Abstract
Here, we investigate how Candida albicans, the most prevalent human fungal pathogen, protects itself from nitric oxide (.NO), an antimicrobial compound produced by the innate immune system. We show that exposure of C. albicans to .NO elicits a reproducible and specific transcriptional response as determined by genome-wide microarray analysis. Many genes are transiently induced or repressed by .NO, whereas a set of nine genes remain at elevated levels during .NO exposure. The most highly induced gene in this latter category is YHB1, a flavohemoglobin that detoxifies .NO in C. albicans and other microbes. We show that C. albicans strains deleted for YHB1 have two phenotypes in vitro; they are hypersensitive to .NO and they are hyperfilamentous. In a mouse model of disseminated candidiasis, a YHB1 deleted C. albicans strain shows moderately attenuated virulence, but the virulence defect is not suppressed by deletion of the host NOS2 gene. These results suggest that .NO production is not a prime determinant of virulence in the mouse tail vein model of candidiasis and that the attenuated virulence of a yhb1Δ/yhb1Δ strain is attributable to a defect other than its reduced ability to detoxify .NO.
INTRODUCTION
Cells of the innate immune system employ several highly effective mechanisms to defend against microbial pathogens. One of the most intriguing is the production of nitric oxide (.NO). At low concentrations, .NO is used as a signaling molecule in both animals and plants to control a diverse set of physiological processes; for example, in mammals it regulates both smooth muscle vasodilation and signaling between nerve cells (Nathan, 1992). At much higher concentrations, .NO is toxic to cells and is used as a defense mechanism against invading pathogens.
.NO is produced from arginine by the enzyme nitric oxide synthase (NOS). In mammals, there are three distinct isoforms of this enzyme, which differ both in their function and expression, and are encoded by separate genes, NOS1, NOS2, and NOS3. The NOS2 gene (often referred to as iNOS) is the only inducible isoform and is predominately expressed in cells of the innate immune system and epithelia upon exposure to microorganisms, resulting in production of toxic levels of .NO (for reviews see Fang, 1997; Nathan, 1997; Nathan and Shiloh, 2000; Chakravortty and Hensel, 2003; Fang, 2004). For example, induction of macrophage NOS2 by the cytokine IFN-γ or microbial components such as lipopolysaccharide (LPS) leads to .NO production (Xie et al., 1992; Chinen et al., 1999).
.NO and its immediate derivatives, termed reactive nitrogen intermediates (RNI), react with many different cellular components, and it is widely recognized that DNA and many classes of proteins and lipids are damaged by exposure to .NO or other RNI species (see reviews cited above). .NO is also known to regulate the catalytic activity of various enzymes primarily by interaction with Fe-S clusters, oxidized copper (Cu2+) centers, hemes, and tyrosyl radicals. For example, .NO reversibly binds to the Cu2+ of cytochrome c oxidase (Cox) thereby shutting down mitochondrial oxidative phosphorylation and cellular respiration (Burney et al., 1999; Carreras et al., 2004). Although high levels of .NO can also damage host cells, its protective value apparently outweighs this incidental damage (Nathan and Shiloh, 2000).
Pathogens have evolved many strategies for combating the adverse affects of .NO; they can detoxify .NO, they can repair .NO-induced damage, and they can modulate host .NO production (see reviews cited above). Prominent among the detoxification enzymes are the flavohemoglobins, an ancient protein family that predates the divergence of bacteria and eukaryotes. Under aerobic conditions, these two-domain proteins can detoxify .NO in a rapid reaction of heme-bound .NO with oxygen to form innocuous nitrate and ferric flavohemoglobin. The enzyme is subsequently reduced back to its ferrous form via electron transfer between NAD(P)H and its flavin-containing FAD oxidoreductase domain. Flavohemoglobins (and closely related proteins) have been suggested, and in many cases definitively shown, to confer resistance to .NO in a wide variety of microbes including Escherichia coli (Vasudevan et al., 1991; Gardner et al., 1998; Hausladen et al., 1998), Salmonella typhimurium (Crawford and Goldberg, 1998), Mycobacterium tuberculosis (Hu et al., 1999; Pathania et al., 2002), Saccharomyces cerevisiae (Liu et al., 2000), Cryptococcus neoformans (de Jesus-Berrios et al., 2003), and C. albicans (Ullmann et al., 2004; for review see Poole and Hughes, 2000).
In this article, we examine the transcriptional response of C. albicans, the major fungal pathogen of humans, to .NO. C. albicans causes mucosal infections in healthy individuals and disseminated infections in immunocompromised patients and is capable of colonizing most tissues in the human body. The available evidence suggests that .NO is likely important in controlling C. albicans infections; it has been implicated in the candidacidal activity of both macrophages and saliva, as determined in vitro (Elahi et al., 2001; Netea et al., 2002). Recently, it was reported that a C. albicans strain deleted for both copies of a putative flavohemoglobin gene (YHB1) is hypersensitive to .NO killing and reduced for virulence in the tail vein model of disseminated candidiasis (Ullmann et al., 2004).
Here, we describe the genome-wide response of C. albicans to .NO and provide an independent characterization of a strain deleted for YHB1, the most highly .NO-induced gene. We show that YHB1 is not required for the transcriptional response of C. albicans to .NO; indeed, the transcriptional response is accentuated in a yhb1Δ/yhb1Δ strain. Finally, in a mouse tail vein model of systemic infection, we show that the virulence defect of the yhb1Δ/yhb1Δ mutant is likely due to a phenotype other than increased sensitivity to .NO.
MATERIALS AND METHODS
Strains and Media
C. albicans strains used in this study are listed in Table 1 and described in greater detail below. CAF2-1, RM1, and RM1000 have been described previously (Fonzi and Irwin, 1993; Negredo et al., 1997). MMY272, BH79, BH94, BH96, BH98, BH113, and BH117 were all derived from RM1000.
Table 1.
C. albicans strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| CAF2-1 | ura3Δ::imm434/URA3, iro1Δ::imm434/IRO1 | Fonzi and Irwin (1993) |
| RM1 | ura3Δ::imm434/URA3, his1Δ::HisG/HIS1, iro1Δ::imm434/IRO1 | Negredo et al. (1997) |
| RM1000 | ura3Δ::imm434/ura3Δ::imm434, his1Δ::HisG/his1Δ::HisG, iro1Δ::imm434/iro1Δ::imm434 | Negredo et al. (1997) |
| BH79 | ura3::URA3/ura3Δ::imm434, his1Δ::HisG/his1Δ::HisG, iro1::IRO1/iro1Δ:: imm434, yhb1Δ::HisG/yhb1Δ::HIS1 | This study |
| BH94 | ura3::URA3/ura3Δ:: imm434, his1Δ::HisG/his1Δ::HisG, iro1::IRO1/iro1Δ:: imm434, yhb1::YHB1/yhb1Δ::HIS1 | This study |
| BH97 | ura3::URA3/ura3Δ:: imm434, his1Δ::HisG/his1Δ::HisG, iro1::IRO1/iro1Δ:: imm434, yhb1::YHB1/yhb1Δ::HIS1 | This study |
| BH98 | ura3::URA3/ura3Δ:: imm434, his1Δ::HisG/his1Δ::HisG, iro1::IRO1/iro1Δ:: imm434, yhb1::YHB1/yhb1Δ::HIS1 | This study |
| BH115 | ura3::URA3/ura3Δ:: imm434, his1::HIS1/his1Δ::HisG, iro1::IRO1/iro1Δ:: imm434, yhb1::HisG/YHB1 | This study |
| BH117 | ura3::URA3/ura3Δ:: imm434, his1::HIS1/his1Δ::HisG, iro1::IRO1/iro1Δ::imm434 | This study |
| MMY272 | ura3Δ::imm434/ura3Δ:: imm434, his1Δ::HisG/his1Δ::HisG, iro1Δ::imm434/iro1Δ:: imm434, yhb1::URA3/yhb1::HIS1 | This study |
Cultures were grown at 30°C in YEPD unless otherwise noted, and C. albicans transformations were performed according to the standard lithium acetate method (Gietz et al., 1995). Transformants were selected on Sabouraud dextrose (SD) medium lacking uracil or histidine (Ura- or His-), depending on the marker used, and correct integration of disruption fragments was confirmed by PCR. For strains manipulated at the YHB1 locus, Southern blot analyses were performed with the Nonradioactive Labeling and Detection kit (Boehringer Mannheim, Indianapolis, IN; Supplementary Figure 1A) and pulsed-field gel electrophoresis was used to verify that none of the newly constructed strains had gross chromosomal abnormalities (Supplementary Figure 1B). The C. albicans URA3 gene was recycled by plating on 5-fluoroorotic acid-containing medium (5-FOA medium) as previously described (Boeke et al., 1984; Alani et al., 1987).
For experiments using DPTA NONOate (see below), C. albicans strains were grown in YEPD broth buffered with 80 mM HEPES, pH 7.5.
Nitric Oxide-releasing and -scavenging Chemicals
Dipropylenetriamine NONOate (DPTA NONOate; Cayman Chemicals, Ann Arbor, MI), which releases two molecules of .NO per amine molecule in a pH-dependent manner (pH 7.0-7.4), was stored at -80°C and resuspended to 0.75 M in 10.0 mM NaOH immediately before use. To activate the DPTA NONOate, the inactive alkaline DPTA NONOate solution was added to buffered YEPD (see above for buffer details) at a final concentration of 1.0 mM DPTA NONOate and ∼13.3 nM NaOH unless otherwise noted. For the .NO scavenging experiment, Carboxy PTIO potassium salt (Carboxy PTIO; Cayman Chemicals), a molecule that scavenges nitric oxide, was solubilized in 1× phosphate-buffered saline (PBS; pH 7.2) and used at a final concentration of 16.0 mM.
Construction of yhb1Δ/yhb1Δ Mutant
All primer sequences are listed in Supplementary Table 3. The C. albicans YHB1 gene was disrupted by a PCR method (Wilson et al., 1999, 2000) in RM1000 (Negredo et al., 1997), which is auxotrophic for uridine and histidine. Primers 4 and 5 (see Supplementary Table 3 for primer sequences) were used in separate PCR reactions with the templates pGEM-HIS1 (Wilson et al., 1999) and pDDB57 (Wilson et al., 2000) to generate the PCR disruption products. Whole-cell PCR, using primers internal to HIS1 (primers 6 and 7) and URA3 (primers 8 and 9), and outside the flanking region of homology to the YHB1 locus (primers 10 and 11), was used to identify a yhb1Δ::URA3/yhb1Δ::HIS1 isolate (MMY272). The absence of a product using primers internal to the open reading frame (primers 12 and 13) confirmed that the YHB1 sequence had not relocated to another position in the genome. Ura+ yhb1Δ/yhb1Δ strains were plated on 5-FOA-containing medium to select for loss of the URA3 gene. To restore a single copy of URA3 and IRO1 to their native loci, Ura- strains were transformed with pLUBP (Fonzi and Irwin, 1993), linearized at PstI and BglII which released a 4.9-kb restriction fragment containing the URA3 and IRO1 genes, and grown on Ura- selective medium. Whole-cell PCR, using primers internal to the URA3-IRO1 sequence (primer 15) and outside the flanking region of homology to the URA3-IRO1 sequence (primer 14) were used to confirm that the URA3-IRO1 open reading frames had reintegrated at their endogenous loci. Southern blot analysis (Supplementary Figure 1A) of BH79 (yhb1Δ::HisG/yhb1Δ::HIS1) with a probe to the YHB1 promoter confirmed the absence of both YHB1 open reading frames, integration of HIS1 at one YHB1 allele, and the excision recombination of URA3 at the other YHB1 allele (see Table 1 for complete strain genotype).
Reintroduction of Wild-type YHB1 to yhb1Δ/yhb1Δ Mutants
The 1194-base pair YHB1 open reading frame, 500 base pairs of upstream promoter sequence, and 300 base pairs of 3′ untranslated region (UTR) were amplified by PCR (primers 16 and 17) with Ex Taq polymerase (TaKaRa), yielding a 2060-base pair sequence containing engineered BamHI sites at both ends. The PCR fragment was gel purified on a 0.8% agarose gel before purification with QIAquick gel extraction kit (Qiagen, Chatsworth, CA), digested with the BamHI enzyme, and ligated into BamHI-linearized pBB510 (Braun and Johnson, 2000), a derivative of pMB7 that contains the HisG-URA3-HisG cassette (Fonzi and Irwin, 1993). The resulting plasmid was named pBH2, and DNA sequencing of the insert confirmed identity to the YHB1 gene reported in the C. albicans Diploid Assembly 19 database of the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida/). The intermediate yhb1Δ/yhb1Δ strain (Ura-, Iro1-, His+) described earlier was transformed with pBH2, which had been linearized at SacII and phosphatase treated, and selected for on Ura- medium. Correct integration of YHB1-URA3 was confirmed by whole-cell PCR using primers 5′ to the integrated sequence (primer 18) and internal to the YHB1 open reading frame (primer 19). Integration at the 3′ end of insertion was confirmed by using primers internal to the inserted plasmid sequence (primer 20) and 3′ to the integrated sequence (primer 21). Correct integrants were plated on 5-FOA to select for loss of the URA3 gene, and the URA3-IRO1 open reading frames were restored to their endogenous loci and confirmed by PCR as described above. Three addback strains, BH94, BH96, and BH98, were obtained and their sensitivity to .NO was assessed by exposing them to DPTA NONOate as described below. Southern blot analysis (Supplementary Figure 1A) with a probe to the YHB1 promoter confirmed reconstitution of YHB1 at one allele, and the presence of HIS1 at the other YHB1 allele.
Construction of Wild-type URA3 Isogenic Strain
As noted earlier in this report, the yhb1Δ/yhb1Δ null, yhb1Δ/YHB1 heterozygous, and yhb1Δ/yhb1::YHB1 addback strains were derived from RM1000. Because virulence differences can result from differential expression of URA3 at heterologous loci (Kirsch and Whitney, 1991; Sundstrom et al., 2002; Cheng et al., 2003; Brand et al., 2004), we created our wild-type comparator (BH117) from RM1000, adding back both URA3 and HIS1 to their endogenous loci. The URA3-IRO1 fragment was restored and confirmed by PCR as described above. To restore HIS1, the resulting Ura+ strain was transformed with pGEMHIS (Wilson et al., 1999), linearized at Nru1. Whole-cell PCR, using primers internal to the HIS1 sequence (primers 23 and 24) and outside the flanking region of homology to the HIS1 sequence (primers 22 and 25), was used to confirm that the HIS1 open reading frame had reintegrated at its endogenous locus.
Construction of C. albicans Microarrays
The C. albicans microarrays used in this article have been previously described (Bennett et al., 2003) and include two YHB1 cDNA features of 1039 and 356 base pairs in length (the entire YHB1 ORF is 1194 base pairs in length). Because YHB1 is part of a gene family including YHB4 and YHB5 (YHB1 has 48 and 57% overall identity to YHB4 and YHB5, respectively), and cross-hybridization of large PCR-generated cDNA features could have obscured our data, we designed three 70-base pairs oligomers, each specific to one YHB gene. The YHB1 oligomer has 14.8 and 33.7% identity, respectively, to the YHB4 and YHB5 oligomers, and each oligomer is spotted in triplicate on our microarray. See Supplementary Table 3 for primer sequences (primers 1-3).
Culture Growth for Nitric Oxide Microarrays
Ten-milliliter cultures of CAF2-1, RM1, RM1000, and MMY272 (yhb1Δ/yhb1Δ) were grown overnight, and each was used to inoculate a 1.8-L culture to an OD (600 nm) of 0.15. The RM1, RM1000, and MMY272 cultures were supplemented with 100 mg/L uridine and 5 g/L histidine. When the OD (600 nm) of the cultures reached ∼1.0, three zero time-point samples were harvested for each strain. The remainder of each culture was then perturbed with (1) DPTA NONOate and NaOH or (2) NaOH alone (mock treated; see above for details on chemicals). Samples were collected by filtration after 10, 40, 70, and 120 min of perturbation and cell pellets were stored at -80°C.
Culture Growth for Wild-type versus yhb1Δ/yhb1Δ Mutant Microarrays
Ten-milliliter cultures of yhb1Δ/yhb1Δ (BH79) and its isogenic wild-type comparator (BH117) were grown overnight and used to inoculate two 150-ml cultures each, one for growth at 30°C and the other at 37°C. When the four cultures reached an OD (600 nm) of ∼1.0, they were harvested by filtration and pellets were stored at -80°C. The experiment was repeated three times at 30°C and twice at 37°C.
RNA Isolation, Microarray Hybridization, and Data Analysis
Total RNA, poly-A RNA, and cDNA were prepared as previously described (Bennett et al., 2003). cDNAs for individual time-points (coupled to Cy5) were hybridized against a reference pool comprising cDNAs from every time point in the series (coupled to Cy3). Microarray hybridizations were incubated for 18-24 h according to the methods described by Bennett et al. (2003). Arrays were scanned on a GenePix 4000 scanner (Axon Instruments, Foster City, CA), and fluorescence signals were assigned to individual features using GENEPIX PRO version 3.0. Data were further processed with NOMAD (available at http://derisilab5.ucsf.edu/NOMAD/nomad-cgi/login.pl). For the nitric oxide time-course experiment, individual microarray fluorescence signals were transformed over the median fluorescence signal of three untreated zero time-point microarrays, and for the wild-type versus yhb1Δ/yhb1Δ mutant experiment, individual yhb1Δ/yhb1Δ microarray fluorescence signals were transformed over wild-type fluorescence signals. The primary data are available on the Johnson laboratory website, http://itsa.ucsf.edu/%7Emicro/Faculty/Johnson/johnson_index.html. Pairwise average linkage clustering analysis was performed with the program CLUSTER (available at http://rana.stanford.edu/software) as previously described (Eisen et al., 1998), and data sets were visualized with the program TREEVIEW (available at http://rana.stanford.edu/software).
Quantitative RT-PCR of YHB1, YHB4, YHB5, and PAT1
A 5-ml culture of CAF2-1 was grown overnight at 30°C, diluted to an OD (600 nm) of 0.15, and upon reaching an OD (600 nm) of ∼1.0, a zero time-point sample was harvested by centrifugation at room temperature (RT) and frozen at -80°C. The remaining culture was split into four 10-ml cultures and treated with 1) DPTA NONOate and Carboxy PTIO (scavenger), 2) DPTA NONOate alone, 3) Carboxy PTIO (scavenger) alone, or 4) mock-treated with NaOH and PBS (see above for details on chemicals). After 10 and 120 min of perturbation, samples were collected by centrifugation. RNA was extracted as previously described (Miller and Johnson, 2002), linearly reverse-transcribed (Superscript), and cDNA was amplified by quantitative PCR in a DNA Engine Opticon 2 (Bio-Rad, Waltham, MA). Signals from each sample were transformed over the average of three untreated zero time-point samples, and the entire experiment was repeated three times and averaged (see Figure 2). Primer sequences for YHB1, YHB4, YHB5, and PAT1 (primers 26-33) are listed in Supplementary Table 3.
Figure 2.
Fold induction of C. albicans flavohemoglobin genes (YHB1, YHB4, YHB5) in response to nitric oxide (.NO). The PAT1 transcript was used as a control and reveals no induction by .NO. Samples were collected 10 (yellow) and 180 (blue) min after exposure to 1.0 mM DPTA NONOate, 8.0 mM Carboxy PTIO (Scavenger), or a combination of both chemicals. Addition of a chemical is indicated by a (+) and no-addition indicated by a (-). Three independent quantitative RT-PCR reactions were performed, and in each experiment, the signals were divided by an untreated zero time-point sample. The plot and error bars represent the average of the three experiments.
In Vitro Growth Inhibition Assay
Five-milliliter cultures of BH79, BH94, BH96, BH98, BH115, and BH117 were grown overnight as described above and used to inoculate a 10-ml culture of each respective strain to an OD (600 nm) of 0.15. Cells were allowed to recover for 1 h and then divided into five 1-ml cultures. DPTA NONOate was added to final concentrations of 0.0, 0.5, 1.0, 2.0, and 3.5 mM, respectively. Cultures were grown for 8 h and serially diluted and plated, and after 24 h of growth at 30°C, colony forming units (CFUs) were counted.
Virulence Tests Using the Mouse Tail Vein Injection Model
Virulence assays were performed in 10-wk-old female BALB/c immunocompetent mice (Charles River Laboratories, Wilmington, MA) and in 9-wk-old male C57BL/6 NOS2+/+ (Jackson ImmunoResearch Laboratories, West Grove, PA) or NOS2-/- mice (B6.129P2-Nos, Jackson ImmunoResearch Laboratories). Ten or 11 mice were tested per strain. Ten-milliliter cultures of BH117, BH98, and BH79 were grown overnight as described earlier and diluted 30-fold into fresh medium. Cultures were allowed to recover for 4-5 h until they reached an OD (600 nm) of ∼3.0. They were washed twice with 10-ml of sterile normal saline, and an aliquot was taken for quantification using a hemacytometer. To confirm the cell number, serial dilutions of the inocula were plated onto YEPD at 30°C, and CFUs were counted. In experiment one (see Figure 6A), the tail veins of BALB/c mice were injected with 3 × 105 CFUs in a total volume of 100-200-μl. In experiment two (Figure 6B), C57BL/6 NOS2 +/+ or NOS2-/- mice were injected with 5 × 105 CFUs in a total volume of 300-400 μl. The mice were fed ad libitum, evaluated twice daily, and sacrificed when moribund as described by Noble and Johnson (2005). Data were analyzed by the Log Rank test (NCSS software) and p ≤ 0.05 was considered significant.
Figure 6.
YHB1 is important for virulence in a mouse tail vein model of systemic candidiasis. (A) Groups of 10 immunocompetent BALB/c mice were tail vein injected with 3 × 105 cells of wild-type (BH117, yellow squares), yhb1Δ/yhb1Δ mutant (BH79, pink squares), or yhb1Δ/yhb1::YHB1 addback (BH98, blue squares) strains, and survival was monitored. (B) Groups of 10 or 11 congenic C57BL/6 NOS2+/+ or NOS2-/- mice were tail vein injected with 5 × 105 cells of wild-type (BH117, yellow squares and green triangles) or yhb1Δ/yhb1Δ mutant (BH79, pink squares and gray triangles) strains, and survival was monitored.
RESULTS
Genome-wide Expression Profile of C. albicans in Response to Nitric Oxide
To identify C. albicans' transcriptional response to nitric oxide (.NO), three different laboratory strains were treated with DPTA NONOate (Caymen Chemicals, Ann Arbor, MI), a chemical that releases .NO in a pH-dependent manner, and the transcriptional profile was monitored over a time course using C. albicans genomic DNA microarrays. The microarrays for this analysis contain 11,325 spots representing ∼6550 protein-encoding nuclear genes, or 95% of the estimated number of C. albicans nuclear genes (Bennett et al., 2003; Jones et al., 2004). Many C. albicans genes are represented by more than one microarray spot, thereby allowing independent evaluation of gene expression in a single experiment. Time courses of .NO response were performed with CAF2-1, RM1, and RM1000 (see Table 1 for strain descriptions, and Materials and Methods for details on media supplementation of nutritionally auxotrophic strains). In all time courses, log-phase cells (OD 600 nm = 1.0) were treated with DPTA NONOate (1.0 mM), and DPTA NONOate-treated and untreated samples were collected 10, 40, 70, and 120 min after treatment. Changes in gene expression were normalized to an untreated zero-time-point sample.
As shown in Figure 1, the expression levels of ∼131 C. albicans genes rapidly changed upon exposure to DPTA NONOate and, based on their response, these genes fall into three categories: 1) those transiently induced by .NO (Figure 1A, blue vertical bar), 2) those whose induction persisted throughout the time course (Figure 1A, yellow vertical bar), and 3) those repressed by .NO (Figure 1B, orange vertical bar). A fourth group of genes was induced in a yhb1Δ/yhb1Δ mutant treated with .NO (Figure 1A, gray vertical bar) and will be discussed later. The 131 .NO induced or repressed genes are graphically displayed in Figure 1 and are listed in Table 2 and Supplementary Table 1.
Figure 1.
Genome-wide expression profile of C. albicans in response to nitric oxide (.NO). Four independent time courses were analyzed (EXPT 1-4) in CAF2-1, RM1, RM1000, and in a yhb1Δ/yhb1Δ mutant strain (MMY272; see Table 1 for strain details). Samples were collected 10, 40, 70, and 120 min (triangles represent increasing time) after exposure to 1.0 mM DPTA NONOate (treated) or 13.3 nM NaOH (mock-treated), and all data were transformed over an untreated zero time-point sample (black column at the beginning of each experiment). Red and green squares indicate induction and repression, respectively, black indicates no change in expression, and gray indicates no data available. (A) Sixty-five genes are up-regulated in response to .NO. Nine genes (yellow vertical bars) are persistently up-regulated throughout the 2-h time course in all strain backgrounds (EXPT 1-4). Fifty-six genes (blue vertical bar) are transiently up-regulated in YHB1 intact strains (EXPT 1-3) and show prolonged induction in a yhb1Δ/yhb1Δ mutant strain (EXPT 4). An additional 34 genes (gray vertical bar) are only up-regulated in a yhb1Δ/yhb1Δ mutant strain (EXPT 4). (B) Sixty-five genes are down-regulated in response to .NO (orange vertical bar). In YHB1 intact strains, these genes are transiently down-regulated at the 10-min time point and return to baseline expression levels by the 40-min time point (EXPT 1-3). In a yhb1Δ/yhb1Δ mutant strain, the majority of these genes are persistently down-regulated throughout the 2-h time course (EXPT 4).
Table 2.
Nitric oxide-induced genes (two-fold or greater in at least one column)
| A. Transcripts persistently induced throughout time course in wild-type and yhb1Δ/yhb1Δ strainsa
| |||||||
|---|---|---|---|---|---|---|---|
| C. albicans ORF19 No.b | C. albicans namec | S. cerevisiae homologd | Description/Functione | 10 min WTf,g | 120 min WTf,g | 10 min yhb1Δ/Δf | |
| orf19.3707 | YHB1 | YHB1 | Flavohemoglobin/nitric oxide dioxygenase | 15.8 | 4.1 | 1.4 | |
| orf19.4773 | AOX2 | AOX1 | Alternative oxidase | 11.6 | 4.0 | 6.1 | |
| orf19.7313 | SSU1 | Sulfite transporter | 7.3 | 3.9 | 9.5 | ||
| orf19.3120 | YOL075C | Ferric cation import ABC transporter | 6.7 | 5.4 | 6.4 | ||
| orf19.4816 | YMR209C | Unknown function | 5.1 | 1.7 | 3.1 | ||
| orf19.4720 | CTR2 | Copper transporter | 3.8 | 2.0 | 2.3 | ||
| orf19.5636 | RBT5 | GPI-linked heme acquisition protein | 3.1 | 4.7 | 2.6 | ||
| orf19.4774 | AOX1 | AOX1 | Alternative oxidase | 2.6 | 1.8 | 2.3 | |
| orf19.3117 | GPI-linked CFEM domain protein | 2.3 | 2.1 | 2.4 | |||
| B. Transcripts induced early in time course, prolonged induction in yhb1Δ/yhb1Δ strain onlyh
| |||||||
|---|---|---|---|---|---|---|---|
| C. albicans ORF19 No.b | C. albicans namec | S. cerevisiae homologd | Description/Functione | 10 min WTf,g | 120 min WTf,g | 10 min yhb1Δ/Δf | |
| orf19.125 | EBP1 | OYE2 | NADPH oxidoreductase | 25.5 | 0.8 | 18.0 | |
| orf19.3132 | MSC2 | Zinc transporter | 18.1 | 1.2 | 64.4 | ||
| orf19.2693 | URE2 | Glutathione s-transferase | 16.7 | 1.2 | 10.8 | ||
| orf19.1149 | ETR1 | Mitochondrial 2-enoyl thioester reductase | 12.5 | 1.6 | 14.4 | ||
| orf19.4290 | TRR1 | NADPH thioredoxin-disulfide reductase | 11.4 | 1.1 | 8.7 | ||
| orf19.2262 | ZTA1 | NADPH quinone oxidoreductase | 10.7 | 1.4 | 4.2 | ||
| orf19.3433 | OYE2 | NADPH oxidoreductase | 10.4 | 0.9 | 3.6 | ||
| orf19.6398 | JLP1 | Iron dependent sulphonate dioxygenase | 10.2 | 2.8 | 1.0 | ||
| orf19.3443 | OYE2 | NADPH oxidoreductase | 9.9 | 1.2 | 9.9 | ||
| orf19.3131 | OYE3 | NADPH oxidoreductase | 8.0 | 0.7 | 6.5 | ||
| orf19.2601 | HEM1 | HEM1 | 5-Aminolevulinate synthase | 6.8 | 1.7 | 10.1 | |
| orf19.5059 | GSH1 | GSH1 | Glutamate-cysteine ligase | 6.8 | 2.0 | 5.2 | |
| orf19.7374 | YAF1 | Zinc finger transcription factor | 6.2 | 1.5 | 4.3 | ||
| orf19.7042 | No good BLAST homology | 6.0 | 1.3 | 3.0 | |||
| orf19.6229 | CAT1 | CTA1 | Catalase | 6.0 | 1.6 | 6.2 | |
| orf19.3395 | YHR048W | Drug transporter | 5.7 | 1.0 | 1.4 | ||
| orf19.3122 | ARR3 | ARR3 | Arsenite transporter | 5.6 | 1.4 | 4.4 | |
| orf19.5785 | No good BLAST homology | 5.2 | 0.9 | 3.5 | |||
| orf19.2356 | CRZ1 | Zinc finger transcription factor | 5.0 | 1.6 | 1.7 | ||
| orf19.5770 | YGL114W | Oligopeptide transporter |
|
4.9 | 1.2 | 4.4 | |
| orf19.5635 | PGA7 | Putative GPI-anchored protein | 4.6 | 2.5 | 1.9 | ||
| orf19.4370 | No good BLAST homology | 4.5 | 1.1 | 2.9 | |||
| orf19.5517 | ADH7 | NADPH alcohol dehydrogenase | 4.2 | 1.2 | 6.1 | ||
| orf19.7091 | No good BLAST homology | 4.0 | 1.0 | 3.4 | |||
| orf19.4147 | GLR1 | Glutathione reductase | 3.9 | 1.4 | 4.2 | ||
| orf19.6586 | YJR115W | Conserved ORF/Function unknown | 3.6 | 1.6 | 1.2 | ||
| orf19.7417 | TSA1 | TSA1 | Thiol peroxidase | 3.6 | 1.1 | 3.7 | |
| orf19.5674 | PGA10 | CSA1 | Heme utilization protein | 3.2 | 1.9 | 1.2 | |
| orf19.6478 | YCF1 | YCF1 | Glutathione s-conjugate transporter | 3.1 | 1.3 | 2.0 | |
| orf19.113 | Cadmium-induced CIP1 like proteini | 3.1 | 0.9 | 3.8 | |||
| orf19.711 | No good BLAST homology | 3.1 | 1.0 | 3.1 | |||
| orf19.1763 | YNL134C | NADPH alcohol dehydrogenase | 3.0 | 0.7 | 3.0 | ||
| orf19.4757 | NAR1 | Nuclear prelamin A recognition factor | 3.0 | 1.4 | 2.6 | ||
| orf19.4907 | YCR061W | Conserved ORF/Function unknown | 2.8 | 1.0 | 3.8 | ||
| orf19.5258 | No good BLAST homology | 2.7 | 1.3 | 1.6 | |||
| orf19.7214 | YBR056W | Glucan 1,3-β-glucosidasej | 2.7 | 1.4 | 1.6 | ||
| orf19.5713 | NDE1 | NADH dehydrogenase | 2.7 | 1.0 | 3.5 | ||
| orf19.8434 | SSY1 | Amino acid binding protein | 2.7 | 1.3 | 2.7 | ||
| orf19.4802 | FTH1 | FTH1 | Iron transporter | 2.6 | 1.3 | 2.4 | |
| orf19.7316 | Conserved ORF/Function unknown | 2.5 | 0.9 | 3.6 | |||
| orf19.6928 | SAP9 | SAP9 | Secreted aspartyl proteinase | 2.5 | 1.4 | 3.6 | |
| orf19.3432 | YCR023C | Conserved ORF/Function unknown | 2.5 | 1.1 | 2.2 | ||
| orf19.3803 | MNN2 | Mannosyltransferase | 2.5 | 1.2 | 1.8 | ||
| orf19.239 | STH1 | ATPase activity/DNA helicase activity | 2.5 | 0.9 | 4.3 | ||
| orf19.1343 | DRE2 | Conserved ORF/Function unknown | 2.5 | 1.6 | 1.8 | ||
| orf19.4754 | ZWF1 | ZWF1 | Glucose-6-phosphate dehydrogenase | 2.5 | 1.1 | 1.6 | |
| orf19.2165 | No good BLAST homology | 2.4 | 1.1 | 1.9 | |||
| orf19.5634 | FRP1 | FRE5 | Ferric reductase | 2.4 | 1.0 | 2.6 | |
| orf19.1027 | PDR16 | Phosphatidylinositol transporter | 2.3 | 1.3 | 2.5 | ||
| orf19.6947 | GTT1 | Glutathione transferase (4e–04)k | 2.3 | 1.2 | 1.5 | ||
| orf19.5604 | MDR1 | FLR1 | Multidrug efflux pump | 2.3 | 1.2 | 4.4 | |
| orf19.2179 | SIT1 | ARN1 | Ferrichrome siderophore transporter | 2.2 | 1.0 | 2.0 | |
| orf19.4150 | YBR014C | Glutaredoxin | 2.2 | 1.0 | 3.0 | ||
| orf19.2175 | CPD1 | NADH or NADPH oxidoreductase | 2.2 | 1.4 | 1.4 | ||
| orf19.7495 | OYE3 | NADPH dehydrogenase | 2.1 | 1.2 | 4.9 | ||
| orf19.2995 | LOT5 (2e–07) | Low temperature responsive protein | 2.1 | 1.1 | 3.5 | ||
| orf19.3538 | CFL1 | Ferric reducatase | 1.9 | 1.1 | 2.7 | ||
Functional Processes:
Oxidative stress 
Iron acquisition
Membrane transport
Transcription
Respiration
Sulfur metabolism
See yellow vertical bars in Figure 1A
Unless noted, all ORF19 number designations were taken from the Candida Genome Database (CGD; http://www.candidagenome.org/)
Gene names taken from CGD for named C. albicans genes
Gene names taken from the Saccharomyces Genome Database (SGD) for named S. cerevisiae homologs (http://www.yeastgenome.org/)
Unless noted, all gene descriptions and functions taken from published sources, or 1) CGD for named C. albicans genes or 2) SGD when no CGD description present and closest homolog is a S. cerevisiae gene
If more than one microarray spot corresponded to an ORF, the average of the fold changes is represented
Average fluorescence signal of CAF2–1, RM1, and RM1000
See blue vertical bar in Figure 1A
Schizosaccharomyces pombe
Similarity to C. albicans protein
BLAST score from Vibrio vulnificus
Genes Up-regulated On Nitric Oxide Exposure
Approximately 65 genes show elevated expression (2- to 25-fold increase) 10 min after the addition of DPTA NONOate and return to baseline expression levels by the 40-min time point (Figure 1A, blue vertical bar). Many (∼19) of these genes encode proteins, homologues of which have previously been implicated in protection against oxidative stress. These include 6 glutathione conjugating and modifying enzymes, 10 NADPH oxidoreductases/dehydrogenases, and the catalase gene (CAT1; see oxidative stress genes, Table 2B). This response makes conceptual sense because, as a potent inhibitor of mitochondrial oxidative phosphorylation, .NO leads to the reduction of O2 and to the production of superoxide anion (O2-·), peroxynitrite (ONOO-), and hydrogen peroxide (H2O2; Poderoso et al., 1996; Carreras et al., 2004). Many of the oxidative stress protection enzymes require iron as a cofactor, and it is perhaps not surprising that several genes that encode iron uptake systems are also transiently induced (see iron acquisition genes, Table 2B). Other genes induced transiently upon exposure to .NO include three genes involved in sulfur assimilation, two zinc finger transcription factors, at least seven transporters for oligopeptides, drugs, or heavy metals, and various other enzymes whose functional roles are not easily discerned (Table 2B). Twelve proteins transiently induced by .NO are either conserved proteins of unknown function or proteins that lack clear homologues in other genome sequences (Table 2B).
Although the expression of the majority of the genes induced by .NO at the 10-minute time point return to baseline levels of expression by 40 min, a group of nine genes remain highly expressed throughout the 2-h time course and in all strain backgrounds tested (Figure 1A, yellow vertical bar, EXPT 1-4, and Table 2A). We hypothesize that this set of genes is involved directly in combating the effects of .NO and that, once these genes are induced and exert their protective effects, the majority of the .NO-induced genes return to their preinduction levels (see accompanying article by Sarver and DeRisi, 2005). Of the nine persistently induced genes, the most highly expressed is YHB1 (orf19.3707), which encodes a putative flavohemoglobin, a protein that has been shown to be important for detoxifying .NO in a variety of pathogens including C. albicans (Ullmann et al., 2004). Although there are three putative flavohemoglobins in the C. albicans genome (YHB1, YHB4, and YHB5), only YHB1 is induced by DPTA NONOate under the conditions tested here (log phase, 30°C, YEPD). Based on its homology to well-characterized flavohemoglobins, Yhb1 is a two-domain protein with an N-terminal globin heme-binding domain and a C-terminal flavin FADH2-binding domain. Yhb1 likely requires heme as a cofactor, and we note that another persistently induced gene, RBT5 (orf19.5636), encodes a heme-acquisition protein (Weissman and Kornitzer, 2004). Also persistently induced is orf19.3117 (called orf19.3119 in the Candida Genome Database), which encodes a protein with a CFEM motif and is homologous to characterized cell surface heme-binding proteins (Weissman and Kornitzer, 2004). Heme-based .NO sensors have been described in signal transduction (Gilles-Gonzalez and Gonzalez, 2005), and it is possible that orf19.3117 is involved in C. albicans detection of .NO. Other transcripts persistently induced by .NO throughout the 2-h time course include SSU1 (orf19.7313), which is homologous to the S. cerevisiae SSU1 that mediates sulfite efflux from the cell (Park and Bakalinsky, 2000), and two alternative oxidases (orf19.4774/AOX1, orf19.4773/AOX2), which accept electrons from the ubiquinone pool of the electron transport chain and reduce molecular oxygen (O2) to water (Huh and Kang, 1999, 2001). These Aox enzymes prevent pools of O2 from being reduced to the toxic superoxide anion (O2-·), and in effect, function to prevent .NO-induced oxidative stress. We also see prolonged induction of CTR2 (orf19.4720), which encodes a putative copper transporter; YOL075C (orf19.3120), which encodes a putative ferric cation transporter; and YMR209C (orf19.4816), which encodes a protein of unknown function that contains a GMP kinase domain.
When the genes induced by .NO in C. albicans are compared with those in S. cerevisiae and Histoplasma capsulatum, some common patterns emerge, although the detailed responses are highly individual. For example, genes encoding catalase and iron acquisition proteins are up-regulated in all three organisms in response to .NO (see accompanying articles by Nittler et al., 2005; Sarver and DeRisi, 2005). In C. albicans and H. capsulatum, alternative oxidase enzymes are up-regulated and are likely functioning to prevent the formation of reactive oxygen intermediates (ROI). In S. cerevisiae and C. albicans, YHB1 is highly induced by .NO, yet H. capsulatum seems to lack this conserved flavohemoglobin altogether. In both C. albicans and S. cerevisiae, exposure to .NO prolonged induction of only a few genes (nine in C. albicans to seven in S. cerevisiae). Of these latter genes, only two (YHB1 and SSU1) are induced in both species.
Genes Down-regulated on Nitric Oxide Exposure
Approximately 65 genes are repressed upon addition of DPTA NONOate and return to normal levels of expression by the 40-min time point (Figure 1B and Supplementary Table 1). Twenty-five of these genes encode proteins of the mitochondrial electron transport chain, many of which contain prosthetic groups that are known to react with .NO, such as heme, Fe-S, and Cu2+ (see respiration genes, Supplementary Table 1). Specifically, .NO has been shown to reversibly inhibit the cytochrome c oxidase enzyme (Cox), to block cellular respiration (Carreras et al., 2004) and to markedly increase the production rate of O2-· and H2O2 (Poderoso et al., 1996). Examples of proteins involved in respiration whose expression was transiently down-regulated by .NO include 2 subunits of the NADH dehydrogenase complex I, 4 subunits of the succinate dehydrogenase complex II, 12 subunits of the ubiquinone-cytochrome c oxidoreductase complex III, including cytochrome c itself, and four subunits of the cytochrome oxidase complex IV (see respiration genes, Supplementary Table 1). Also included in the set of .NO repressed genes are 17 ribosomal proteins, representing about one third of the total number of C. albicans annotated ribosomal genes (see ribosomal components, Supplementary Table 1). The repression of ribosomal genes was also observed upon phagocytosis of C. albicans by macrophages (Lorenz et al., 2004) and has also been observed in S. cerevisiae upon exposure to various stresses and has been characterized as part of the general stress response in that organism (Gasch et al., 2000).
The Induction of YHB1 Is a Result of Nitric Oxide
We further investigated the induction of YHB1 because, of all the genes persistently induced by .NO, YHB1 showed the highest induction ratio and is known to have a role in detoxifying .NO in many microorganisms. To verify that induction of the YHB1 transcript results from exposure to the .NO released by DPTA NONOate and not some other part of the molecule, we added the .NO scavenging molecule Carboxy PTIO to the induction experiments. For the experiment in Figure 2, DPTA NONOate alone, scavenger alone, or DPTA NONOate and scavenger together (at eightfold molar excess over DPTA NONOate) were added to log phase cultures (OD 600 nm = 1.0) of the C. albicans wild-type strain CAF2-1. Samples were collected 10 min and 2 h after treatment and analyzed by quantitative RT-PCR. As shown in Figure 2, the induction of YHB1 mRNA was substantially blocked upon addition of the scavenger, supporting the conclusion that YHB1 induction results from exposure to .NO (or another RNI product) and not the DPTA NONOate backbone. mRNA of PAT1 (orf19.3792), which is homologous to a topoisomerase II-associated protein in S. cerevisiae, was used as a control in this experiment and shows no induction by .NO. These data confirm a previous report showing that YHB1 is induced by .NO, as demonstrated by its induction in response to three different sources of .NO: gaseous .NO, DETA NONOate, and sodium nitrite (Ullmann et al., 2004). Figure 2 also confirms that, of the three putative C. albicans flavohemoglobins, only the YHB1 gene is induced by nitric oxide; under the conditions tested, expression of YHB4 and YHB5 is detectable, but is not affected by the presence of .NO.
The C. albicans yhb1Δ/yhb1Δ Mutant Displays Increased Sensitivity to Nitric Oxide In Vitro
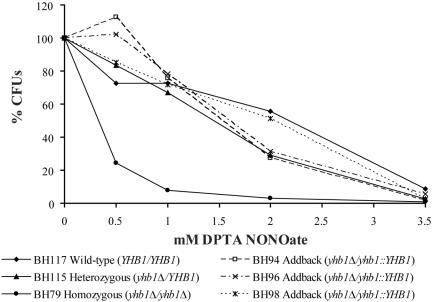
To study the biological role of YHB1, we deleted both copies of the gene in the lab strain RM1000. We also constructed a heterozygous strain, as well as a YHB1 addback strain in which a wild-type allele of YHB1 was reintroduced into the homozygous deletion mutant. All strains are described in the Materials and Methods section and listed in Table 1. Parental and mutant cultures were exposed to five different concentrations of DPTA NONOate (0.0, 0.5, 1.0, 2.0, and 3.5 mM) and after 8 h of exposure were serially diluted and plated. After 24 h of incubation at 30°C, CFUs were counted. As shown in Figure 3, the yhb1Δ/yhb1Δ strain (BH79) was significantly more sensitive to .NO than a genetically matched wild-type strain (BH117), a heterozygous yhb1Δ/YHB1 strain (BH115), and three different gene addback strains yhb1Δ/yhb1::YHB1 (BH94, BH96, BH98). The results of Figure 3 show that the YHB1 addback strains, where one copy of YHB1 has been added back to its endogenous locus, have reacquired the ability to survive nitrosative challenge and that expression of one copy of YHB1 is sufficient for protection against .NO in vitro. We note that growth of the yhb1Δ/yhb1Δ strain (BH79) was not significantly sensitive to oxidative stress, as measured by exposure to both hydrogen peroxide (H2O2) and the superoxide anion (O2-·) generator menadione (unpublished data). Consistent with this observation, Ullmann et al. (2004) reported that YHB1 is not induced by H2O or O2-·.
Figure 3.
The yhb1Δ/yhb1Δ mutant is hypersensitive to nitric oxide (.NO) in vitro. C. albicans cultures were exposed to five different concentrations of DPTA NONOate (0.0, 0.5, 1.0, 2.0, 3.5 mM) and after 8 h of exposure, were serial diluted and plated. After 24 h of incubation, colony-forming units were counted. Wild-type (BH117), yhb1Δ/YHB1 heterozygous (BH115), and yhb1Δ/yhb1::YHB1 addback (BH94, BH96, BH98) strains showed comparable survival at low concentrations of DPTA NONOate (0.5 and 1.0 mM); the yhb1Δ/yhb1Δ mutant (BH79, •), however, is hypersensitive to .NO at these concentrations. At higher concentrations of DPTA NONOate (3.5 mM), all strains are sensitive.
Genome-wide Expression Profile of the yhb1Δ/yhb1Δ Mutant in Response to Nitric Oxide
We next investigated the transcriptional response of the yhb1Δ/yhb1Δ deletion strain to .NO (Figure 1A, EXPT 4). Although the response of the yhb1Δ/yhb1Δ strain generally resembles that of wild-type cells, it differs in three important respects: 1) genes that show only transient induction or repression in wild-type cells show prolonged changes in the yhb1Δ/yhb1Δ strain, 2) genes that are persistently induced in wild-type cells continue to increase in expression throughout the time course in the yhb1Δ/yhb1Δ strain, whereas they level off in the wild-type strains, and 3) a new set of 34 genes is induced in the yhb1Δ/yhb1Δ strain. We believe that most of these effects arise because deletion of the YHB1 gene removes a major source of .NO detoxification; hence the deletion mutants are in essence subject to significantly higher intracellular levels of .NO. These results also indicate that .NO is produced throughout the time course of the experiment and that in wild-type cells the protective effects of YHB1 are responsible for the rapid return of the transiently induced and repressed genes to their normal levels. The results with the yhb1Δ/yhb1Δ mutant are also consistent with the observation made in S. cerevisiae that the magnitude of an environmental stress is correlated with the time required to transcriptionally adapt to it (Gasch et al., 2000).
The new class of 34 genes encodes proteins required for repair of DNA damage, two transcription factors that may regulate genes involved in protecting against .NO, multiple transporters, iron acquisition proteins, and additional oxidative stress proteins (Figure 1A, gray vertical bar, and Supplementary Table 2). It is possible that these genes are subtly induced in the wild-type time courses and that loss of YHB1 simply enhances this effect. The induction pattern reveals that YHB1 itself is not required for either the transient or persistent gene induction produced by .NO. It should be noted that, in the yhb1Δ/yhb1Δ strain, no induction of the YHB4 or YHB5 transcripts was observed by microarray analysis or quantitative RT-PCR (unpublished data), indicating that loss of the Yhb1 protein is not compensated by enhanced expression of the other YHB genes.
The C. albicans yhb1Δ/yhb1Δ Mutant Is Hyperfilamentous under Nonfilamenting Conditions
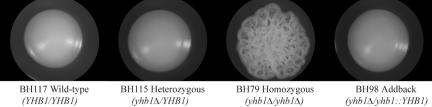
Wild-type (BH117), yhb1Δ/yhb1Δ (BH79), a strain heterozygous for YHB1 (BH115), and three YHB1 addback strains (BH94, BH96, BH98) were tested for a series of additional phenotypes, and a defect in filamentation was observed for the yhb1Δ/yhb1Δ strain. All strains appeared similar to wild type on YEPD + 10% serum at 37°C and Spider at 30°C, two conditions that strongly induce filamentous growth (unpublished data). However, ∼90% of the yhb1Δ/yhb1Δ (BH79) colonies exhibited filamentous growth on YEPD at 37°C, a condition where the wild type and a heterozygous deletion strain did not form filaments (Figure 4).
Figure 4.
The yhb1Δ/yhb1Δ mutant is hyperfilamentous under nonfilamenting conditions. Wild-type (BH117), yhb1Δ/YHB1 heterozygous (BH115), yhb1Δ/yhb1Δ mutant (BH79), and yhb1Δ/yhb1::YHB1 addback (BH98) strains were tested for filamentation defects on YEPD at 37°C.
We note that only one of the three YHB1 addback strains tested (BH98) showed complete reversal of the filamentation defect (Figure 4). Using an RT-PCR-based methodology, we determined that, under a variety of conditions (YEPD at 30°C, YEPD at 37°C, and YEPD + .NO at 30°C) the addback strains had consistently lower levels of YHB1 than either the YHB1/YHB1 or yhb1Δ/YHB1 strains (unpublished data). Of the three addback strains, BH98 had the highest levels of YHB1 expression (unpublished data). We therefore chose BH98, an addback strain that did show full complementation of both the filamentation and .NO induced lethality defects, for subsequent analyses in mice.
Genome-wide Expression Profile of the yhb1Δ/yhb1Δ Mutant Compared with Wild-type in the Absence of Nitric Oxide
The hyperfilamentous phenotype of the yhb1Δ/yhb1Δ strain was further investigated using microarray analysis. We compared the transcriptional profile of the yhb1Δ/yhb1Δ mutant (BH79) and the otherwise isogenic wild-type strain (BH117) under two conditions, YEPD at 30°C and YEPD at 37°C. Competitive hybridizations were performed on microarrays, and the results are shown in Figure 5. Compared with wild-type cells grown in YEPD at 37°C, eight genes were induced greater than sixfold in the yhb1Δ/yhb1Δ mutant grown under the same conditions. The majority of these have been shown to be part of the filamentous growth program in C. albicans (Nantel et al., 2002; Kadosh and Johnson, 2005), supporting the observation that at least part of the filamentous pathway is inappropriately activated in the yhb1Δ/yhb1Δ strain.
Figure 5.
Hyphal-specific genes are induced in the yhb1Δ/yhb1Δ mutant in YEPD at 37° in the absence of nitric oxide (.NO). Genes induced over sixfold in the yhb1Δ/yhb1Δ mutant (BH79) are shown (mutant (BH79) transformed over wild type (BH117)). The induction of hyphal-specific transcripts supports the hyperfilamentous plate phenotype of the yhb1Δ/yhb1Δ mutant (see Figure 4).
The yhb1Δ/yhb1Δ Mutant Is Attenuated in Virulence
We utilized a murine model of systemic candidiasis to test whether the yhb1Δ/yhb1Δ strain has a virulence defect compared with its parental strain and the YHB1 addback strain. In all strains (BH79, BH98, BH117), URA3 was restored to its native locus, thereby controlling for complications that can arise from URA3 position effects (Lay et al., 1998; Cheng et al., 2003; Brand et al., 2004).
Ten immunocompetent female BALB/c mice per strain (BH79, BH98, and BH117) were injected with 3 × 105 C. albicans cells. Figure 6A shows that mice infected with the yhb1Δ/yhb1Δ mutant strain (BH79) lived significantly longer than mice infected with either the YHB1 addback strain (BH98) or wild-type strain (BH117; p = 0.0015 and 0.0002, respectively). At day 28 the experiment was ended and the six remaining mice infected with the yhb1Δ/yhb1Δ strain (BH79) were sacrificed. Although some of these remaining mice had initially shown signs of infection, by the end of the experiment all appeared healthy and some had gained weight. Two additional experiments revealed similar trends in virulence (unpublished data); that is, in all three experiments, the yhb1Δ/yhb1Δ strain showed reduced virulence compared with the wild-type (BH117) and YHB1 addback (BH98) strains. We note that the virulence defect of the yhb1Δ/yhb1Δ mutant is moderate compared with those of other published virulence mutants (see, for example Buurman et al., 1998; Braun et al., 2000). Nevertheless, the defect is statistically significant and reproducible. Complementation of the virulence defect by restoration of a single wild-type copy of YHB1 confirms that the virulence defect of the yhb1Δ/yhb1Δ knockout strain is dependent on YHB1. The doubling times of the yhb1Δ/yhb1Δ strains grown at 30°C in YEPD medium were comparable to that of wild-type when measured in vitro (unpublished data), suggesting that the virulence attenuation does not result from a nonspecific growth defect. Attenuated virulence of a yhb1Δ/yhb1Δ mutant relative to wild type was also observed by Ullmann et al. (2004) using a set of independently derived strains.
The Virulence Defect of the yhb1Δ/yhb1Δ Mutant in the Tail Vein Model of Systemic Infection May Be Unrelated to Its Increased Sensitivity to Nitric Oxide
To determine whether the attenuated virulence of the yhb1Δ/yhb1Δ mutant was due to its hypersensitivity to nitrosative stress, we infected male knockout mice deleted for both copies of the NOS2 gene, which encodes the inducible nitric oxide synthase (iNOS; NOS2; see Materials and Methods for experimental details). The increased susceptibility of NOS2-/- mice to numerous pathogens has been extensively documented (for reviews see Nathan and Shiloh, 2000; Fang, 2004). Furthermore, NOS2 is the only isoform that can be induced to yield high levels of .NO for sustained periods in immune cells (Lowenstein and Padalko, 2004). Thus, NOS2-/- mice have been used to determine the importance of .NO production for the control of various pathogens (see cited reviews above). The mice used in this experiment were of the C57BL/6 background, which is different from the BALB/c background used for the virulence experiment described above. Although the great majority of knockout mouse strains have the C57BL/6 genetic background, most published C. albicans virulence experiments have been performed in the BALB/c background. Genetically matched NOS2 +/+ mice were used as controls.
The results of this experiment are shown in Figure 6B. First, we consider whether NOS2-/- and NOS2+/+ mice show any difference in their susceptibility to wild-type C. albicans. As shown in Figure 6B (green triangles vs. yellow squares), there is no statistically significant difference between the two mouse strains in their susceptibility to C. albicans using the tail vein injection model (p = 0.5088, with p < 0.05 considered significant). We next consider the effects of deleting the YHB1 gene from C. albicans. Both NOS2+/+ and NOS2-/- mice (Figure 6B, pink squares and gray triangles) show a statistically significant increase in survival when infected with the yhb1Δ/yhb1Δ knockout strain compared with wild-type (p = 0.0399 and 0.0166, respectively, for NOS2+/+ and NOS2-/-). In other words, loss of YHB1 results in a virulence defect that is not suppressed by removing the primary .NO synthetic machinery from the immune cells of the host. This observation implies that YHB1 has an important function in virulence, measured by the tail vein model, which is distinct from protection against .NO. Given that other hyperfilamentous strains of C. albicans show virulence defects in the mouse tail vein injection model (Braun et al., 2000, 2001; Kadosh and Johnson, 2001; Murad et al., 2001), it is plausible that this property, as opposed to the increased sensitivity to .NO, is responsible for the decreased virulence of the yhb1Δ/yhb1Δ strain.
DISCUSSION
A major defense mechanism mounted against invading pathogens is the production of nitric oxide (.NO), a free radical that rapidly diffuses across cell membranes and is capable of reacting with a variety of molecules and causing multiple types of cell damage. The relationship between .NO and individual human pathogens is complex and has not been systematically investigated for C. albicans. However, available evidence supports the view that .NO is important for the control of C. albicans infections. For example, it has been reported that mice deficient in .NO production are hypersensitive to C. albicans infections as judged by organ load (Netea et al., 2002). In addition, killing of C. albicans by murine saliva and macrophages has been shown, in some cases, to require NOS2 (Elahi et al., 2001; Netea et al., 2002; Balish et al., 2005).
Although the physiological importance of .NO in control of C. albicans requires further investigation, it has been well established that .NO production by the NOS2 enzyme is crucial for the control of other pathogens, including Mycobacterium tuberculosis, Leishmania spp. (for review see Nathan and Shiloh, 2000), and Cryptococcus neoformans (de Jesus-Berrios et al., 2003). In the latter organism, it was shown that deletion of a flavohemoglobin gene (FHB1) results in attenuated virulence in wild-type mice that can be suppressed in infections of NOS2-/- mice (de Jesus-Berrios et al., 2003).
Because of the established importance of .NO in the control of numerous other pathogens, we examined the genome-wide response of C. albicans to .NO. In this article, we identified 131 genes that are induced or repressed in wild-type C. albicans in response to .NO (see Figure 1, Table 2, and Supplementary Table 1). Based on their kinetic profiles, these genes fall into three classes: 1) transiently induced genes, 2) transiently repressed genes, and 3) persistently induced genes. As described in Results, we propose that the transiently induced and repressed genes primarily encode proteins involved in counteracting the secondary .NO-induced effects such as oxidative stress, whereas the persistently induced genes encode proteins that specifically protect against .NO. Transiently induced transcripts encode oxidative stress proteins such as glutathione-conjugating and -modifying enzymes, NADPH oxidoreductases/dehydrogenases, catalase, iron acquisition proteins, transcription factors, sulfur assimilation enzymes, transporters of oligopeptides, drugs, and heavy metals, and heme-binding proteins (see Results and Table 2B). Transiently repressed transcripts encode subunits of the mitochondrial electron transport chain and ribosomal proteins (see Results and Supplementary Table 1). Only nine genes are persistently induced by .NO; these include YHB1, two alternative oxidases, two putative cell surface heme-binding proteins, putative transporters for copper, sulfite, and iron, and a conserved protein of unknown function (see Table 2A). As described in the accompanying article (Sarver and DeRisi, 2005), two of these nine genes (YHB1 and SSU1, a putative sulfite transporter) are also persistently induced when S. cerevisiae is exposed to .NO.
On deletion of the YHB1 gene in C. albicans, the genome-wide expression profile in response to .NO is more pronounced. That is, the genes that show only transient induction or repression in wild-type strains show prolonged and enhanced changes in expression in the yhb1Δ/yhb1Δ deletion strain (Figure 1, EXPT 4). In addition, a cluster of 34 new genes showed significant induction only in the yhb1Δ/yhb1Δ strain (Figure 1, gray vertical bar); these genes encode proteins involved in the repair of DNA damage, as well as additional proteins involved in processes such as oxidative stress, iron acquisition, and transport (Supplementary Table 2). The prolonged and enhanced changes in .NO-induced gene expression observed upon removal of the YHB1 gene, strongly supports the idea that Yhb1 is indeed functioning to detoxify .NO in C. albicans. Consistent with this idea, growth of the yhb1Δ/yhb1Δ mutant in vitro is much more sensitive to .NO than is wild type (Figure 3). This latter result was also reported by Ullmann et al. (2004), using an independently derived yhb1Δ/yhb1Δ mutant.
Also in agreement with Ullmann et al. (2004), we demonstrated that the yhb1Δ/yhb1Δ strain is moderately attenuated for virulence in BALB/c mice, as assessed by the mouse tail vein model of disseminated candidiasis (Figure 6A). To our surprise, however, the C. albicans yhb1Δ/yhb1Δ virulence defect was not suppressed in mice deleted for the NOS2 gene (Figure 6B). This result implies that the virulence defect of the yhb1Δ/yhb1Δ mutant is not solely due to an inability to detoxify .NO and implicates an additional function as being responsible. One possibility is a role for YHB1 in the control of filamentous growth. As described in the Results section, the yhb1Δ/yhb1Δ strain is hyperfilamentous, as observed as altered colony morphologies on laboratory media (Figure 4) and as the expression of “filament-specific” genes under conditions where they should be repressed (Figure 5). Several C. albicans hyperfilamentous mutants have been shown to have defects in virulence; these include tup1Δ/tup1Δ and nrg1Δ/nrg1Δ (both of which are severely hyperfilamentous) (Braun et al., 2000, 2001; Murad et al., 2001), as well as rfg1Δ/rfg1Δ (which has a mild hyperfilamentous phenotype similar to that of yhb1Δ/yhb1Δ; Kadosh and Johnson, 2001). It is also possible that the yhb1Δ/yhb1Δ strain has additional defects that render it less virulent. For example, YHB1 is also induced in media rich in iron (Lan et al., 2004), upon phagocytosis by macrophages (Lorenz et al., 2004), and by sodium sulfite (our unpublished result). Thus, Yhb1 likely has roles in addition to protecting against nitrosative stress, and at least one of these additional functions must be required for full virulence in the mouse tail vein model of candidiasis.
Another unanticipated result concerns the susceptibility of mice lacking the NOS2 gene to infection by C. albicans. We found that NOS2-/- mice are not significantly more susceptible than immunocompetent mice to killing by C. albicans, as assessed by the tail vein model of infection (Figure 6B). These results indicate that NOS2 production of .NO has little or no effect on the susceptibility of mice to C. albicans infection through this route. By introducing C. albicans directly into the venous system, this model bypasses, for example, mucosal and epithelial barriers, and thereby escapes a step where .NO production may have a significant effect on the outcome of the infection. These observations suggest that the yhb1Δ/yhb1Δ mutant strain could be a useful tool to identify models of infection that do require .NO for containing infections caused by C. albicans and, by implication, to identify the steps of infection at which .NO is important for defense against this pathogen. Given the high conservation of the .NO transcriptional response in C. albicans and S. cerevisiae (see accompanying article [Sarver and DeRisi, 2005]), it seems likely that both fungi routinely encounter concentrated levels of .NO in their environment. Further studies employing the C. albicans yhb1Δ/yhb1Δ mutant strain should illuminate the host microenvironment where .NO presents a serious threat for C. albicans.
Supplementary Material
Acknowledgments
We thank Greg Petsko and Matt Miller for discussions that lead to this project and Matt Miller for valuable help with the grant proposal that funded this study. We are grateful to Carly Klein for strain construction of one of the yhb1Δ/yhb1Δ mutant strains (MMY272) used in this study. We also thank Richard Bennett and other members of the Johnson lab for careful reading of this manuscript and valuable discussions. We are grateful to Joe DeRisi and Adam Carroll for help with the microarray printing and analyses, and we thank Joe DeRisi, Anita Sil, Aaron Sarver, and Paige Nittler for communicating results before publication and for advice in experimental design of the nitric oxide induction experiments. We thank Diane Inglis, Mike Lorenz, and Gerald Fink for the collaboration that produced the DNA microarrays used in this article, and we are also grateful to the Stanford Genome Technology Center (http://www.sequence.stanford.edu/group/candida/search.html) for providing sequence data for C. albicans. Sequencing of the C. albicans genome was supported by National Institute of Dental and Craniofacial Research and the Burroughs Wellcome Fund. The work described in this article was supported by grants from the Sandler Foundation (Mechanisms of Nitric Oxide Resistance in C. albicans) and the National Institutes of Health (R01 AI49187) to A.D.J. S.M.N. was supported by a Burroughs Wellcome Career Award in the Biomedical Sciences.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-05-0435) on July 19, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alani, E., Cao, L., and Kleckner, N. (1987). A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116, 541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish, E., Warner, T. F., Nicholas, P. J., Paulling, E. E., Westwater, C., and Schofield, D. A. (2005). Susceptibility of germfree phagocyte oxidase- and nitric oxide synthase 2-deficient mice, defective in the production of reactive metabolites of both oxygen and nitrogen, to mucosal and systemic candidiasis of endogenous origin. Infect. Immun. 73, 1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R. J., Uhl, M. A., Miller, M. G., and Johnson, A. D. (2003). Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 23, 8189-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., LaCroute, F., and Fink, G. R. (1984). A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345-346. [DOI] [PubMed] [Google Scholar]
- Brand, A., MacCallum, D. M., Brown, A. J., Gow, N. A., and Odds, F. C. (2004). Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3, 900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B. R., Head, W. S., Wang, M. X., and Johnson, A. D. (2000). Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156, 31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B. R., and Johnson, A. D. (2000). TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155, 57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B. R., Kadosh, D., and Johnson, A. D. (2001). NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20, 4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney, S., Caulfield, J. L., Niles, J. C., Wishnok, J. S., and Tannenbaum, S. R. (1999). The chemistry of DNA damage from nitric oxide and peroxynitrite. Mutat. Res. 424, 37-49. [DOI] [PubMed] [Google Scholar]
- Buurman, E. T., Westwater, C., Hube, B., Brown, A. J., Odds, F. C., and Gow, N. A. (1998). Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 95, 7670-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras, M. C., Franco, M. C., Peralta, J. G., and Poderoso, J. J. (2004). Nitric oxide, complex I, and the modulation of mitochondrial reactive species in biology and disease. Mol. Aspects Med. 25, 125-139. [DOI] [PubMed] [Google Scholar]
- Chakravortty, D., and Hensel, M. (2003). Inducible nitric oxide synthase and control of intracellular bacterial pathogens. Microbes Infect. 5, 621-627. [DOI] [PubMed] [Google Scholar]
- Cheng, S., Nguyen, M. H., Zhang, Z., Jia, H., Handfield, M., and Clancy, C. J. (2003). Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 71, 6101-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen, T., Qureshi, M. H., Koguchi, Y., and Kawakami, K. (1999). Candida albicans suppresses nitric oxide (NO) production by interferon-gamma (IFN-gamma) and lipopolysaccharide (LPS)-stimulated murine peritoneal macrophages. Clin. Exp. Immunol. 115, 491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, M. J., and Goldberg, D. E. (1998). Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273, 12543-12547. [DOI] [PubMed] [Google Scholar]
- de Jesus-Berrios, M., Liu, L., Nussbaum, J. C., Cox, G. M., Stamler, J. S., and Heitman, J. (2003). Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13, 1963-1968. [DOI] [PubMed] [Google Scholar]
- Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi, S., Pang, G., Ashman, R. B., and Clancy, R. (2001). Nitric oxide-enhanced resistance to oral candidiasis. Immunology 104, 447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, F. C. (1997). Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 99, 2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820-832. [DOI] [PubMed] [Google Scholar]
- Fonzi, W. A., and Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, P. R., Costantino, G., and Salzman, A. L. (1998). Constitutive and adaptive detoxification of nitric oxide in Escherichia coli. Role of nitric-oxide dioxygenase in the protection of aconitase. J. Biol. Chem. 273, 26528-26533. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355-360. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez, M. A., and Gonzalez, G. (2005). Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 99, 1-22. [DOI] [PubMed] [Google Scholar]
- Hausladen, A., Gow, A. J., and Stamler, J. S. (1998). Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 95, 14100-14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Butcher, P. D., Mangan, J. A., Rajandream, M. A., and Coates, A. R. (1999). Regulation of hmp gene transcription in Mycobacterium tuberculosis: effects of oxygen limitation and nitrosative and oxidative stress. J. Bacteriol. 181, 3486-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., and Kang, S. O. (1999). Molecular cloning and functional expression of alternative oxidase from Candida albicans. J. Bacteriol. 181, 4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., and Kang, S. O. (2001). Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356, 595-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. et al. (2004). The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101, 7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh, D., and Johnson, A. D. (2001). Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21, 2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh, D., and Johnson, A. D. (2005). Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16, 2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch, D. R., and Whitney, R. R. (1991). Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect. Immun. 59, 3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, C. Y., Rodarte, G., Murillo, L. A., Jones, T., Davis, R. W., Dungan, J., Newport, G., and Agabian, N. (2004). Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 53, 1451-1469. [DOI] [PubMed] [Google Scholar]
- Lay, J., Henry, L. K., Clifford, J., Koltin, Y., Bulawa, C. E., and Becker, J. M. (1998). Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66, 5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Zeng, M., Hausladen, A., Heitman, J., and Stamler, J. S. (2000). Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. USA 97, 4672-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., Bender, J. A., and Fink, G. R. (2004). Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3, 1076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein, C. J., and Padalko, E. (2004). iNOS (NOS2) at a glance. J. Cell Sci. 117, 2865-2867. [DOI] [PubMed] [Google Scholar]
- Miller, M. G., and Johnson, A. D. (2002). White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110, 293-302. [DOI] [PubMed] [Google Scholar]
- Murad, A. M. et al. (2001). NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20, 4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel, A. et al. (2002). Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13, 3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, C. (1992). Nitric oxide as a secretory product of mammalian cells. FASEB J. 6, 3051-3064. [PubMed] [Google Scholar]
- Nathan, C. (1997). Inducible nitric oxide synthase: what difference does it make? J. Clin. Invest. 100, 2417-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, C., and Shiloh, M. U. (2000). Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97, 8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo, A., Monteoliva, L., Gil, C., Pla, J., and Nombela, C. (1997). Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143(Pt 2), 297-302. [DOI] [PubMed] [Google Scholar]
- Netea, M. G., Meer, J. W., Verschueren, I., and Kullberg, B. J. (2002). CD40/CD40 ligand interactions in the host defense against disseminated Candida albicans infection: the role of macrophage-derived nitric oxide. Eur. J. Immunol. 32, 1455-1463. [DOI] [PubMed] [Google Scholar]
- Nittler, M. P., Murray, D. H., Foo, C., and Sil, A. (2005). Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol. Biol. Cell 16, 4792-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, S. M., and Johnson, A. D. (2005). Strains and strategies for large-scale Gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4, 298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., and Bakalinsky, A. T. (2000). SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 16, 881-888. [DOI] [PubMed] [Google Scholar]
- Pathania, R., Navani, N. K., Gardner, A. M., Gardner, P. R., and Dikshit, K. L. (2002). Nitric oxide scavenging and detoxification by the Mycobacterium tuberculosis haemoglobin, HbN in Escherichia coli. Mol. Microbiol. 45, 1303-1314. [DOI] [PubMed] [Google Scholar]
- Poderoso, J. J., Carreras, M. C., Lisdero, C., Riobo, N., Schopfer, F., and Boveris, A. (1996). Nitric oxide inhibits electron transfer and increases superoxide radical production in rat heart mitochondria and submitochondrial particles. Arch. Biochem. Biophys. 328, 85-92. [DOI] [PubMed] [Google Scholar]
- Poole, R. K., and Hughes, M. N. (2000). New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36, 775-783. [DOI] [PubMed] [Google Scholar]
- Sarver, A., and DeRisi, J. (2005). Fzf1p regulates an inducible response to nitrosative stress in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 4781-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom, P., Cutler, J. E., and Staab, J. F. (2002). Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect. Immun. 70, 3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann, B. D., Myers, H., Chiranand, W., Lazzell, A. L., Zhao, Q., Vega, L. A., Lopez-Ribot, J. L., Gardner, P. R., and Gustin, M. C. (2004). Inducible defense mechanism against nitric oxide in Candida albicans. Eukaryot. Cell 3, 715-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, S. G., Armarego, W. L., Shaw, D. C., Lilley, P. E., Dixon, N. E., and Poole, R. K. (1991). Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol. Gen. Genet. 226, 49-58. [DOI] [PubMed] [Google Scholar]
- Weissman, Z., and Kornitzer, D. (2004). A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53, 1209-1220. [DOI] [PubMed] [Google Scholar]
- Wilson, R. B., Davis, D., Enloe, B. M., and Mitchell, A. P. (2000). A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16, 65-70. [DOI] [PubMed] [Google Scholar]
- Wilson, R. B., Davis, D., and Mitchell, A. P. (1999). Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q. W., Cho, H. J., Calaycay, J., Mumford, R. A., Swiderek, K. M., Lee, T. D., Ding, A., Troso, T., and Nathan, C. (1992). Cloning and characterization of inducible nitric oxide synthase from mouse macrophages. Science 256, 225-228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.