Abstract
All eukaryotic cells sense extracellular stimuli and activate intracellular signaling cascades via G protein-coupled receptors (GPCR) and associated heterotrimeric G proteins. The Saccharomyces cerevisiae GPCR Gpr1 and associated Gα subunit Gpa2 sense extracellular carbon sources (including glucose) to govern filamentous growth. In contrast to conventional Gα subunits, Gpa2 forms an atypical G protein complex with the kelch repeat Gβ mimic proteins Gpb1 and Gpb2. Gpb1/2 negatively regulate cAMP signaling by inhibiting Gpa2 and an as yet unidentified target. Here we show that Gpa2 requires lipid modifications of its N-terminus for membrane localization but association with the Gpr1 receptor or Gpb1/2 subunits is dispensable for membrane targeting. Instead, Gpa2 promotes membrane localization of its associated Gβ mimic subunit Gpb2. We also show that the Gpa2 N-terminus binds both to Gpb2 and to the C-terminal tail of the Gpr1 receptor and that Gpb1/2 binding interferes with Gpr1 receptor coupling to Gpa2. Our studies invoke novel mechanisms involving GPCR-G protein modules that may be conserved in multicellular eukaryotes.
INTRODUCTION
All eukaryotic cells deploy on their surface signaling modules composed of G protein-coupled receptors (GPCR) and heterotrimeric G proteins to sense extracellular cues. GPCRs are conserved from yeasts to humans and constitute a family of cell surface receptors that contain seven transmembrane domains and sense myriad extracellular ligands including nutrients, odorants, hormones and pheromones, and photons (Gilman, 1987; Strader et al., 1994; Lefkowitz, 2000; Mombaerts, 2004). Heterotrimeric G proteins consist of α, β, and γ subunits, in which the Gα subunits are guanine nucleotide binding proteins and the Gβγ subunits form a membrane-tethered heterodimer (Bourne, 1997; Sprang, 1997; Gautam et al., 1998; Schwindinger and Robishaw, 2001; Cabrera-Vera et al., 2003). Ligand binding triggers conformational changes in the GPCR that stimulate GDP-GTP exchange on Gα and release of the Gβγ dimer. Released Gα-GTP, Gβγ, or both signal downstream effectors. GTP-to-GDP hydrolysis (either intrinsic or RGS protein-stimulated) induces reassociation of the Gα-GDP subunit with Gβγ, extinguishing the signal (De Vries and Gist Farquhar, 1999; Guan and Han, 1999; Ross and Wilkie, 2000).
The yeast Saccharomyces cerevisiae expresses 3 GPCRs (Ste2, Ste3, and Gpr1) and 2 Gα subunits (Gpa1 and Gpa2), comprising two signaling modules: one that senses pheromones during mating and the other that senses nutrients and controls filamentous growth (Lengeler et al., 2000; Harashima and Heitman, 2004). S. cerevisiae exists in two haploid mating types, a and α, which communicate via mating pheromones. a haploid cells express a pheromone and the GPCR Ste2 to sense extracellular α pheromone. α haploid cells express α pheromone and the GPCR Ste3 that senses a pheromone. In both cell types, Ste2 and Ste3 are coupled to the Gα subunit Gpa1, which forms a conventional heterotrimeric G protein with the Gβγ subunits Ste4/18. On pheromone binding to either receptor, GDP-GTP exchange occurs on Gpa1 and the Ste4/18 Gβγ complex dissociates. The liberated Ste4/18 dimer activates the pheromone responsive MAP kinase cascade culminating in mating (for reviews, see Dohlman and Thorner, 2001; Dohlman, 2002; Schwartz and Madhani, 2004).
In contrast to the pheromone GPCRs that are haploid- and mating-type-specific, a distinct GPCR, Gpr1, is expressed in both diploid and haploid cells. The Gpr1 receptor activates cAMP-PKA signaling and governs diploid pseudohyphal differentiation and haploid invasive growth via the coupled Gα subunit Gpa2 (for reviews, see Lengeler et al., 2000; Pan et al., 2000; Gancedo, 2001; Harashima and Heitman 2004). gpr1 and gpa2 mutants are defective in both pseudohyphal growth and transient cAMP production in response to glucose (Kübler et al., 1997; Lorenz and Heitman, 1997; Colombo et al., 1998; Yun et al., 1998; Kraakman et al., 1999; Lorenz et al., 2000; Rolland et al., 2000; Tamaki et al., 2000; Lemaire et al., 2004). Recent studies provide evidence that glucose and structurally related sugars serve as ligands for the GPCR Gpr1 (Kraakman et al., 1999; Lorenz et al., 2000; Rolland et al., 2000; Lemaire et al., 2004).
The yeast Gα subunit Gpa2 shares 35–55% identity with other fungal and mammalian Gα subunits, and the predicted secondary structures are highly conserved between Gpa2 and canonical Gα subunits (Harashima and Heitman, 2004). Amino acid residues that confer dominant phenotypes when mutated are also conserved. For instance, a mutation of Gln300 to Leu (Q300L) in Gpa2 is analogous to the Giα1 Q204L mutation that abolishes the intrinsic GTPase activity and functions as an activated form of Gpa2 (Harashima and Heitman, 2002). A mutation of Gly299 to Ala (Gpa2 G299A) is analogous to Giα1 G203A and Gαs G226A that fail to undergo the GTP-induced conformational change and thereby serves as a dominant negative allele and interacts with Gpb1/2 and Gpr1 more strongly compared with the wild-type Gpa2 (Lorenz and Heitman 1997; Harashima and Heitman, 2002).
Nevertheless, Gpa2 does not form a heterotrimeric complex with the known yeast Gβγ subunits Ste4/18 (Lorenz et al., 2000; Harashima and Heitman, 2002, 2004). Recent studies identified two novel Gpa2 associated proteins, the kelch proteins Gpb1 and Gpb2, which are functionally redundant and share ∼35% identity (Harashima and Heitman, 2002; Batlle et al., 2003). The kelch motif is known to mediate protein-protein interactions (Adams et al., 2000). Gpb1 and Gpb2 each contain seven kelch repeats, which share no sequence homology with the seven WD40 repeats of canonical Gβ subunits. The crystal structure of the kelch repeat enzyme galactose oxidase reveals that the seven kelch repeats can adopt a seven-bladed β-propeller structure strikingly similar to Gβ subunits (Ito et al., 1991, 1994; Wall et al., 1995; Lambright et al., 1996; Sondek et al., 1996; Adams et al., 2000; Harashima and Heitman, 2002).
gpb1,2 mutants exhibit enhanced PKA phenotypes, including increased filamentous growth, sensitivity to nitrogen starvation and heat shock, reduced glycogen accumulation, and reduced sporulation (Harashima and Heitman, 2002; Batlle et al., 2003). The gpb1,2 mutant phenotypes are partially alleviated by gpa2 mutations and abolished by mutation of the TPK2 gene that encodes one of the three PKA catalytic subunits. These genetic findings support a model in which the kelch proteins Gpb1/2 negatively regulate the cAMP signaling pathway by inhibiting Gpa2 and an unidentified target that may be an upstream element of the PKA pathway including adenylyl cyclase or its regulator Ras or regulatory proteins of Ras (Harashima and Heitman, 2002).
In contrast to canonical Gα subunits, Gα Gpa2 has an extended N-terminus (Figure 1). This region shares no homology with known Gα subunits, whereas the remainder of Gpa2 shares >60% identity with Gα subunits in closely related yeasts and >40% identity with mammalian Gα subunits. The N-terminal regions of Gα subunits are known to mediate membrane localization and physical interactions with the cognate GPCR and Gβγ dimer (Navon and Fung, 1987; Hamm et al., 1988; Journot et al., 1991; Lambright et al., 1996; Wall et al., 1998; Yamaguchi et al., 2003; Herrmann et al., 2004).
Figure 1.
N-terminal alpha helix of Gα subunits (αN domain) is involved in receptor and Gβγ dimer coupling. (A) The αN domain provides one of the binding interfaces between Gα and Gβ and the receptor. This image shows a hypothetical model (PDB file 1BOK) for a GPCR-G protein module (GPCR; Rhodopsin, PDB file 1F88, G protein; PDB file 1GOT). The αN domain of the Gα subunit that is required for Gβ subunit and receptor coupling is shown (modified from Cabrera-Vera et al., 2003). (B) The predicted secondary structures of the conventional rat Gαi subunit and the yeast Gα Gpa2 protein based on PHD (Rost et al., 1993). Gpa2 shares 34% identity with the rat Gαi subunit and the predicted secondary structure is highly conserved between the two, except for the extended Gpa2 N-terminus. Secondary structure assignments were based on those of Gαt/αI (Lambright et al., 1996). (C) An alignment of the amino acid sequence of the N-terminus of Gpa2 homologues from S. cerevisiae and the related yeasts C. glabrata and S. castellii. C. glabrata and S. castellii express homologues of the S. cerevisiae GPCR Gpr1 and Gβ mimic Gpb1/2 proteins as well as a Gpa2 homologue, yet the N-termini of their Gpa2 homologues share no significant homology. Amino acids forming a potential alpha helix in the N-termini are indicated by red rectangles. Identical amino acids are marked (*) and shaded in gray, and conserved amino acids are also indicated (•). The 100th amino acid (R) of Gpa2 is shown in red. The β1 and α1 domains assigned in Figure 1B are shown. Alignments were obtained using Clustal W (Thompson et al., 1994).
All Gα subunits of heterotrimeric G proteins bear N-terminal lipid modifications (myristoylation and palmitoylation) necessary for membrane targeting (for reviews, see Chen and Manning, 2001; Cabrera-Vera et al., 2003). Myristoylation involves the irreversible cotranslational addition of a 14-carbon myristoyl group on glycine at the second position in the consensus sequence MGXXXS and this occurs via an amide linkage after proteolytic removal of the initiating methionine (Johnson et al., 1994; Ashrafi et al., 1998; Farazi et al., 2001). Palmitoylation occurs on all Gα subunits with the exception of Gαt (transducin) and involves posttranslational attachment of a saturated 16-carbon fatty acid, palmitate, via thioester linkage to cysteine residue(s) near the N-terminus. There is no palmitoylation consensus sequence, and palmitoylation is reversible and may be regulated. Both palmitoylation and myristoylation may play roles in addition to membrane localization (Linder et al., 1991; Gallego et al., 1992; Wedegaertner et al., 1993; Wilson and Bourne, 1995; Wise et al., 1997; Morales et al., 1998; Evanko et al., 2000; Fishburn et al., 2000).
S. cerevisiae serves as a powerful model to study GPCR-G protein signaling (for reviews, see Jeansonne, 1994; Lengeler et al., 2000; Dohlman and Thorner, 2001; Dohlman, 2002; Harashima and Heitman, 2004). The Gα subunit Gpa1 is myristoylated at the Gly2 residue and palmitoylated at the Cys3 residue (Song and Dohlman, 1996; Song et al., 1996). Myristoylation is required for Gpa1 membrane targeting and palmitoylation, yet not for interaction with Gβγ (Song et al., 1996). On the other hand, a Gpa1 palmitoylation-site mutant protein (Gpa1C3A) is still partially localized to the plasma membrane, partially functional, and bound to Gβγ (Song and Dohlman, 1996). The Gβγ dimer, the associated GPCR Ste2/3, or components of the Gpa1 mediated MAP kinase cascade are not required for Gpa1 membrane localization (Song and Dohlman, 1996), but the Ste4/18 Gβγ dimer does promote receptor-Gpa1 coupling (Blumer and Thorner, 1990).
The distinct Gα subunit Gpa2 forms an unusual protein complex with the atypical binding partner kelch Gβ mimics Gpb1/2 and contains an extended N-terminus. Thus novel regulatory mechanisms may direct Gpa2 to the plasma membrane and enable Gpa2 to function as a molecular switch. Here we show that Gpa2 shares similar characteristics with Gpa1 involving lipid modifications and their function. Gpa2 interacting proteins are dispensable for Gpa2 membrane localization. However, unexpectedly, Gpa2 is required for membrane targeting of the kelch Gβ mimic Gpb2, in striking contrast to conventional heterotrimeric G proteins. Furthermore, the kelch Gβ mimic proteins Gpb1/2 were found to interfere with Gpr1 receptor-Gα Gpa2 coupling.
MATERIALS AND METHODS
Strains, Media, and Plasmids
Media and standard yeast experimental procedures were as described (Sherman, 1991). To express genes heterologously in yeast cells, an attenuated ADH1 promoter and an ADH1 terminator from the yeast two-hybrid vector pGBT9 were amplified by fusion PCR using primers, GCTTGCATGCAACTTCTTTT/CGACGGATCCCCGGGAATTCCATCTTTCAGGAGGCTTGCT and AGCAAGCCTCCTGAAAGATGGAATTCCCGGGGATCCGTCG/CGGCATGCCGGTAGAGGTGT, for the 1st round PCR and primers, GCTTGCATGCAACTTCTTTT/CGGCATGCCGGTAGAGGTGT for the second round PCR. The resulting PCR products were blunted with T4 DNA polymerase and cloned into the 2μ plasmid YEplac195 that was digested with HindIII and EcoRI and then blunted with T4 DNA polymerase to create a yeast expression vector pTH19 (URA3 2μ). pTH171 (LEU2 2μ), pTH172 (TRP1 2μ), and pTH173 (LYS5 2μ) are pTH19 derivatives. The nuclear localization signal (NLS) derived from the SV40 T antigen (PPKKKRKVA) was used to direct fusion proteins into the nucleus (Arévalo-Rodríguez and Heitman, 2005). pFA6a-GFP(S65T)-kanMX6 was used as the substrate for PCR to amplify GFP (Longtine et al., 1998). Plasmids and yeast strains used in this study are listed in Tables 1 and 2. Details of plasmids and strains are available upon request.
Table 1.
S. cerevisiae strains
| Strain | Genotype | Source/Reference |
|---|---|---|
| Σ1278b congenic strains | ||
| MLY40α | MATα ura3-52 | Lorenz and Heitman (1997) |
| MLY61a/ α | MATa / α ura3-52 / ura3-52 | Lorenz and Heitman (1997) |
| MLY97a/ α | MATa / α ura3-52 / ura3-52 leu2Δ::hisG/leu2Δ::hisG | Lorenz and Heitman (1997) |
| MLY132α | MATα gpa2Δ::G418 ura3-52 | Lorenz and Heitman (1997) |
| MLY132a/ α | MATa / α gpa2Δ::G418 / gpa2Δ::G418 ura3-52 / ura3-52 | Lorenz and Heitman (1997) |
| MLY212a/ α | MATa / α gpa2Δ::G418 / gpa2Δ::G418 ura3-52 / ura3-52 leu2Δ::hisG/leu2Δ::hisG | Lorenz and Heitman (1997) |
| MLY232a/ α | MATa / α gpr1Δ::G418 / gpr1Δ::G418 ura3-52 / ura3-52 | Lorenz et al. (2000) |
| MLY277a/ α | MATa / α gpa2Δ::G418 / gpa2Δ::G418 gpr1Δ::G418 / gpr1Δ::G418 ura3-52 / ura3-52 | Laboratory stock |
| THY212a/ α | MATa / α gpb1Δ::hph / gpb1Δ::hph gpb2Δ::G418 / gpb2Δ::G418 ura3-52 / ura3-52 | Harashima and Heitman (2002) |
| THY224a/ α | MATa/α gpg1Δ::hph/gpg1Δ::hph ura3-52/ura3-52 | This study |
| THY243a/ α | MATa / α gpb1Δ::hph / gpb1Δ::hph gpb2Δ::G418 / gpb2Δ::G418 gpr1Δ::hph / gpr1Δ::hph ura3-52 / ura3-52 | Harashima and Heitman (2002) |
| THY246a/ α | MATa / α gpb1Δ::hph / gpb1Δ::hph gpb2Δ::G418 / gpb2Δ::G418 gpg1Δ::nat / gpg1Δ::nat ura3-52 / ura3-52 | Harashima and Heitman (2002) |
| S288C background strains | ||
| S1338 | MATaura3Δ::loxP leu2Δ::loxP trp1Δ::loxP gal2 | Ito-Harashima |
| THY452 | MATaura3Δ::loxP leu2Δ::loxP trp1Δ::loxP lys5Δ::loxP gal2 | This study |
Table 2.
Plasmids
| Plasmid | Description | Source/Reference |
|---|---|---|
| pTH19 | PADH1URA3 2μ | This study |
| pTH26 | PADH1-GPB1 URA3 2μ (pTH19) | This study |
| pTH27 | PADH1-GPB2 URA3 2μ (pTH19) | This study |
| pTH47 | PADH1-GPA2 URA3 2μ (pTH19) | This study |
| pTH48 | PADH1-GPA2Q300L URA3 2μ (pTH19) | This study |
| pTH49 | PADH1-GPA2G299A URA3 2μ (pTH19) | This study |
| pTH62 | PADH1-GPA2G2A URA3 2μ (pTH19) | This study |
| pTH65 | PADH1-GPA21-30 aa::GFP URA3 2μ (pTH19) | This study |
| pTH68 | PADH1-GPA2C4A URA3 2μ (pTH19) | This study |
| pTH69 | PADH1-GPA2S6Y URA3 2μ (pTH19) | This study |
| pTH71 | PADH1-GPA21-10 aa::GFP URA3 2μ (pTH19) | This study |
| pTH73 | PADH1-GFP URA3 2μ (pTH19) | This study |
| pTH75 | PADH1-GFP-GPB2 URA3 2μ (pTH19) | This study |
| pTH80 | PADH1-GPA21-10::GFP::GPA24-449 URA3 2μ (pTH19) | This study |
| pTH81 | PADH1-GPA21-20 aa::GFP-FLAG URA3 2μ (pTH19) | This study |
| pTH84 | PADH1-GFP-GPB2 LEU2 2μ (pTH171) | This study |
| pTH91 | PADH1-GPA21-20 aa G2A::GFP-FLAG URA3 2μ (pTH19) | This study |
| pTH92 | PADH1-GPA21-20 aa C4A::GFP-FLAG URA3 2μ (pTH19) | This study |
| pTH93 | PADH1-GPA21-20 aa S6Y::GFP-FLAG URA3 2μ (pTH19) | This study |
| pTH100 | PADH1- GFP-FLAG URA3 2μ (pTH19) | This study |
| pTH103 | PADH1-GPA11-20 aa::GFP-FLAG URA3 2μ (pTH19) | This study |
| pTH106 | PADH1-GFP-GPB1 URA3 2μ (pTH19) | This study |
| pTH114 | PADH1-GPB2 LEU2 2μ (pTH171) | This study |
| pTH127 | PADH1-GPA11-10-GPA2Δ1-100 URA3 2μ (pTH19) | This study |
| pTH128 | PADH1-GPA11-10-GPA2Δ1-100 G299A URA3 2μ (pTH19) | This study |
| pTH130 | PADH1-GPA2Δα (51-57) G299A URA3 2μ (pTH19) | This study |
| pTH133 | PADH1-GPA2Δα (51-57) URA3 2μ (pTH19) | This study |
| pTH134 | PADH1-GPA11-10-GPA2Δ1-29 G299A URA3 2μ (pTH19) | This study |
| pTH136 | PADH1-GPA2Δ16-84 G299A URA3 2μ (pTH19) | This study |
| pTH144 | PADH1-GPA11-10-GPA2Δ1-14 G299A URA3 2μ (pTH19) | This study |
| pTH145 | PADH1-GPA2Δ46-84 G299A URA3 2μ (pTH19) | This study |
| pTH149 | PADH1-GPA2G2A-NLS URA3 2μ (pTH19) | This study |
| pTH155 | PADH1-GPA2Δ46-100 URA3 2μ (pTH19) | This study |
| pTH157 | PADH1-GPA11-10-GPA2Δ1-29 URA3 2μ (pTH19) | This study |
| pTH158 | PADH1-GPA2Δ46-84 URA3 2μ (pTH19) | This study |
| pTH159 | PADH1-GPA2Δ31-84 G299A URA3 2μ (pTH19) | This study |
| pTH160 | PADH1-GPA2Δ31-84 URA3 2μ (pTH19) | This study |
| pTH161 | PADH1-GPA2Δ16-84 URA3 2μ (pTH19) | This study |
| pTH163 | PADH1-MLS-GFP-GPB2 URA3 2μ (pTH19) | This study |
| pTH164 | PADH1-MLS-GFP-GPB1 URA3 2μ (pTH19) | This study |
| pTH166 | PADH1-NLS-GFP-GPB2 URA3 2μ (pTH19) | This study |
| pTH167 | PADH1-NLS-GFP-GPB1 URA3 2μ (pTH19) | This study |
| pTH168 | PADH1-GPA2Δ46-100 G299A URA3 2μ (pTH19) | This study |
| pTH169 | PADH1-GPA11-10-GPA2Δ1-14 URA3 2μ (pTH19) | This study |
| pTH170 | PADH1-GFP-GPR1C TRP1 2μ (pTH172) | This study |
| pTH171 | PADH1LEU2 2μ | This study |
| pTH172 | PADH1TRP1 2μ | This study |
| pTH173 | PADH1LYS5 2μ | This study |
| pTH174 | PADH1-GPB1 LYS5 2μ (pTH173) | This study |
| pTH178 | PADH1-GPA2Δ46-449 URA3 2μ (pTH19) | This study |
| pTH191 | PADH1-GPA11-10-GPA2Δ1-44 URA3 2μ (pTH19) | This study |
| pTH192 | PADH1-GPA11-10-GPA2Δ1-44 G299A URA3 2μ (pTH19) | This study |
Pseudohyphal and Invasive Growth
Pseudohyphal and invasive growth assays were investigated as described previously (Harashima and Heitman, 2002).
Microscopic Studies
If not specifically described in figure legends, growth conditions were as follows. For protein localization study, cells were grown in synthetic minimal media to stationary phase and examined for protein localization under a fluorescent microscope (Zeiss Axioskop2 plus, Thornwood, NY) or a confocal microscope (Zeiss LSM 410).
Preparation of Crude Cell Extracts and Immunoprecipitation
Total cell extracts from yeast cells that were grown to midlog phase (OD600 ≅ 0.8) in synthetic dropout media were prepared in lysis buffer (50 mM HEPES, pH 7.6, 120 mM NaCl, 0.3% CHAPS, 1 mM EDTA, 20 mM NaF, 20 mM β-glycerophosphate, 0.1 mM Na-orthovanadate, 0.5 mM dithiothreitol, protease inhibitors (Calbiochem, La Jolla, CA; cocktail IV), and 0.5 mM phenylmethylsulfonyl fluoride) using a bead-beater. After centrifugation (25,000 × g, 20 min), crude extracts (2 mg) were mixed with anti-FLAG M2 affinity gel (Sigma, St. Louis, MO) to precipitate FLAG tagged proteins.
In Vivo Lipid Modifications
Cells were grown in 10 ml of SD-Ura medium to OD600 = 0.6–0.7, collected, and resuspended into 5 ml of fresh SD-Ura medium. After 10 min, cerulenin was added at a final concentration of 2 μg/ml, and cells were incubated for an additional 15 min under the same conditions. Subsequently, [3H]myristic acid or [3H]palmitic acid was added to the cultures at a final concentration of 50 μCi/ml for myristoylation analysis or 500 μCi/ml for palmitoylation analysis. After 3 h, cells were collected and washed once with H2O and twice with phosphate-buffered saline. Preparation of crude cell extracts and immunoprecipitation of FLAG tagged proteins were performed as above. The bound FLAG tagged proteins were eluted by boiling for 5 min in SDS-PAGE sample buffer in the presence of β-mercaptoethanol for the myristoylation analysis and in the absence of β-mercaptoethanol for the palmitoylation analysis (Song and Dohlman, 1996). After SDS-PAGE, gels were fixed in H2O/2-propanol/acetic acid (65:25:10 vol/vol/vol) for 30 min and then soaked at room temperature for 18 h either in 1 M hydroxylamine (pH 7.0) to cleave thioester-linked fatty acids or 1 M Tris-HCl (pH 7.0) as a control. The gels were fixed again, treated with Amplify (Amersham, Piscataway, NJ) for 30 min, dried, and then exposed to an x-ray film (BioMax MS film, Eastman Kodak, Rochester, NY) with an intensifying screen (BioMax Transcreen LE, Kodak) at –80°C for 1–2 mo. Expression of the FLAG-tagged proteins was verified by Western blot analysis using anti-FLAG M2 antibody (Sigma).
cAMP Assay
cAMP assay was as described in Lorenz et al. (2000) with some modifications. Briefly, at the time points indicated, 0.5 ml of cell suspension was transferred into a microfuge tube containing 0.5 ml of 10% ice-cold trichloroacetic acid and was immediately frozen in liquid nitrogen. To prepare intracellular cAMP, cells were permeabilized by defrosting at 4°C overnight. Cell extracts were neutralized by ether extraction and lyophilized. Intracellular cAMP levels were determined by using a cAMP enzyme immunoassay kit (Amersham).
RESULTS
Gα Subunit Gpa2 Is Myristoylated and Palmitoylated
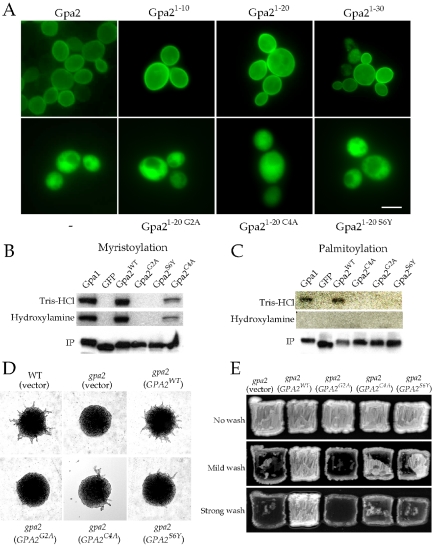
The Gα protein Gpa2 is coupled to the GPCR Gpr1 and signals to activate the downstream effector adenylyl cyclase in response to glucose. Based on analogy to other GPCR-Gα systems, we hypothesized that Gpa2 would be localized to the cell membrane for function. To address this, Gpa2 was fused to green fluorescent protein (GFP). To avoid perturbing protein localization or receptor coupling sequences typically linked to the amino and carboxy terminal regions of Gα proteins (Figure 1A), GFP was fused between the first 10 amino acids (1–10) of Gpa2 and the remainder of the protein (amino acids 4–449) to produce a Gpa21–10-GFP-Gpa24–449 internal fusion protein. This Gpa2-GFP fusion protein was functional based on its ability to complement the pseudohyphal defect of gpa2 mutant cells (unpublished data). As shown in Figure 2A, the Gpa2-GFP fusion protein was localized to the cell membrane. A C-terminally GFP tagged Gpa2 protein was nonfunctional (unpublished data), in accord with the known role of the Gα C-terminal domain in receptor coupling (Slessareva et al., 2003; Herrmann et al., 2004).
Figure 2.
Myristoylation and palmitoylation are required for membrane localization and function of the Gα subunit Gpa2. (A) The first 10 amino acids from Gpa2 are sufficient for membrane localization. A functionally, internally GFP-tagged Gpa2 (Gpa2, pTH80), truncated GFP-tagged Gpa2 proteins, Gpa21–10-GFP (Gpa21–10, pTH71), Gpa21–20-GFP-FLAG (Gpa21–20, pTH81), and Gpa21–30-GFP (Gpa21–30, pTH65), or mutant truncated GFP-tagged Gpa2 proteins, Gpa21–20 G2A-GFP-FLAG (Gpa21–20 G2A, pTH91), Gpa21–20 C4A-GFP-FLAG (Gpa21–20 C4A, pTH92), and Gpa21–20 S6Y-GFP-FLAG (Gpa21–20 S6Y, pTH93), were expressed from a 2μ plasmid in wild-type yeast cells (MLY61a/α) to test for protein localization. The GFP cassette alone (–, pTH73) was also expressed as a control. Scale bar, 5 μm. (B and C) Gpa2 is myristoylated (B) and palmitoylated (C). gpa2 mutant cells (MLY132a/α) expressing the Gpa21–20-GFP-FLAG (Gpa2WT, pTH81), Gpa21–20 G2A-GFP-FLAG (Gpa2G2A, pTH91), Gpa21–20 S6Y-GFP-FLAG (Gpa2S6Y, pTH93), Gpa21–20 C4A-GFP-FLAG (Gpa2C4A, pTH92), GFP-FLAG (GFP, pTH100), or Gpa11–20-GFP-FLAG (Gpa1, pTH103) proteins were metabolically labeled with [3H]myristic acid or [3H]palmitic acid. FLAG-tagged proteins were purified using anti-FLAG affinity gel and subjected to SDS-PAGE. Gels were treated with 1 M Tris-HCl, 1 M hydroxylamine that cleaves the palmitoyl moiety of fatty acids, or subjected to Western blot using an anti-FLAG antibody to verify purified protein levels. Radiolabeled purified proteins were visualized by autoradiography. (D and E) Myristoylation and palmitoylation are required for Gpa2 function. Full-length wild-type (Gpa2WT, pTH47) or mutant Gpa2 proteins (Gpa2G2A (pTH62), Gpa2C4A (pTH68), and Gpa2S6Y (pTH69)) were expressed in gpa2 mutant cells (MLY132a/α or MLY132α) to test for diploid filamentous growth (D) and haploid invasive growth (E). gpa2 mutant cells containing an empty plasmid (pTH19) served as control.
To establish the minimal Gpa2 domain required for membrane localization, the first 10 (Gpa21–10), 20 (Gpa21–20), or 30 (Gpa21–30) amino acids of Gpa2 were fused to a GFP cassette and expressed in vivo. All three C-terminally tagged Gpa2-GFP proteins were localized to the plasma membrane (Figure 2A). Therefore, as few as the first 10 amino acids of Gpa2 suffice for plasma membrane targeting.
In conventional Gα subunits, lipid modifications of the N-terminus mediate membrane localization (Chen and Manning, 2001). Myristoylation occurs at Gly2 in the myristoylation consensus sequence G2XXXS6 (Johnson et al., 1994). Palmitoylation can occur at any cysteine residue near the N-terminus. Gpa2 contains glycine and serine in the second and sixth positions for myristoylation and cysteine at the fourth position from the N-terminus. To examine whether these sites are lipid modified, a Gpa21–20-GFP-FLAG protein in which the first 20 amino acids of Gpa2 were fused to a GFP-FLAG cassette was expressed in yeast cells and assessed for lipid modifications. Gpa21–20-GFP-FLAG variants containing mutations in the potential lipid modification sites (G2A, C4A, or S6Y) were also analyzed.
As a positive control for lipid modification experiments, an equivalent Gpa11–20-GFP-FLAG protein was constructed, which was derived from the Gpa1 Gα subunit coupled to the Ste2/3 pheromone receptors (Figure 2B). Gpa1 is known to be myristoylated at the second position on glycine (Gly2) and palmitoylated on cysteine in the third position (Cys3) (Song and Dohlman, 1996; Song et al., 1996). Gpa1 myristoylation is essential for membrane localization and function and required for palmitoylation, and palmitoylation also promotes membrane localization and function. In addition, the first 9 amino acids of Gpa1 suffice for membrane localization of a Gpa1-GST fusion protein (Gillen et al., 1998).
As shown in Figure 2B, the wild-type Gpa2 fusion protein was myristoylated and the myristoylation site and myristoylation consensus sequence mutant proteins, Gpa2G2A and Gpa2S6Y, were not, suggesting that Gpa2 is subject to myristoylation at Gly2. Gpa2 was also palmitoylated and a mutation in the putative palmitoylation site (Gpa2C4A) abolished this modification (Figure 2C). Therefore, Gpa2 is also subject to palmitoylation at Cys4. We note that the Gpa2C4A fusion protein exhibited a decreased level of myristoylation compared with the wild-type protein. Interestingly, reduced myristoylation was also observed with the Gpa1C3S mutant (Song and Dohlman, 1996). These results are indicative of either a sequence preference in the myristoylation consensus sequence (G2XXXS6) or a role for palmitoylation in promoting myristoylation or its maintenance.
Similar to Gpa1, Gpa2 requires myristoylation for palmitoylation because the G2A and S6Y mutations, which abolish myristoylation, also blocked palmitoylation. Consistent with these results, the Gpa2-GFP-FLAG proteins bearing the G2A, C4A, or S6Y mutations failed to localize to the plasma membrane, and thus myristoylation and palmitoylation are required for Gpa2 plasma membrane localization (Figure 2A).
To address the physiological roles of these lipid modifications, the G2A, C4A, and S6Y mutations were introduced into the GPA2 gene and expressed in a Σ1278b gpa2/gpa2 diploid or gpa2 haploid mutant strain. As shown in Figure 2, D and E, the GPA2G2A myristoylation site mutant failed to complement either the pseudohyphal or the invasive growth defects. The GPA2S6Y and GPA2C4A myristoylation consensus sequence or palmitoylation site mutants showed severe defects in both assays. Furthermore, introduction of a dominant active mutation (Q300L) that abolishes Gpa2 GTPase activity failed to restore activity of the GPA2G2A mutant protein (Gpa2G2A, Q300L, unpublished data). Thus, myristoylation and palmitoylation both play critical roles in Gpa2 membrane localization and signaling. Importantly, the unusual Gα subunit Gpa2 shares common features with the conventional Gα subunit Gpa1 with respect to lipid modifications and their physiological roles.
Gpa2 Binding Partners Are Not Required for Gpa2 Membrane Localization
In heterotrimeric G proteins, Gβγ subunits can promote membrane localization of their associated Gα subunits. Therefore, the localization of Gpa2 was examined in the absence of Gpb1/2 or when Gpb1/2 were overexpressed. As shown in Figure 3, A and B, Gpa2 membrane localization was unchanged under both conditions. Furthermore, deletion of other known Gpa2 associated proteins, namely the GPCR Gpr1 or the Gγ subunit mimic Gpg1, or even the elimination of multiple binding partners (Gpb1/2 and Gpr1 or Gpb1/2 and Gpg1), did not perturb Gpa2 plasma membrane localization, suggesting these binding partners are not required for membrane targeting (Figure 3A).
Figure 3.
The Gα subunit Gpa2 is localized to the plasma membrane independent of its known binding partners. (A) Gpa2-GFP protein (pTH80) was expressed in gpr1 (MLY232a/α), gpg1 (THY224a/α), gpb1,2 (THY212a/α), gpb1,2 gpr1 (THY243a/α), and gpb1,2 gpg1 (THY246a/α) mutant cells and protein localization was analyzed. (B) Overexpression of the kelch Gβ mimic proteins Gpb1/2 has no effect on Gpa2 membrane localization. The Gpa2-GFP protein was coexpressed with Gpb1 (pTH26), Gpb2 (pTH27), or both (pTH26 and pTH114) in wild-type cells (MLY97a/α). (C) Membrane localization of Gpa2 was not altered by carbon sources. gpa2 mutant cells (MLY132a/α) expressing the Gpa2-GFP protein were grown in synthetic media containing different carbon sources and Gpa2 protein localization was assessed. Scale bars, 5 μm.
Because Gpa2 is a component of the glucose sensing cAMP signaling pathway and the agonist induced redistribution of Gαs has been reported in mammalian cells (Wedegaertner et al., 1996; Thiyagarajan et al., 2002), we examined if carbon source affects Gpa2 protein localization (Figure 3C). Glucose serves as a ligand for Gpr1 (Yun et al., 1998; Kraakman et al., 1999; Lorenz et al., 2000; Rolland et al., 2000; Lemaire et al., 2004). Glucose, fructose, and galactose are structurally related hexoses, yet galactose is not a ligand for Gpr1 (Lorenz et al., 2000; Lemaire et al., 2004). Fructose is controversial, although fructose can induce cAMP production when added to glucose-starved cells (Yun et al., 1998; Lemaire et al., 2004). Maltose and galactose induce filamentous growth in a Gpr1-Gpa2-independent manner (Lorenz et al., 2000). Ethanol and glycerol are structurally unrelated nonfermentable carbon sources. As shown in Figure 3C, Gpa2 was localized to the plasma membrane to the same extent under all conditions tested. Therefore, the carbon sources examined do not influence Gpa2 protein localization and Gpa2 is localized to the cell membrane irrespective of activity of the Gpr1-Gpa2 signaling pathway.
Kelch Gβ Mimic Gpb2 Is Recruited to the Plasma Membrane by Gpa2
If the kelch proteins Gpb1/2 function as Gβ mimics, we hypothesized that Gpb1/2 should also be membrane localized. To examine protein localization, a functional GFP-Gpb2 protein was expressed in gpa2Δ cells (Figure 4). When GFP-Gpb2 was expressed alone, Gpb2 was found to be cytoplasmic. However, when GFP-Gpb2 was coexpressed with either wild-type Gpa2 or a dominant negative Gpa2 (Gpa2G299A), GFP-Gpb2 was directed to the plasma membrane (Figure 4). Confocal microscopic analysis revealed that Gpb2 was localized to the plasma membrane more extensively when coexpressed with the Gpa2G299A mutant protein that is unable to undergo the GTP-induced conformational change when compared with wild-type Gpa2 (Figure 4A). This finding is in accord with previous data showing that Gpb2 binds to Gpa2 in vivo and preferentially associates with Gpa2-GDP (Harashima and Heitman, 2002).
Figure 4.
Gα subunit Gpa2 recruits the kelch Gβ subunit mimic Gpb2 to the plasma membrane. (A) A functional GFP-Gpb2 protein (pTH84) was coexpressed with Gpa2 (pTH47), Gpa2G299A (pTH49), Gpa2G2A (pTH62), or Gpa2G2A-NLS (pTH149) proteins in gpa2Δ mutant cells (MLY212a/α), and protein localization was investigated by confocal (A) or direct fluorescence microscopy (B). The empty vector pTH19 (–) served as control. Nuclear localization was confirmed by DAPI staining (unpublished data). Scale bar, 5 μm.
When GFP-Gpb2 was coexpressed with the nonfunctional Gpa2G2A mutant that is no longer directed to the plasma membrane, GFP-Gpb2 was no longer localized to the plasma membrane (Figure 4). To exclude the possibility that the observed Gpb2 membrane localization is an indirect secondary consequence due to overexpression of the functional wild-type Gpa2 protein, GFP-Gpb2 was coexpressed with a nuclear localization signal (NLS) containing Gpa2G2A mutant protein (Gpa2G2A-NLS). Strikingly, Gpa2G2A-NLS now misdirected Gpb2 to the nucleus (Figure 4B). Therefore, the Gα protein Gpa2 forms a stable complex with the kelch Gβ mimic protein Gpb2 and serves to recruit Gpb2 to the plasma membrane. That Gpa2G2A-NLS directs Gpb2 to the nucleus also demonstrates that lipid modifications are not required for the Gpa2-Gpb2 interaction. This is consistent with findings regarding interaction of the yeast Gα subunit Gpa1 and the mammalian Gα subunit Gαi with their respective Gβ subunits (Jones et al., 1990; Song et al., 1996).
Kelch Gβ Mimic Gpb2 and the C-terminal Tail of the Gpr1 Receptor Bind to the N-terminal Region of Gpa2
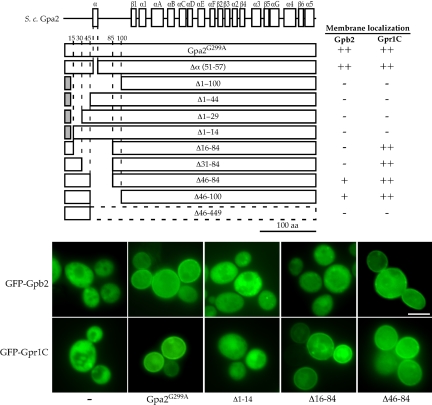
In canonical Gα subunits, an N-terminal alpha helix called the αN domain provides a binding surface for the Gβ subunit and the coupled receptor (Lambright et al., 1996; Wall et al., 1998). Because the αN domain is less conserved among Gα subunits, we searched for any related alpha helical domain in the extended N-terminus of Gpa2 using the PHD secondary structure prediction method (Rost and Sander, 1993). A sequence spanning amino acid residues 49–57 was identified that is predicted to form an alpha helix, although this region does not share any significant identity with known αN domains (Figure 1).
To examine if this candidate alpha helical domain of Gpa2 is involved in the interaction with Gpb2, the domain was deleted in the dominant negative Gpa2G299A mutant (Gpa2Δα (51–57)) and the resulting mutant derivative was coexpressed with the GFP-Gpb2 protein to test for protein localization. As noted above, Gpa2G299A recruits GFP-Gpb2 to the plasma membrane (Figure 5). Similarly, Gpa2Δα (51–57) also brought GFP-Gpb2 to the plasma membrane (Figure 5). Therefore, the sequence spanning amino acids 51–57, which is predicted to be an N-terminal alpha helical region, is not required for Gpa2-Gpb2 binding.
Figure 5.
N-terminus of Gα Gpa2 is required for binding to the kelch protein Gpb2 and the GPCR Gpr1. A series of deletions was created in the N-terminal region of Gpa2G299A, and these deletion constructs were coexpressed with the GFP-Gpb2 protein (pTH84) in gpa2Δ cells (MLY212a/α) or with GFP-Gpr1C (pTH170) in wild-type cells (S1338) to determine roles of the N-terminal region of Gpa2 on interaction with Gpb2 and the C-terminal tail of Gpr1. Deletion mutant Gpa2G299A proteins constructed and results are shown schematically. Δ1–14, Δ1–29, Δ1–44, and Δ1–100 mutant proteins were fused to the first 10 amino acids from the yeast Gα subunit Gpa1 to restore targeting to the plasma membrane and Gpa1 residues are depicted as a gray box. Scale bar, 5 μm.
We next addressed whether other sequences in the Gpa2 N-terminal extension are required for Gpb2 interaction. For this purpose, deletions were introduced into the N-terminal region of the GPA2G299A allele to create Δ1–14, Δ1–29, Δ1–44, and Δ1–100 derivatives of Gpa2G299A, which were also then fused to the first 10 amino acids from the S. cerevisiae Gα subunit Gpa1 that are sufficient for membrane localization (unpublished data; Gillen et al., 1998). Internal deletions were also created (Δ16–84, Δ31–84, Δ46–84, and Δ46–100, Figure 5). This deletion mutant series was coexpressed with GFP-Gpb2 to examine which Gpa2 mutants are capable of recruiting GFP-Gpb2 to the plasma membrane (Figure 5). All deletions generated for this study (except for the Δ46–449 Gpa2 mutant) are predicted to have no significant impact on the secondary structure of Gpa2, based on PHD analysis, and the function and expression of these alleles of GPA2G299A were confirmed by introducing these alleles into wild-type diploid cells and examining pseudohyphal growth (unpublished data). All deletion constructs and representative results are shown in Figure 5.
GFP-Gpb2 did not associate with the plasma membrane when coexpressed with the Δ1–14, Δ1–29, Δ1–44, or Δ1–100 Gpa2 derivatives, indicating that the N-terminus of Gpa2 plays an important role in Gpb2 binding (Figure 5). However, the first 15 or 30 amino acids were not sufficient for Gpb2 binding because neither the Gpa2 Δ16–84 nor the Δ31–84 mutant was able to recruit Gpb2 to the plasma membrane. On the other hand, membrane localization of GFP-Gpb2 was observed when it was coexpressed with the Gpa2 Δ46–84 and Δ46–100 mutants. Taken together, these findings indicate that the first 45 amino acids are necessary for Gpb2 interaction. This N-terminal region alone (1–45 aa) was not sufficient because GFP-Gpb2 was cytoplasmic with the Gpa2Δ46–449 variant. Structural analyses have revealed that Gβ binding interfaces are present not only in the N-terminus (the αN domain) but also in the central region (β2 to α2 domain) of conventional Gα molecules (Figure 1 and Lambright et al., 1996; Wall et al., 1998). Therefore, by analogy Gpa2 may also require the corresponding internal conserved region in conjunction with the N-terminal 1–45 aa to bind Gpb2, although we cannot exclude a possibility that the Gpa2Δ46–449 variant failed to recruit Gpb2 to the plasma membrane because of instability. Note that the deletions examined were also introduced into a wild-type Gpa2 construct and tested for GFP-Gpb2 interaction as above, and results were essentially equivalent to the ones with the Gpa2G299A deletion variants with the minor difference that plasma membrane localization of GFP-Gpb2 was weaker when the wild-type Gpa2 deletion variant were coexpressed. This is consistent with the fact that Gpa2G299A binds to Gpb2 more strongly than does wild-type Gpa2 (Figure 4, Harashima and Heitman, 2002, 2004).
We next addressed regions of the Gpa2 molecule involved in association with the Gpr1 receptor. Previously, the Gpr1 C-terminal tail composed of 99 amino acids was isolated in a yeast two-hybrid screen that identified Gpa2 interacting proteins (Xue et al., 1998). Because Gpr1 that is C-terminally tagged with GFP is nonfunctional (unpublished data), likely because of interference with Gpr1-Gpa2 coupling, we fused GFP to the N-terminus of the 99 amino acid soluble C-terminal tail of Gpr1. The resulting GFP fusion protein (GFP-Gpr1C) was coexpressed with the Gpa2G299A variants to examine roles of the N-terminal extension on interactions with the coupled receptor Gpr1, as above (Figure 5, also see Figure 8).
Figure 8.
Kelch Gβ mimic proteins Gpb1/2 interfere with the interaction between Gpa2 and the C-terminal tail of Gpr1. (A) The GFP-Gpr1C fusion protein (pTH170) was expressed alone or coexpressed with Gpa2 variants, wild-type Gpa2 (pTH47), Gpa2Q300L (pTH48), Gpa2G299A (pTH49), or NLS-Gpa2G2A (pTH149) with or without Gpb1/2 (pTH174/pTH114) in wild-type cells (THY452). Empty vectors (pTH171 and pTH173) were used as controls for the Gpb1/2 plasmids, pTH174 and pTH114. The location of nuclei were confirmed by DAPI staining.
As shown in Figure 5, any variant of Gpa2 lacking the first 15 amino acids failed to recruit GFP-Gpr1C to the plasma membrane (Gpa2Δ1–14, Gpa2Δ1–29, Gpa2Δ1–44, and Gpa2Δ1–100), whereas all of the variants containing amino acids 1–15 (Gpa2Δ16–84, Gpa2Δ31–84, Gpa2Δ46–84, and Gpa2Δ46–100) recruited GFP-Gpr1C, similar to full length Gpa2G299A. The only exception was Gpa2Δ46–449, which failed to recruit the GFP-Gpr1C to the plasma membrane. These observations indicate that the N-terminal region of Gpa2 participates in associating with the receptor C-terminal tail, but that C-terminal regions of Gpa2 likely also participate. Importantly, the C-terminal tail of other Gα subunits is known to be involved in receptor coupling (Slessareva et al., 2003; Herrmann et al., 2004). Consistent with this model, Gpa2Δ1–100 still interacted with the C-terminal tail of Gpr1 in the yeast two-hybrid assay and Gpa2 function was perturbed by a C-terminal GFP tag (unpublished data). In summary, these data indicate that both the N-terminal and more C-terminal regions of the Gα protein Gpa2 are required for interactions with both Gpb2 and Gpr1.
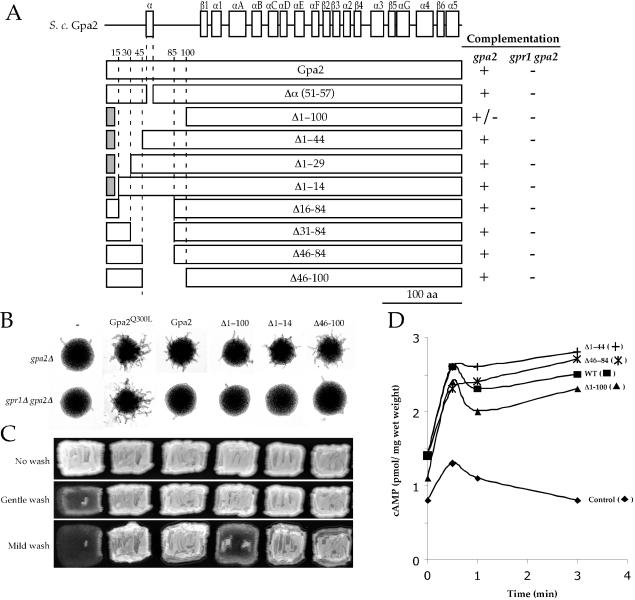
Functional Roles of the Gpa2 N-terminus
To address roles of the Gpa2 amino terminus, N-terminal deletions were introduced into wild-type Gpa2. The resulting deletion alleles were expressed in diploid or haploid gpa2 mutant cells to examine whether these mutants complement gpa2 defects in pseudohyphal growth, invasive growth, and glucose-induced cAMP production (Figure 6). These mutant alleles were also introduced into diploid gpr1 gpa2 mutant cells to examine whether they require Gpr1 for function or act as dominant alleles that bypass the receptor. Cells expressing Gpa2Δ1–100 exhibited reduced pseudohyphal and invasive growth and reduced levels of basal and glucose-induced cAMP, indicating that the N-terminal region plays an important functional role or that deletion of the 1–100 amino acids might result in misfolding of Gpa2 (Figures 6). Gpa2Δ46–84, Gpa2Δ46–100, and Gpa2Δα (51–57) all functioned as wild-type Gpa2, likely because Gpb2 and the C-terminal tail of Gpr1 still bind to these deletion proteins (Figure 6 and unpublished data). The Δ1–14, Δ1–29, Δ1–44, Δ16–84, or Δ31–84 GPA2 mutant genes were largely able to complement gpa2 mutant phenotypes. One interpretation of these results is that these deletion proteins still functionally interact with Gpr1 and Gpb2 via other Gpa2 domains and are capable of functioning, similar to wild-type Gpa2. Or expression of the deletion Gpa2 proteins from a multicopy plasmid might mask their reduced activity so that expression from a low copy plasmid could elicit altered mutant phenotypes. Alternatively, these results could be due to counterbalancing defects in Gpa2 interaction with Gpr1 and Gpb2 because Gpr1/Gpa2 and Gpb2 control the cAMP signaling pathway positively and negatively, respectively (see Discussion).
Figure 6.
Function of the N-terminal deletion Gpa2 proteins in vivo. (A) Schematic of N-terminal deletion Gpa2 variants and complementation results in gpa2 or gpr1 gpa2 mutant cells. N-terminal deletions were created in the wild-type GPA2 gene and introduced into gpa2 (MLY132α for invasive growth assay and MLY132a/α for pseudohyphal growth assay) or gpr1 gpa2 (MLY277a/α) mutant cells and ability to complement pseudohyphal and invasive growth defects was examined. Representative data are shown in B for pseudohyphal growth and in C for invasive growth. (D) Glucose-induced cAMP production in gpa2 (MLY132α) mutant cells expressing the N-terminal deletion Gpa2 derivatives. The values shown are the mean of two independent experiments, except the control, which is representative of cells carrying the empty vector (pTH19).
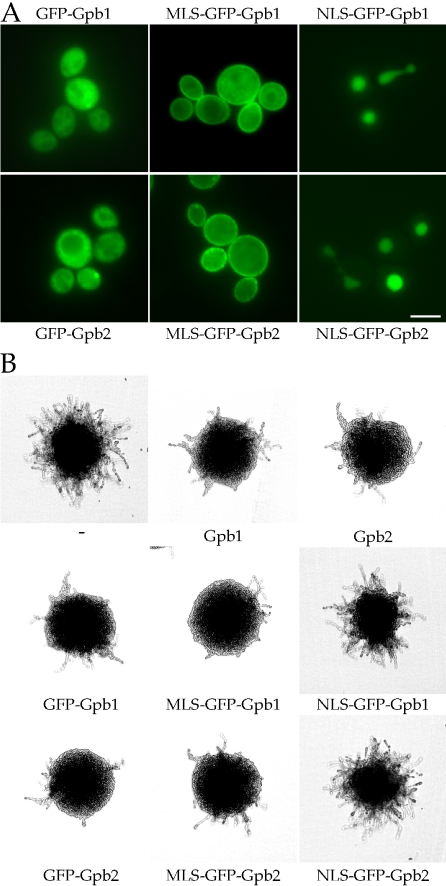
Kelch Gβ Mimic Proteins Gpb1/2 Function on the Plasma Membrane
Gpb2 is directed to the plasma membrane in a Gpa2 dependent manner, indicating that the kelch Gβ mimic proteins Gpb1/2 may function on the plasma membrane. To examine this hypothesis, the first 10 amino acids of Gpa2 (hereafter, the membrane localization sequence [MLS]) that suffice for membrane localization were fused to the N-terminus of the GFP-Gpb1 or GFP-Gpb2 protein. The resulting fusion proteins were tested for protein localization and complementation of the elevated filamentous phenotype of gpb1,2 mutant cells (Figure 7). We also tested the effects of fusing a nuclear localization signal (NLS) from the SV40 T antigen to the N-terminus of the GFP-Gpb1 or GFP-Gpb2 protein (Figure 7).
Figure 7.
Kelch Gβ mimic proteins Gpb1/2 function on the plasma membrane. A membrane localization sequence (MLS) or nuclear localization signal (NLS) was fused to the N-terminus of the functional GFP-Gpb1/2 proteins (pTH106/pTH75) and the resulting fusion proteins (pTH163, pTH164, pTH166, or pTH167) were expressed in diploid gpb1,2 double mutant cells (THY212a/α) to test for protein localization (A) and function (B). The MLS-GFP-Gpb1/2 fusion proteins were recruited to the plasma membrane and were as functional as the wild-type Gpb1/2 proteins, whereas the NLS-GFP-Gpb1/2 fusion proteins were directed to the nucleus and nonfunctional. Cells bearing the empty vector (pTH19) or the GPB1 (pTH26) or GPB2 (pTH27) plasmid served as controls. Scale bar, 5 μm.
The MLS- and NLS-fused GFP-Gpb1/2 proteins were predominantly localized to the plasma membrane and the nucleus, respectively (Figure 7A). Furthermore, the MLS-GFP-Gpb1/2 fusion proteins complemented the gpb1,2 double mutant phenotype and restored wild-type pseudohyphal growth (Figure 7B). In contrast, the nuclear localized Gpb1/2 proteins (NLS-GFP-Gpb1/2) were nonfunctional (Figure 7B). These findings provide evidence that Gpb1/2 can function when heterologously targeted to the plasma membrane. These results also indicate that the as yet unidentified second target of Gpb1/2 might be membrane associated.
Kelch Gβ Mimic Proteins Gpb1/2 Inhibit Gpr1-Gpa2 Coupling
Gpa2 interacts with the C-terminal tail of the Gpr1 receptor and recruits the GFP-Gpr1 C-tail fusion protein to the plasma membrane. Here we used this assay to analyze Gpr1-Gpa2 coupling in further detail. GFP-Gpr1C is localized to the plasma membrane when coexpressed with the dominant negative Gpa2G299A allele. Additionally, membrane localization of GFP-Gpr1C was less pronounced when coexpressed with wild-type Gpa2, suggesting that the C-terminal tail of Gpr1 binds more strongly to Gpa2G299A compared to wild-type Gpa2 (Figure 8). On the other hand, interaction of Gpa2 with the C-terminal tail of Gpr1 was reduced even further with the dominant Gpa2Q300L allele (Figure 8). This is consistent with the widely accepted model in which the Gα-GDP complex binds to the cognate GPCR, whereas the Gα-GTP complex dissociates from the GPCR. To confirm the interaction between GFP-Gpr1C and Gpa2, the nonfunctional nuclear localized Gpa2G2A-NLS was coexpressed with GFP-Gpr1C. In this case, GFP-Gpr1C was now misdirected to the nucleus (Figure 8).
Because Gpb2 is directed to the plasma membrane in a Gpa2-dependent manner and binds to the N-terminus of Gpa2 where the C-terminal tail of Gpr1 also binds, we hypothesized that Gpb1/2 could negatively regulate Gpa2 function by inhibiting the Gpr1-Gpa2 interaction. To address this hypothesis, the wild-type Gpb1/2 proteins were simultaneously coexpressed with the GFP-Gpr1C and Gpa2G299A proteins. As shown in Figure 8, the membrane localization of GFP-Gpr1 was significantly reduced by coexpression of Gpb1/2, indicating that Gpb1/2 compete with the C-terminal tail of Gpr1 for binding to the N-terminus of Gpa2. Gpb1/2 may thereby control Gpa2 function by impairing receptor coupling. This is in contrast to canonical Gβ subunits, which function to promote interactions of the Gα subunit with the associated GPCR.
DISCUSSION
The Roles of the N-terminal Region of Gpa2
The MG2XXXS6 sequence in open reading frames and the glycine residue of the consensus sequence are well defined as a myristoylation consensus sequence and the myristoylation site. On the other hand, no obvious consensus sequence is established for palmitoylation, yet palmitoylation mostly occurs in a cysteine residue(s) near the N-terminus. The Gα subunit Gpa2 contains the MG2XXXS6 myristoylation consensus sequence and a cysteine at the fourth position of its N-terminus. A cysteine after the N-terminal cysteine appears at the 189th position of the Gpa2 protein. Our biochemical studies revealed that Gpa2 is myristoylated and palmitoylated. Furthermore, the labeling and site-directed mutagenesis studies shown in Figure 2 provide evidence that Gpa2 is myristoylated at Gly2 and, most likely, also palmitoylated at Cys4.
Introduction of site-specific mutations (G2A, C4A, and S6Y) into the GPA2 and GPA2-GFP fusion genes demonstrates that myristoylation and palmitoylation are critical for plasma membrane targeting and function of Gpa2. Although it still remains to be established why myristoylation is essential for Gα function, recent studies demonstrate that GPCR-Gα fusion proteins, in which Gα is localized to the plasma membrane yet no longer lipid modified, are functional in vivo (for review, see Seifert et al., 1999). Furthermore, a nonmyristoylated Gαi2Q205L protein is unable to signal and fails to transform rat fibroblasts (Gallego et al., 1992). Consistently, we also found that a nonmyristoylated dominant Gpa2Q300L mutant (equivalent to Gαi2 Q205L) is incapable of enhancing filamentous growth in wild-type cells. These findings support a model in which lipid modifications are necessary for plasma membrane targeting that is a prerequisite for Gα function. Alternatively, myristoylation may play an important role in Gα structure that is required for receptor coupling (Preininger et al., 2003).
In heterotrimeric G proteins, the N-terminus is also involved in interactions with Gβγ dimer, receptors, and effectors. Structural and biochemical studies implicate the N-terminal alpha helix (αN domain) in Gβγ dimer and receptor coupling (Lambright et al., 1996; Wall et al., 1998). Gpa2 contains an alpha helix in the extended N-terminus, yet the position of this helix is not conserved (Figure 1). More strikingly, the alpha helix is not involved in coupling to the kelch subunit Gpb2 or to the Gpr1 C-terminal tail. Studies using Gpa2 variants that carry a series of deletions in the Gpa2 N-terminus identified binding domains for the Gpr1 C-terminal tail and Gpb2 that map to amino acids 1–15 and 1–45 and are not predicted to form an alpha helix.
Lipid modifications alone are not sufficient to restore these interactions as the Gpa2 Δ1–14 mutant that is lipid modified on an appended Gpa11–10 peptide did not direct the binding partners to the plasma membrane. Rather, amino acid sequences that lie between residues 1–45 are important for the interactions. Interestingly, the non-alpha helical N-terminus (spanning amino acids 1–6) of Gαq is known to be involved in receptor selectivity (Kostenis et al., 1997). Therefore, the N-terminus may play a direct role in receptor coupling by providing a binding interface or an indirect role by influencing overall structure. Either possibility is novel and further studies, especially structural studies, should address the role of the N-terminus of Gpa2.
The Role of the Gpr1 C-terminal Tail
Previous studies suggest the presence of preactivation complexes in which an unoccupied, inactive GPCR is coupled to the Gα subunit (Samama et al., 1993; Stefan et al., 1998; Dosil et al., 2000). Such preactivation complexes are not necessarily required for formation of the activated ternary complex in which a ligand bound, activated receptor forms a complex with a G protein to stimulate GDP-GTP exchange on Gα, yet the preactivation complexes are involved in regulation of specificity and intensity of G-protein mediated signaling (Neubig, 1994; Shea and Linderman, 1997). In S. cerevisiae, the C-terminal tail of the α-factor receptor Ste2 is implicated in the formation of the preactivation complex with its associated Gα Gpa1 (Dosil et al., 2000). Although no direct evidence has been reported for a preactivation complex between the Gpr1 receptor and Gpa2, our data support the existence of one. First, the cytoplasmic C-terminal tail of Gpr1 binds to wild-type Gpa2 and a nuclear localized Gpa2G2A-NLS. Second, Gpr1 and Gpa2 are still functional in the absence of the Gβ mimic subunits Gpb1/2, suggesting a promiscuous coupling between Gpr1 and Gpa2.
These observations may be relevant to our finding that N-terminal deletion variants of Gpa2 (Δ1–14, Δ1–29, Δ1–44, and Δ1–100) that are unable to bind to the Gpr1 C-terminal tail are still functional and can respond to glucose to stimulate cAMP production. This interpretation may also explain why cells expressing these Gpa2 variants exhibited near wild-type phenotypes. It is conceivable that a reduced affinity of the Gpa2 variants with the Gpr1 receptor could result in a decrease in signaling leading to a low-PKA phenotype. However, these Gpa2 variants also show decreased binding to the kelch subunits Gpb1/2 that negatively control cAMP signaling, affecting Gpb1/2 function to activate the as yet unidentified second target that inhibits cAMP signaling.
Kelch Subunits Gpb1/2 Inhibit Gpr1-Gpa2 Coupling
G-protein activity is controlled at multiple steps including expression, protein localization, GDP-GTP exchange, and GTPase activity. GPCRs activate G proteins by stimulating GDP dissociation from Gα and acting as guanine nucleotide exchange factors, thereby leading to Gα in the active Gα-GTP form. On the other hand, the GoLoco family protein AGS3 functions as a guanine nucleotide dissociation inhibitor (GDI) by inhibiting GDP-GTP exchange (De Vries et al., 2000). Although GoLoco homologues are conserved in multicellular eukaryotes, no such homolog is apparent in the yeast genome.
Our previous studies revealed that the kelch subunits Gpb1 and Gpb2 negatively control Gpa2 and preferentially associate with Gpa2-GDP (Harashima and Heitman, 2002). However, neither loss nor overexpression of Gpb1/2 perturbed Gpa2 membrane localization or expression. In addition, Gpb1/2 did not exhibit GDI activity under standard in vitro conditions (unpublished data). Here we show that Gpb1/2 inhibit Gpa2-Gpr1 coupling. A model governing how the kelch Gpb1/2 subunits control Gpa2 is that Gpb1/2 bind to the Gpa2 N-terminal region spanning amino acids 1–45 and occlude binding of the Gpr1 C-terminal tail to the first fifteen amino acids of Gpa2 (Figure 9).
Figure 9.
Model of canonical heterotrimeric and atypical G protein signaling in budding yeast. The canonical heterotrimeric G protein composed of the Gpa1/Ste4/Ste18 subunits regulates the pheromone responsive MAPK cascade, whereas the atypical heteromeric G protein consisting of the Gpa2/Gpb1/2 subunits controls the nutrient sensing cAMP-PKA signaling pathway. For details, see Discussion.
In canonical heterotrimeric G proteins, Gβγ subunits are required for receptor-Gα coupling. In S. cerevisiae, the Gβγ dimer plays an essential role in pheromone receptor-Gα Gpa1 coupling (Blumer and Thorner, 1990). In mammalian systems, a role for the Gβγ subunits in coupling of β2-adrenergic receptor-Gαs, M2-muscarinic receptor-Gαo, A1-adenosine and 5-HT1A receptors-Gαi, and β2-adrenergic receptor-Gαi has been established (Richardson and Robishaw, 1999; Hou et al., 2001; Lim et al., 2001; Kühn et al., 2002). This function is opposite to the role of the kelch subunits, yet importantly, yeast and mammalian WD40 repeat Gβγ subunits and the kelch subunits all converge to modulate receptor-Gα coupling. That receptor-Gα coupling is oppositely regulated may depend on how tightly and specifically a given Gα binds to its associated receptor. In yeast, the pheromone receptor Ste2 is functionally coupled to the Gα protein Gpa1 and not to the Gpa2 Gα subunit (Blumer and Thorner, 1990). During diploid filamentation, the glucose receptor Gpr1 is associated with Gpa2 and not with the haploid specific Gα Gpa1. Importantly, the Gpa2 Gα subunit is still partially functional and able to signal in response to the agonist glucose via Gpr1 in the absence of Gpb1/2, suggesting that Gpa2 can functionally couple to its receptor in the absence of Gpb1/2 (Harashima and Heitman, 2002). Therefore, Gpa2 may normally be tightly associated with the Gpr1 receptor, and Gpb1/2 function to compete with this association to reduce signaling in the absence of glucose.
Generally, the intracellular third loop of GPCRs plays a crucial role in interactions with the Gα subunit. Although S. cerevisiae Gpa2 has been reported to interact with the intracellular third loop of Gpr1 in the yeast two-hybrid assay (Yun et al., 1997), we were unable to recapitulate this result (unpublished data). This could be attributable to a weak interaction between Gpa2 and the third loop of Gpr1. In contrast, the Gpr1 C-terminal tail avidly binds to Gpa2 in two-hybrid assays (Yun et al., 1997; Xue et al., 1998; Kraakman et al., 1999; Harashima and Heitman, 2002). We also showed that the Gpa2-Gpr1 C-terminal tail interaction can be detected using the GFP tagged C-terminal tail of Gpr1 in vivo (Figures 5 and 8). These data indicate that the Gpr1 C-terminus plays an important role in Gpa2 binding. This atypical feature of the Gpr1 receptor-Gpa2 Gα complex may mirror the unusual aspects by which the kelch subunits Gpb1/2 inhibit the signaling complex.
Is Gpa2 an Unusual Gα or an Ancestral Gα Subunit?
Our studies provide evidence that lipid modifications (myristoylation and palmitoylation) of Gα Gpa2 are necessary and sufficient for Gpa2 plasma membrane targeting but are not required for interaction with the kelch Gβ mimic subunit Gpb2. Instead, Gpa2 directs Gpb2 to the plasma membrane. Mammalian Gα subunits as well as the yeast canonical Gα subunit Gpa1 share similar features. Like Gpa2, lipid modifications but not the Gβγ dimer are required for plasma membrane localization of yeast Gpa1 and mammalian Gα (Song et al., 1996; Gillen et al., 1998; Galbiati et al., 1999). It has also been reported that a nonlipidated Gα still binds to Gβγ subunits in yeast and mammals (Jones et al., 1990; Degtyarev et al., 1994; Song et al., 1996). Studies also provide evidence that Gα, at least in part, directs Gβγ subunits to the plasma membrane in vivo (Song et al., 1996; Takida and Wedegaertner, 2003). Although Gpa2 shares similar features with canonical Gα subunits, a striking contrast is the inability of Gpa2 to form a heterotrimeric G protein. The Gα subunit Gpa1 in the fission yeast Schizosaccharomyces pombe, which functions in pheromone-mediated signaling, also fails to form a heterotrimeric G protein with the known Gβγ subunits Git5/11. The kelch protein Ral2 has been proposed as a possible Gpa1-associated subunit based on genetic studies (Fukui et al., 1989; Harashima and Heitman, 2002; Hoffman, 2005).
Another contrast between canonical Gα subunits and Gpa2 is that Gβγ subunits typically promote receptor-Gα coupling, whereas Gpb1/2 inhibit receptor-Gpa2 coupling (Figure 9). The receptor Gpr1 and Gα Gpa2 can still in part function and signal in response to glucose without the Gβ mimic subunits Gpb1/2, indicating a promiscuous and specific coupling between Gpr1 and Gpa2 even in the absence of Gpb1/2 (Harashima and Heitman, 2002). In S. cerevisiae, the cAMP-PKA signaling pathway is essential for cell growth and determines cell fates in response to extracellular nutrients (Harashima and Heitman, 2004). Therefore the cAMP-PKA signaling pathway should be strictly controlled, and for this reason, Gpb1/2 may interfere with promiscuous Gpr1-Gpa2 coupling to facilitate responses to extracellular nutrients. On the other hand, in canonical G proteins, the Gβγ dimer may control Gα function by increasing the specificity of receptor coupling (Richardson and Robishaw, 1999; Hou et al., 2001; Lim et al., 2001; Kühn et al., 2002). Importantly, the kelch Gβ mimic subunits Gpb1/2 and canonical Gβγ dimer both regulate receptor-Gα coupling. Thus, the Gpa2/Gpb1/2 protein complex shares features with canonical heterotrimeric G proteins, and we propose Gpa2 is an ancestral subunit rather than an unusual Gα subunit. In this model, eukaryotic cells first acquired a GPCR and associated Gα subunit to sense and signal extracellular cues. Later, seven-bladed β-propeller-type subunits (kelch or WD40 based) were recruited to the GPCR-Gα signaling complex. Finally, farnesylated Gγ subunits were recruited to promote membrane localization. In this model, the atypical features of the nutrient and pheromone GPCR-Gα signaling modules in budding and fission yeasts might mirror features of their ancestral signaling modules from which they derive.
Alternatively, yeasts might uniquely have evolved an “alternative” Gα subunit and established a novel G protein signaling system to sense extracellular stimuli, in which an atypical Gα subunit forms a complex and functions with an unusual binding-partner kelch Gβ mimic protein. Further studies in both unicellular and multicellular organisms would distinguish these possibilities.
Acknowledgments
We thank Sayoko Ito-Harashima for providing a yeast strain and Cristl Arndt and Emily Wenink for assistance. We also thank Yong-Sun Bahn, Alex Idnurm, Julian Rutherford, Chaoyang Xue, Andy Alspaugh, Pat Casey, Henrik Dohlman, and Bob Lefkowitz for critical reading. This study was supported by the Department of Defense Neurofibromatosis program (W81xwh-04-01-0208). T.H. was supported by a fellowship from the Children's Tumor Foundation and J.H. is an investigator of the Howard Hughes Medical Institute.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–05–0403) on July 19, 2005.
References
- Adams, J., Kelso, R., and Cooley, L. (2000). The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10, 17–24. [DOI] [PubMed] [Google Scholar]
- Arévalo-Rodríguez, M., and Heitman, J. (2005). Cyclophilin A is localized to the nucleus and controls meiosis in Saccharomyces cerevisiae. Eukaryot. Cell 4, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi, K., Farazi, T. A., and Gordon, J. I. (1998). A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 273, 25864–25874. [DOI] [PubMed] [Google Scholar]
- Batlle, M., Lu, A. L., Green, D. A., Xue, Y., and Hirsch, J. P. (2003). Krh1p and Krh2p act downstream of the Gpa2p Gα subunit to negatively regulate haploid invasive growth. J. Cell Sci. 116, 701–710. [DOI] [PubMed] [Google Scholar]
- Blumer, K. J., and Thorner, J. (1990). β and γ subunits of a yeast guanine nucleotide-binding protein are not essential for membrane association of the α subunit but are required for receptor coupling. Proc. Natl. Acad. Sci. USA 87, 4363–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, H. R. (1997). How receptors talk to trimeric G proteins. Curr. Opin. Cell Biol. 9, 134–142. [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera, T. M., Vanhauwe, J., Thomas, T. O., Medkova, M., Preininger, A., Mazzoni, M. R., and Hamm, H. E. (2003). Insights into G protein structure, function, and regulation. Endocr. Rev. 24, 765–781. [DOI] [PubMed] [Google Scholar]
- Chen, C. A., and Manning, D. R. (2001). Regulation of G proteins by covalent modification. Oncogene 20, 1643–1652. [DOI] [PubMed] [Google Scholar]
- Colombo, S. et al. (1998). Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17, 3326–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries, L., Fischer, T., Tronchère, H., Brothers, G. M., Strockbine, B., Siderovski, D. P., and Farquhar, M. G. (2000). Activator of G protein signaling 3 is a guanine dissociation inhibitor for Gαi subunits. Proc. Natl. Acad. Sci. USA 97, 14364–14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries, L., and Gist Farquhar, M. (1999). RGS proteins: more than just GAPs for heterotrimeric G proteins. Trends Cell Biol. 9, 138–144.10203790 [Google Scholar]
- Degtyarev, M. Y., Spiegel, A. M., and Jones, T. L. (1994). Palmitoylation of a G protein αi subunit requires membrane localization not myristoylation. J. Biol. Chem. 269, 30898–30903. [PubMed] [Google Scholar]
- Dohlman, H. G. (2002). G proteins and pheromone signaling. Annu. Rev. Physiol. 64, 129–152. [DOI] [PubMed] [Google Scholar]
- Dohlman, H. G., and Thorner, J. W. (2001). Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70, 703–754. [DOI] [PubMed] [Google Scholar]
- Dosil, M., Schandel, K. A., Gupta, E., Jenness, D. D., and Konopka, J. B. (2000). The C terminus of the Saccharomyces cerevisiae α-factor receptor contributes to the formation of preactivation complexes with its cognate G protein. Mol. Cell. Biol. 20, 5321–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanko, D. S., Thiyagarajan, M. M., and Wedegaertner, P. B. (2000). Interaction with Gβγ is required for membrane targeting and palmitoylation of Gαs and Gαq. J. Biol. Chem. 275, 1327–1336. [DOI] [PubMed] [Google Scholar]
- Farazi, T. A., Waksman, G., and Gordon, J. I. (2001). The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276, 39501–39504. [DOI] [PubMed] [Google Scholar]
- Fishburn, C. S., Pollitt, S. K., and Bourne, H. R. (2000). Localization of a peripheral membrane protein: Gβγ targets GαZ. Proc. Natl. Acad. Sci. USA 97, 1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui, Y., Miyake, S., Satoh, M., and Yamamoto, M. (1989). Characterization of the Schizosaccharomyces pombe ral2 gene implicated in activation of the ras1 gene product. Mol. Cell. Biol. 9, 5617–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati, F., Volonté, D., Meani, D., Milligan, G., Lublin, D. M., Lisanti, M. P., and Parenti, M. (1999). The dually acylated NH2-terminal domain of Gi1α is sufficient to target a green fluorescent protein reporter to caveolin-enriched plasma membrane domains. Palmitoylation of caveolin-1 is required for the recognition of dually acylated G-protein α subunits in vivo. J. Biol. Chem. 274, 5843–5850. [DOI] [PubMed] [Google Scholar]
- Gallego, C., Gupta, S. K., Winitz, S., Eisfelder, B. J., and Johnson, G. L. (1992). Myristoylation of the Gαi2 polypeptide, a G protein α subunit, is required for its signaling and transformation functions. Proc. Natl. Acad. Sci. USA 89, 9695–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo, J. M. (2001). Control of pseudohyphae formation in Saccharomyces cerevisiae. Fems Microbiol. Rev. 25, 107–123. [DOI] [PubMed] [Google Scholar]
- Gautam, N., Downes, G. B., Yan, K., and Kisselev, O. (1998). The G-protein βγ complex. Cell Signal 10, 447–455. [DOI] [PubMed] [Google Scholar]
- Gillen, K. M., Pausch, M., and Dohlman, H. G. (1998). N-terminal domain of Gpa1 (G protein α subunit) is sufficient for plasma membrane targeting in yeast Saccharomyces cerevisiae. J. Cell Sci. 111, 3235–3244. [DOI] [PubMed] [Google Scholar]
- Gilman, A. G. (1987). G-Proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615–649. [DOI] [PubMed] [Google Scholar]
- Guan, K. L., and Han, M. (1999). A G-protein signaling network mediated by an RGS protein. Genes Dev. 13, 1763–1767. [DOI] [PubMed] [Google Scholar]
- Hamm, H. E., Deretic, D., Arendt, A., Hargrave, P. A., Koenig, B., and Hofmann, K. P. (1988). Site of G protein binding to rhodopsin mapped with synthetic peptides from the α subunit. Science 241, 832–835. [DOI] [PubMed] [Google Scholar]
- Harashima, T., and Heitman, J. (2002). The Gα protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol. Cell 10, 163–173. [DOI] [PubMed] [Google Scholar]
- Harashima, T., and Heitman, J. (2004). Nutrient control of dimorphic growth in Saccharomyces cerevisiae. In: Topics in Current Genetics, Vol. 7, ed. J Winderickx and P. M. Taylor, Heidelberg: Springer-Verlag, 131–169. [Google Scholar]
- Herrmann, R., Heck, M., Henklein, P., Henklein, P., Kleuss, C., Hofmann, K. P., and Ernst, O. P. (2004). Sequence of interactions in receptor-G protein coupling. J. Biol. Chem. 279, 24283–24290. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S. (2005). Except in every detail: comparing and contrasting G-protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryot. Cell 4, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., Chang, V., Capper, A. B., Taussig, R., and Gautam, N. (2001). G Protein β subunit types differentially interact with a muscarinic receptor but not adenylyl cyclase type II or phospholipase C-β2/3. J. Biol. Chem. 276, 19982–19988. [DOI] [PubMed] [Google Scholar]
- Ito, N., Phillips, S. E., Stevens, C., Ogel, Z. B., McPherson, M. J., Keen, J. N., Yadav, K. D., and Knowles, P. F. (1991). Novel thioether bond revealed by a 1.7 Å crystal structure of galactose oxidase. Nature 350, 87–90. [DOI] [PubMed] [Google Scholar]
- Ito, N., Phillips, S.E.V., Yadav, K.D.S., and Knowles, P. F. (1994). Crystal structure of a free radical enzyme, galactose oxidase. J. Mol. Biol. 238, 794–814. [DOI] [PubMed] [Google Scholar]
- Jeansonne, N. E. (1994). Yeast as a model system for mammalian seven-transmembrane segment receptors. Proc. Soc. Exp. Biol. Med. 206, 35–44. [DOI] [PubMed] [Google Scholar]
- Johnson, D. R., Bhatnagar, R. S., Knoll, L. J., and Gordon, J. I. (1994). Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem. 63, 869–914. [DOI] [PubMed] [Google Scholar]
- Jones, T. L., Simonds, W. F., Merendino, J. J., Jr., Brann, M. R., and Spiegel, A. M. (1990). Myristoylation of an inhibitory GTP-binding protein α subunit is essential for its membrane attachment. Proc. Natl. Acad. Sci. USA 87, 568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot, L., Pantaloni, C., Bockaert, J., and Audigier, Y. (1991). Deletion within the amino-terminal region of Gsα impairs its ability to interact with βγ subunits and to activate adenylate cyclase. J. Biol. Chem. 266, 9009–9015. [PubMed] [Google Scholar]
- Kostenis, E., Degtyarev, M. Y., Conklin, B. R., and Wess, J. (1997). The N-terminal extension of Gαq is critical for constraining the selectivity of receptor coupling. J. Biol. Chem. 272, 19107–19110. [DOI] [PubMed] [Google Scholar]
- Kraakman, L., Lemaire, K., Ma, P. S., Teunissen, A.W.R.H., Donaton, M.C.V., Van Dijck, P., Winderickx, J., de Winde, J. H., and Thevelein, J. M. (1999). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32, 1002–1012. [DOI] [PubMed] [Google Scholar]
- Kübler, E., Mösch, H. U., Rupp, S., and Lisanti, M. P. (1997). Gpa2p, a G-protein α-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272, 20321–20323. [DOI] [PubMed] [Google Scholar]
- Kühn, B., Christel, C., Wieland, T., Schultz, G., and Gudermann, T. (2002). G-protein βγ-subunits contribute to the coupling specificity of the β2-adrenergic receptor to Gs. Naunyn Schmiedebergs Arch. Pharmacol. 365, 231–241. [DOI] [PubMed] [Google Scholar]
- Lambright, D. G., Sondek, J., Bohm, A., Skiba, N. P., Hamm, H. E., and Sigler, P. B. (1996). The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379, 311–319. [DOI] [PubMed] [Google Scholar]
- Lefkowitz, R. J. (2000). The superfamily of heptahelical receptors. Nat. Cell Biol. 2, E133–E136. [DOI] [PubMed] [Google Scholar]
- Lemaire, K., Van de Velde, S., Van Dijck, P., and Thevelein, J. M. (2004). Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16, 293–299. [DOI] [PubMed] [Google Scholar]
- Lengeler, K. B., Davidson, R. C., D'Souza, C., Harashima, T., Shen, W. C., Wang, P., Pan, X. W., Waugh, M., and Heitman, J. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, W. K., Myung, C. S., Garrison, J. C., and Neubig, R. R. (2001). Receptor-G protein γ specificity: γ11 shows unique potency for A1 adenosine and 5-HT1A receptors. Biochemistry 40, 10532–10541. [DOI] [PubMed] [Google Scholar]
- Linder, M. E., Pang, I. H., Duronio, R. J., Gordon, J. I., Sternweis, P. C., and Gilman, A. G. (1991). Lipid modifications of G protein subunits. Myristoylation of Goα increases its affinity for βγ. J. Biol. Chem. 266, 4654–4659. [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie III, A., Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lorenz, M. C., and Heitman, J. (1997). Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 16, 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., Pan, X. W., Harashima, T., Cardenas, M. E., Xue, Y., Hirsch, J. P., and Heitman, J. (2000). The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts, P. (2004). Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 5, 263–278. [DOI] [PubMed] [Google Scholar]
- Morales, J., Fishburn, C. S., Wilson, P. T., and Bourne, H. R. (1998). Plasma membrane localization of Gαz requires two signals. Mol. Biol. Cell 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon, S. E., and Fung, B. K. (1987). Characterization of transducin from bovine retinal rod outer segments. Participation of the amino-terminal region of Tα in subunit interaction. J. Biol. Chem. 262, 15746–15751. [PubMed] [Google Scholar]
- Neubig, R. R. (1994). Membrane organization in G-protein mechanisms. FASEB J. 8, 939–946. [DOI] [PubMed] [Google Scholar]
- Pan, X., Harashima, T., and Heitman, J. (2000). Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3, 567–572. [DOI] [PubMed] [Google Scholar]
- Preininger, A. M., Van Eps, N., Yu, N. J., Medkova, M., Hubbell, W. L., and Hamm, H. E. (2003). The myristoylated amino terminus of Gαi1 plays a critical role in the structure and function of Gαi1 subunits in solution. Biochemistry 42, 7931–7941. [DOI] [PubMed] [Google Scholar]
- Richardson, M., and Robishaw, J. D. (1999). The α2A-adrenergic receptor discriminates between Gi heterotrimers of different βγ subunit composition in Sf9 insect cell membranes. J. Biol. Chem. 274, 13525–13533. [DOI] [PubMed] [Google Scholar]
- Rolland, F., de Winde, J. H., Lemaire, K., Boles, E., Thevelein, J. M., and Winderickx, J. (2000). Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38, 348–358. [DOI] [PubMed] [Google Scholar]
- Ross, E. M., and Wilkie, T. M. (2000). GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827. [DOI] [PubMed] [Google Scholar]
- Rost, B., and Sander, C. (1993). Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232, 584–599. [DOI] [PubMed] [Google Scholar]
- Samama, P., Cotecchia, S., Costa, T., and Lefkowitz, R. J. (1993). A mutation-induced activated state of the β2-adrenergic receptor. Extending the ternary complex model. J. Biol. Chem. 268, 4625–4636. [PubMed] [Google Scholar]
- Schwartz, M. A., and Madhani, H. D. (2004). Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38, 725–748. [DOI] [PubMed] [Google Scholar]
- Schwindinger, W. F., and Robishaw, J. D. (2001). Heterotrimeric G-protein βγ-dimers in growth and differentiation. Oncogene 20, 1653–1660. [DOI] [PubMed] [Google Scholar]
- Seifert, R., Wenzel-Seifert, K., and Kobilka, B. K. (1999). GPCR-Gα fusion proteins: molecular analysis of receptor-G-protein coupling. Trends Pharmacol. Sci. 20, 383–389. [DOI] [PubMed] [Google Scholar]
- Shea, L., and Linderman, J. J. (1997). Mechanistic model of G-protein signal transduction. Determinants of efficacy and effect of precoupled receptors. Biochem. Pharmacol. 53, 519–530. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Slessareva, J. E., Ma, H., Depree, K. M., Flood, L. A., Bae, H., Cabrera-Vera, T. M., Hamm, H. E., and Graber, S. G. (2003). Closely related G-protein-coupled receptors use multiple and distinct domains on G-protein α-subunits for selective coupling. J. Biol. Chem. 278, 50530–50536. [DOI] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D. G., Hamm, H. E., and Sigler, P. B. (1996). Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature 379, 369–374. [DOI] [PubMed] [Google Scholar]
- Song, J., and Dohlman, H. G. (1996). Partial constitutive activation of pheromone responses by a palmitoylation-site mutant of a G protein α subunit in yeast. Biochemistry 35, 14806–14817. [DOI] [PubMed] [Google Scholar]
- Song, J., Hirschman, J., Gunn, K., and Dohlman, H. G. (1996). Regulation of membrane and subunit interactions by N-myristoylation of a G protein α subunit in yeast. J. Biol. Chem. 271, 20273–20283. [DOI] [PubMed] [Google Scholar]
- Sprang, S. R. (1997). G protein mechanisms: Insights from structural analysis. Annu. Rev. Biochem. 66, 639–678. [DOI] [PubMed] [Google Scholar]
- Stefan, C. J., Overton, M. C., and Blumer, K. J. (1998). Mechanisms governing the activation and trafficking of yeast G protein-coupled receptors. Mol. Biol. Cell 9, 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader, C. D., Fong, T. M., Tota, M. R., Underwood, D., and Dixon, R. A. (1994). Structure and function of G protein-coupled receptors. Annu. Rev. Biochem. 63, 101–132. [DOI] [PubMed] [Google Scholar]
- Takida, S., and Wedegaertner, P. B. (2003). Heterotrimer formation, together with isoprenylation, is required for plasma membrane targeting of Gβγ. J. Biol. Chem. 278, 17284–17290. [DOI] [PubMed] [Google Scholar]
- Tamaki, H., Miwa, T., Shinozaki, M., Saito, M., Yun, C. W., Yamamoto, K., and Kumagai, H. (2000). GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 267, 164–168. [DOI] [PubMed] [Google Scholar]
- Thiyagarajan, M. M., Bigras, E., Van Tol, H. H., Hébert, T. E., Evanko, D. S., and Wedegaertner, P. B. (2002). Activation-induced subcellular redistribution of Gαs is dependent upon its unique N-terminus. Biochemistry 41, 9470–9484. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, M. A., Coleman, D. E., Lee, E., Iñiguez-Lluhi, J. A., Posner, B. A., Gilman, A. G., and Sprang, S. R. (1995). The structure of the G protein heterotrimer Giα1β1γ2. Cell 83, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Wall, M. A., Posner, B. A., and Sprang, S. R. (1998). Structural basis of activity and subunit recognition in G protein heterotrimers. Structure 6, 1169–1183. [DOI] [PubMed] [Google Scholar]
- Wedegaertner, P. B., Bourne, H. R., and von Zastrow, M. (1996). Activation-induced subcellular redistribution of Gsα. Mol. Biol. Cell 7, 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedegaertner, P. B., Chu, D. H., Wilson, P. T., Levis, M. J., and Bourne, H. R. (1993). Palmitoylation is required for signaling functions and membrane attachment of Gqα and Gsα. J. Biol. Chem. 268, 25001–25008. [PubMed] [Google Scholar]
- Wilson, P. T., and Bourne, H. R. (1995). Fatty acylation of az. Effects of palmitoylation and myristoylation on αz signaling. J. Biol. Chem. 270, 9667–9675. [DOI] [PubMed] [Google Scholar]
- Wise, A., Grassie, M. A., Parenti, M., Lee, M., Rees, S., and Milligan, G. (1997). A cysteine-3 to serine mutation of the G-protein Gi1α abrogates functional activation by the α2A-adrenoceptor but not interactions with the βγ complex. Biochemistry 36, 10620–10629. [DOI] [PubMed] [Google Scholar]
- Xue, Y., Batlle, M., and Hirsch, J. P. (1998). GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17, 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y., Katoh, H., and Negishi, M. (2003). N-terminal short sequences of α subunits of the G12 family determine selective coupling to receptors. J. Biol. Chem. 278, 14936–14939. [DOI] [PubMed] [Google Scholar]
- Yun, C. W., Tamaki, H., Nakayama, R., Yamamoto, K., and Kumagai, H. (1997). G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 240, 287–292. [DOI] [PubMed] [Google Scholar]
- Yun, C. W., Tamaki, H., Nakayama, R., Yamamoto, K., and Kumagai, H. (1998). Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252, 29–33. [DOI] [PubMed] [Google Scholar]