. The relevance of these observations to clinically significant situations is indicated by noting that male mice develop more severe cardiac hypertrophy than females when the gene coding for NPRA (the active receptor for all three of natriuretic peptides) is inactivated (16), and that a deletion polymorphism in the promoter of the human gene for NPRA has been shown to be significantly associated with essential hypertension and/or cardiac hypertrophy in a recent Japanese study (17). The marked gender-related differences that we observe in wild-type mice and in response to modest increases in Agt expression and in BP emphasize the need to look for similar differences in humans.
Two completely unexpected findings from our study were detected because the molecular phenotyping procedure is sufficiently sensitive for the expression of multiple genes to be determined in several tissues from each individual animal; it is not necessary to pool samples from individual mice. In addition, the precision of the data is such that differences among individual animals can be detected and measured well enough to lead to meaningful conclusions. The first of these findings is that seen in females between expression in the kidney of the Atr1a and Klk1 genes (P < 0.0001, squares D6 and F4 in Fig. 2). To ascertain that this correlation is not a type 1 error, we tested for the presence of this correlation in a completely independent set of mice (our unpublished data from animals in which the expression of Atr1a was genetically varied over a range from 100 to 130% of the wild-type levels). A strong correlation between the renal Atr1a and Klk1 gene expressions was again observed in the females (P = 0.0014, n = 17) but not in the males (P = 0.8, n = 21). Thus, these data strongly suggest that the Atr1a and Klk1 genes share a female-restricted regulatory mechanism related to their expression in the kidney. A computer search for regulatory sequences in the 5′ flanking DNA of these two genes identified the sequence GGTATATGACC approximately 400 bases upstream of the start of transcription of the mouse Klk1 gene (18), and the sequence GGTTGGAACC approximately 70 bases upstream of the start of transcription of the mouse Atr1a gene. These sequences are similar to a consensus sequence, GGTCANNNTGACC, identified as an estrogen-responsive element (19–21), which suggests a possible role of estrogen in the expression of these two genes. However, preliminary tests (unpublished data) indicate that the expression levels of the two genes are still correlated in ovarectomized females with or without administration of exogenous estradiol.
The second finding is that the BPs of male mice are very significantly altered when the number of Agt genes is varied from 2 through 4 (P = 0.0015), but in females this effect is borderline (P = 0.064) and is obscured by a highly significant correlation between BP and liver Agt expression (P = 0.0005) that is unrelated to Agt genotype. The direct relevance of this finding to humans is made clear by noting that the genetic linkage between hypertension and the 235T variant described by Jeunemaitre et al. (1) was significant in male sib-pairs but not in female sib-pairs, even though the 235T variant was found significantly more often in female hypertensives than in control subjects. Indeed, in discussing the absence of the genetic linkage in females, the authors speculated that “AGT contributes to hypertensive risk directly in males but indirectly in females, as another estrogen-modulated factor may mediate the impact of the AGT-associated predisposition.” A putative estrogen-responsive element sequence, GGTCAGAAGGCC, approximately 300 bases upstream of the promoter region of the human AGT gene (22), and GGGTAAATGTGTAACCC, at the homologous region of the mouse Agt gene (23), support a role of estrogen in regulating Agt gene expression. Additionally, plasma AGT levels in humans are clearly increased after estrogen treatment and during pregnancy (24–26). However, our observed variation in Agt mRNA levels is unlikely to be accounted for by the estrous cycle (3-day) of female mice, because the BP measurements and the isolation of tissue for RNA preparation were at different times. We conclude that in female mice (and probably in humans) either hepatic Agt gene expression is an important factor in determining BP or both are coregulated. The correlation between these factors is so strong in the female mice (Fig. 4D) that it suggests the possibility that the level of hepatic Agt gene expression could be used to predict BP. In male mice there are additional factors involved, such as adrenal AS and cardiac Nppb expression.
In summary, we have demonstrated the statistical and experimental power of molecular phenotyping animals individually by using high-precision quantitative RT-PCR. When applied to mice with genetically determined differences in expression of the Agt gene, the resulting modest alterations in liver Agt expression were positively correlated with adrenal expression of aldosterone synthase and negatively correlated with renal renin expression. The analyses also revealed a surprising number of significant gender-related differences in the expression and regulation of genes related to the control of BP and cardiac hypertrophy.
Supplementary Material
Acknowledgments
We thank Thomas M. Coffman, Howard A. Rockman, Mei-Hong Lin, Emily Riggs, Hui-Ying Li, and Jenny Holt for their help and advice. This work was supported by National Institutes of Health Grants GM20069, HL49277, and HL65184 (to O.S.).
Abbreviations
- RAS
renin-angiotensin system
- Ang II
angiotensin II
- BP
blood pressure
- AGT
angiotensinogen
- RT
reverse transcription
References
- 1.Jeunemaitre X, Soubrier F, Kotelevtsev Y V, Lifton R P, Williams C S, Charru A, Hunt S C, Hopkins P N, Williams R R, Lalouel J-M, Corvol P. Cell. 1992;71:169–180. doi: 10.1016/0092-8674(92)90275-h. [DOI] [PubMed] [Google Scholar]
- 2.Smithies O, Kim H-S. Proc Natl Acad Sci USA. 1994;91:3612–3615. doi: 10.1073/pnas.91.9.3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watt G C M, Harrap S B, Foy C J W, Holton D W, Edwards H V, Davidson H R, Connor J M, Lever A F, Fraser R. J Hypertens. 1992;10:473–482. doi: 10.1097/00004872-199205000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Ohkubo H, Kawakami H, Kakehi Y, Takumi T, Arai H, Yokota Y, Iwai M, Tanabe Y, Masu M, Hata J, et al. Proc Natl Acad Sci USA. 1990;87:5153–5157. doi: 10.1073/pnas.87.13.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura S, Mullins J J, Bunnemann B, Metzger R, Hilgenfeldt U, Zimmermann F, Jacob H, Fuxe K, Ganten D, Kaling M. EMBO J. 1992;11:821–827. doi: 10.1002/j.1460-2075.1992.tb05119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H-S, Krege J H, Kluckman K D, Hagaman J R, Hodgin J B, Best C F, Jennette J C, Coffman T M, Maeda N, Smithies O. Proc Natl Acad Sci USA. 1995;92:2735–2739. doi: 10.1073/pnas.92.7.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H-S, Maeda N, Oh G T, Fernandez L G, Gomez R A, Smithies O. J Biol Chem. 1999;274:14210–14217. doi: 10.1074/jbc.274.20.14210. [DOI] [PubMed] [Google Scholar]
- 8.Gibson U E M, Heid C A, Williams P M. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 9.Mckenna T J, Island D P, Nicholson W E, Liddle G W. J Steroid Biochem. 1978;9:967–972. doi: 10.1016/0022-4731(78)90059-6. [DOI] [PubMed] [Google Scholar]
- 10.Inoue I, Nakajima T, Williams C S, Quachenbush J, Puryear R, Power M, Cheng T, Ludwig E H, Sharma A M, Hata A, et al. J Clin Invest. 1997;99:1786–1797. doi: 10.1172/JCI119343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeunemaitre X, Inoue I, Williams C, Charru A, Tichet J, Powers M, Sharma A M, Gimenez-Roqueplo A P, Hata A, Corvol P, Lalouel J M. Am J Hum Genet. 1997;60:1448–1460. doi: 10.1086/515452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotchen T A, Guthrie G P., Jr Endocr Rev. 1980;1:78–99. doi: 10.1210/edrv-1-1-78. [DOI] [PubMed] [Google Scholar]
- 13.Lifton R P, Dluhy R G, Powers M, Rich G M, Cook S, Ulick S, Lalouel J-M. Nature (London) 1992;355:262–265. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- 14.Stowasser M, Bachmann A W, Huggard P R, Rossetti T R, Gordon R D. J Clin Endocrinol Metab. 2000;85:2160–2166. doi: 10.1210/jcem.85.6.6651. [DOI] [PubMed] [Google Scholar]
- 15.Hirata Y, Matsumoto A, Aoyagi T, Yamaoki K, Komuro I, Suzuki T, Ashida T, Sugiyama T, Hada Y, Kuwajima I, et al. Cardiovasc Res. 2001;51:585–591. doi: 10.1016/s0008-6363(01)00320-0. [DOI] [PubMed] [Google Scholar]
- 16.Oliver P M, Fox J E, Kim R, Rockman H A, Kim H-S, Reddick R L, Pandey K M, Milgram S L, Smithies O, Maeda N. Proc Natl Acad Sci USA. 1997;94:14730–14735. doi: 10.1073/pnas.94.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama T, Soma M, Takahashi Y, Rehemudula D, Kanmatsuse K, Furuya K. Circ Res. 2000;86:841–845. doi: 10.1161/01.res.86.8.841. [DOI] [PubMed] [Google Scholar]
- 18.Leeuwen van B H, Evans B A, Tregear G W, Richards R I. J Biol Chem. 1986;261:5529–5535. [PubMed] [Google Scholar]
- 19.Jost J P, Seldran M, Geiser M. Proc Natl Acad Sci USA. 1984;81:429–433. doi: 10.1073/pnas.81.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saluz H P, Jiricny I, Jost J P. Proc Natl Acad Sci USA. 1986;83:7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peale F V, Ludwig L B, Zain S, Hilf R, Bambara R A. Proc Natl Acad Sci USA. 1988;85:1038–1042. doi: 10.1073/pnas.85.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaillard I, Clauser E, Corvol P. DNA. 1989;8:87–99. doi: 10.1089/dna.1.1989.8.87. [DOI] [PubMed] [Google Scholar]
- 23.Clouston W M, Evans B A, Haralambidis J, Richards R I. Genomics. 1988;2:240–248. doi: 10.1016/0888-7543(88)90008-0. [DOI] [PubMed] [Google Scholar]
- 24.Lynch K R, Peach M J. Hypertension. 1991;17:263–269. doi: 10.1161/01.hyp.17.3.263. [DOI] [PubMed] [Google Scholar]
- 25.Ward K, Hata A, Jeunemaitre X, Helin C, Nelson L, Namikawa C, Farrington P F, Ogasawara M, Suzumori K, Tomoda S, et al. Nat Genet. 1993;4:59–61. doi: 10.1038/ng0593-59. [DOI] [PubMed] [Google Scholar]
- 26.Ohkubo H, Nakayama K, Tanaka T, Nakanishi S. J Biol Chem. 1986;261:319–323. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.