Abstract
Objective
G protein-coupled receptor 126 (GPR126) gene has been implicated as a potential susceptibility factor for aggressive periodontitis in Japanese patients. This study aimed to investigate the presence of the GPR126 [c.3086 G > A] (rs536714306) polymorphism in patients with periodontitis in a Greek population and periodontal cases of European ancestry. A total of 82 subjects were recruited: 53 patients periodontally compromised (P) and 29 healthy controls (H). GPR126 genotyping was performed using Sanger sequencing. Additionally, data from the Gene-Lifestyle Interactions in Dental Endpoints (GLIDE) consortium were included in this study.
Results
No variants (rs536714306) in the GPR126 gene were detected in any of the samples. The homozygous for the reference allele GG genotype was observed in 100% of participants across all groups examined. Absence of GPR126 [c.3086 G > A] polymorphism indicates no association with susceptibility to periodontitis in a Greek cohort and periodontally compromised cases of European ancestry. This is the first focused report evaluating the presence of this polymorphism in periodontitis patients in a European population. Further genome-wide studies in larger sample and diverse populations are warranted to fully elucidate the potential role of GPR126 polymorphisms in periodontal disease susceptibility.
Keywords: Periodontitis, GPR126 gene, Polymorphism
Background
Periodontitis is a complex inflammatory disease characterised by the progressive destruction of the supporting tissues around teeth and tooth loss if left untreated [1]. Apart from the well-established role of the periodontopathogens, individuals’ susceptibility to the periodontal disease may be genetically determined [2, 3]. A number of studies have been conducted to investigate the possible role of genetic polymorphisms in the pathogenesis of periodontitis, especially of genes of immunoregulatory molecules, such as interleukins, chemokines and membrane receptors [4].
G protein-coupled receptors (GPCRs) represent the largest and most diverse family of membrane receptors in vertebrates. GPCRs transduce a great variety of extracellular messages such as hormones, growth factors, neurotransmitters and sensory messages of light and odors and subsequently trigger second messenger cascade mechanisms from the adjacent microenvironment or by other cells [5]. These receptors are grouped in five distinct families, including glutamate, rhodopsin, adhesion, frizzled/taste2, and secretin [6]. The adhesion family of GPCRs (aGPCR) contains the typical heptahelical domain and is characterized by a conserved GPCR proteolytic site and a long extracellular glycosylated N-terminal region with adhesion-like motifs, which promote cell-to-cell and cell-to-matrix interactions [7].
GPR126, a member of the aGPCR family, is encoded by the ADGRG6 gene, is located at 6q24.2, consists of 26 exons [8] and was first described by Fredriksson et al. [9]. The precise function of GPR126 is not yet fully understood, however it seems that this receptor participates in various developmental processes, including the formation of the segmental body plan, trabeculation of the developing heart and formation of the semicircular canals of the inner ear [10–12]. It is considered a determining factor in endothelial cell biology and angiogenesis, in stimulating vascular endothelial growth factor (VEGF) signaling [13]. Furthermore, GPR126 is required for myelination of nerve axons by Schwann cells [14]. Along with a significant number of G protein-coupled receptors family members [15], GPR126 was until recently considered an orphan receptor, but it has been shown that collagen type IV [16], laminin-211 [17] and the prion protein [18] are activating ligands of GPR126.
To date, several variations in the GPR126 gene have been described by genome-wide association studies. Single nucleotide polymorphisms (SNPs) of GPR126 have been implicated in adolescent idiopathic scoliosis [19], determination of body and trunk height [20], pulmonary function [21], Gorlin syndrome [22] and arthrogryposis multiplex congenital [23]. Moreover, a GPR126 rs536714306 polymorphism was reported that may be associated with aggressive periodontitis in the Japanese population, indicating possible involvement with mechanisms regarding the homeostasis of the periodontal ligament tissues and suggesting that differences between ethnic populations may be present when it comes to periodontitis-related genetic factor [24].
Based on these findings, we designed a study to investigate whether the GPR126 SNP (rs536714306) was present in Greek individuals with and without periodontitis. The results were then compared with previous genome-wide association studies (GWAS) focusing on cases of European ancestry.
Methods
Study design and sample collection
All subjects participating in this study were referred to the Postgraduate Clinic of the Department of Periodontology, School of Dentistry, National and Kapodistrian University of Athens. All ethical requirements have been fulfilled and all relevant documents are available. Τhe Ethics and Research Committee of the School of Dentistry approved the study protocol (196/01-11-2012). All participants signed an informed consent, according to the general recommendations of the Declaration of Helsinki.
Participants were recruited from 2013 to 2015. They were classified according to the 2018 classification of periodontitis [25] and assigned into two groups: periodontally healthy individuals (H) and individuals with periodontitis (stages I, II, III and IV) (P). All participants were of Caucasian/Greek origin, non smokers and systemically healthy.
Gingival tissue samples and oral mucosa swab samples were obtained from the subjects and were processed for genomic DNA analysis. Briefly, 21 P patients and 12 H subjects, provided gingival tissue samples during conventional periodontal surgery and crown lengthening surgical procedure, respectively. We further recruited 32 P patients and 17 H subjects, who all provided an oral mucosa swab sample, by using a cotton stick against the buccal mucosa for 1 min. Gingival tissue samples were collected during routine periodontal surgical procedures to avoid additional invasive sampling, while buccal mucosa swabs were used in patients not undergoing surgery, in order to provide a non-invasive method for genetic analysis. It is noteworthy that both biological materials (i.e. gingival tissue and oral mucosa swabs) yielded high-quality genomic DNA, suitable for downstream molecular techniques and that the DNA source did not impact the detection of the SNP under investigation [26, 27].
Genotyping
DNA was extracted with the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) following manufacturer’s instructions. The DNA concentration was estimated by measurement of OD260 using a spectrophotometer (BioSpec-nano, Shimadzu, Japan). The target sequence of 454 bp was amplified, using previously reported primers [24]. Polymerase chain reaction (PCR) was carried out in a total volume of 25µL, containing 100ng genomic DNA, 5 pmol of each primer, 0.2 mM of each dNTP, 2.0mM of MgCl2, 2 µL of 10x PCR buffer and 0.5 U Taq DNA polymerase (KAPA Biosystems). Cycling conditions were 2 min at 95 °C followed by 40 cycles of 30 s at 95 °C, 30 s at 57 °C and 40 s at 72 °C, with a final extension of 72 °C for 7 min. PCR products were purified with a PureLink PCR purification kit (Invitrogen Life Technologies).
Direct sequencing
Direct sequencing was carried out on purified PCR products. Sequence data were visualized with Chromas software (version 2.6.4) and analyzed using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology (NCBI).
Data sources
The Gene-Lifestyle Interactions in Dental Endpoints (GLIDE) consortium [28] is a unique collection of epidemiological cohorts with detailed information on clinical diseases and summary statistics for periodontitis. The study, among others, includes 17,353 cases and 28,210 controls, all from European ancestry.
Results
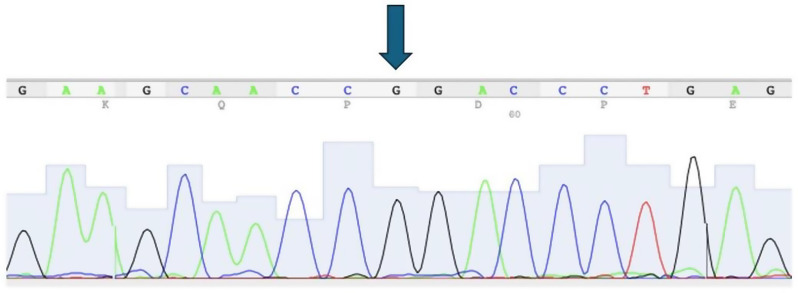
In the present study, 53 patients were diagnosed with periodontitis, and 29 individuals were periodontally healthy. Gender proportion was similar in all study groups. In the total of the 82 participants, the minor allele A was absent in all samples. The homozygous GG genotype (Fig. 1) was observed in 100% of P and H groups (Table 1).
Fig. 1.
Sequence chromatogram
Table 1.
Frequency distribution of alleles
|
GPR126 [c.3086] GG genotype |
GPR126 [c.3086] GA / AA genotype |
|
|---|---|---|
| Periodontitis (n = 53) | 53 (100%) | 0 |
| Healthy periodontium (n = 29) | 29 (100%) | 0 |
Similar results were obtained when exploring the GLIDE database, indicating that a correlation between the development of periodontitis and the presence of the GPR126 rs536714306 polymorphism could not be established, at least in European populations.
The arrow indicates the G allele of the GG genotype of GPR126 [c.3086 G > A] in a P patient.
Discussion
The precise role of GPR126 in periodontal tissues remains unclear. In vitro studies have shown that wild-type GPR126 significantly increases the mRNA expression of bone sialoprotein, osteopontin, and Runx2 genes [24]. This effect is mediated through GPR126-induced upregulation of bone morphogenetic protein-2, inhibitor of DNA binding (ID) 2, and ID4 expression. These findings suggest that GPR126 may play a crucial role in maintaining periodontal ligament tissue homeostasis through the cytodifferentiation of human periodontal ligament (HPDL) cells [24]. Notably, the rs536714306 variant appears to negate these effects. In addition, GPR126’s unique combination of adhesion domains, including CUB (C1r-C1s, Uegf and Bmp1) and pentraxin domains [29], which are components of complement activation and acute phase plasma proteins respectively, suggests a potential role in innate immunity and inflammation [30]. This multifaceted functionality underscores the complexity of GPR126’s potential involvement in periodontal health and disease.
The present study investigated the potential association between the GPR126 SNP rs536714306 [c.3086 G > A] and periodontitis in a Greek cohort. This missense SNP, located in exon 20, results in an arginine to glutamine substitution at position 1029 between transmembrane helices V and VI. Our analysis revealed no SNP alterations in any of the samples, with the homozygous for the reference allele GG genotype consistently present across all groups (P and H). These findings contrast with those of Kitagaki et al. [24], who reported a minor A allele frequency of 2.44% in Japanese patients with aggressive periodontitis, compared to 0.27% in controls.
Our research work represents the first examination focusing specifically on the GPR126 SNP in a European population with periodontitis. An analysis of the NCBI dbSNP repository (https://www.ncbi.nlm.nih.gov/snp/rs536714306) revealed intriguing population differences in the prevalence of this variant. East Asian populations, particularly Japanese and Korean, show a higher prevalence of the variant compared to European and mixed populations. The variant appears to be extremely rare or absent in populations of African descent. The discrepancies between our findings and those in Asian populations highlight the importance of considering ethnic backgrounds in genetic risk factors for periodontitis. As some genes identified in Western genome-wide association studies are not detected in Asian GWAS, there may be ethnicity-specific genetic risk factors for the initiation and progression of periodontitis [24, 31, 32].
This study has potential limitations. A larger pool of periodontal compromised patients and healthy subjects would support our finding, if verified. Moreover, our analysis of the GPR126 gene could have been extended throughout the whole gene and not be limited in detecting the rs536714306 polymorphism, with possible outcome the discovery of one or more polymorphisms, since GPR126 seems to be involved in mechanisms that could affect the periodontal health.
However, while our sample size is relatively small, this study provides solid, albeit negative, results. These findings contribute to the emerging field of precision dentistry, which aims to tailor diagnostics and therapy to individual patients. By leveraging diverse data sources and rigorously testing prediction models, we can move towards more personalized oral healthcare [33].
Conclusions
In conclusion, although this study found no presence of the GPR126 SNP rs536714306 in periodontal patients in our Greek cohort, nor in periodontal cases of European ancestry, it serves as an additional step towards further investigation of GPR126 rs536714306 in diverse populations worldwide. Extensive genome-wide association studies in Caucasian populations are warranted to further explore the potential role of this polymorphism in periodontal pathogenesis and its association with periodontal tissues.
Acknowledgements
The authors would like to thank all the participants in this research for their generous contribution. Additionally, the authors would like to express sincere thanks to Professor Phoebus Madianos and the Department of Periodontology, along with all its personnel, for the valuable support during the patients’ recruitment. Last, but not least, the authors would like to thank Vasiliki Papantoniou for her technical assistance.
Abbreviations
- GPCRs
G protein-coupled receptors
- GPR126
G protein-coupled receptor 126
- VEGF
Vascular endothelial growth factor
- SNP
Single nucleotide polymorphism
- GLIDE
Gene-Lifestyle Interactions in Dental Endpoints
- H
Periodontally healthy individuals
- P
Periodontal patients
- PCR
Polymerase chain reaction
- BLAST
Basic Local Alignment Search Tool
- NCBI
National Center for Biotechnology
- GWAS
Genome-wide association studies
Author contributions
E.C., G.F. and H.V. conceived the study; E.C., G.F., N.P. and X.D. contributed to data collection and analysis; E.C. and G.F. wrote the draft of the paper; I.C. and H.V. revised and refined the manuscript; I.C., X.D. and H.V. contributed to data interpretation; All authors approved the final draft and accepted the decision to submit the manuscript for publication.
Funding
This paper was partially supported by the School of Dentistry, National and Kapodistrian University of Athens research fund and T. Koulourides Foundation.
Data availability
Sequence data that support the findings of the current study are available in the GenBank repository, GenBank accession numbers PQ509770-PQ509851.
Declarations
Ethics approval and consent to participate
All participants signed an informed consent, according to the general recommendations of the Declaration of Helsinki. Τhe Ethics and Research Committee of the School of Dentistry approved the study protocol (196/01-11-2012). All related documents are available on request.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Eirini Chatzopoulou and Galinos Fanourakis contributed equally to this work.
References
- 1.Van Dyke TE, Sheilesh D. Risk factors for periodontitis. J Int Acad Periodontol. 2005;7:3–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Laine ML, Crielaard W, Loos BG. Genetic susceptibility to periodontitis. Periodontol 2000. 2012;58:37–68. [DOI] [PubMed] [Google Scholar]
- 3.Nibali L, Bayliss-Chapman J, Almofareh SA, Zhou Y, Divaris K, Vieira AR. What is the heritability of periodontitis? A systematic review. J Dent Res. 2019;98:632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Sun X, Xiao L, Xie C, Xuan D, Luo G. Gene polymorphisms and periodontitis. Periodontol 2000. 2011;56:102–24. [DOI] [PubMed] [Google Scholar]
- 5.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. [DOI] [PubMed] [Google Scholar]
- 6.Fredriksson R, Lagerström MC, Lundin L-G, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnadóttir TK, Geirardsdóttir K, Ingemansson M, Mirza MAI, Fredriksson R, Schiöth HB. Identification of novel splice variants of adhesion G protein-coupled receptors. Gene. 2007;387:38–48. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Huo A, Li M, Wang J, Yin Q, Chen L, et al. Structure, ligands, and roles of GPR126/ADGRG6 in the development and diseases. Genes Dis. 2024;11:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fredriksson R, Gloriam DEI, Höglund PJ, Lagerström MC, Schiöth HB. There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem Biophys Res Commun. 2003;301:725–34. [DOI] [PubMed] [Google Scholar]
- 10.Glenn TD, Talbot WS. Analysis of Gpr126 function defines distinct mechanisms controlling the initiation and maturation of Myelin. Dev. 2013;140:3167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waller-Evans H, Prömel S, Langenhan T, Dixon J, Zahn D, Colledge WH, et al. The orphan adhesion-GPCR GPR126 is required for embryonic development in the mouse. PLoS ONE. 2010;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng F-S, Abbas L, Baxendale S, Holdsworth CJ, Swanson AG, Slanchev K, et al. Semicircular Canal morphogenesis in the zebrafish inner ear requires the function of gpr126 (lauscher), an adhesion class G protein-coupled receptor gene. Development. 2013;140:4362–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui H, Wang Y, Huang H, Yu W, Bai M, Zhang L, et al. GPR126 protein regulates developmental and pathological angiogenesis through modulation of VEGFR2 receptor signaling. J Biol Chem. 2014;289:34871–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, Dominguez C, et al. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325:1402–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jobe A, Vijayan R. Orphan G protein-coupled receptors: the ongoing search for a home. Front Pharmacol. 2024;15:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paavola KJ, Sidik H, Zuchero JB, Eckart M, Talbot WS. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci Signal. 2014;7:ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen SC, Luo R, Liebscher I, Giera S, Jeong S-J, Mogha A, et al. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron. 2015;85:755–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Küffer A, Lakkaraju AKK, Mogha A, Petersen SC, Airich K, Doucerain C, et al. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature. 2016;536:464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kou I, Takahashi Y, Johnson TA, Takahashi A, Guo L, Dai J, et al. Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat Genet. 2013;45:676–9. [DOI] [PubMed] [Google Scholar]
- 20.Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, Hammond N et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5. [DOI] [PMC free article] [PubMed]
- 21.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu N, Wang J, Zhu B, Zhang M, Qi F, Wang X, et al. Whole-exome sequencing to identify novel mutations of nevoid basal cell carcinoma syndrome in a Chinese population. Cancer Biomark. 2017;21:161–8. [DOI] [PubMed] [Google Scholar]
- 23.Ravenscroft G, Nolent F, Rajagopalan S, Meireles AM, Paavola KJ, Gaillard D, et al. Mutations of GPR126 are responsible for severe arthrogryposis multiplex congenita. Am J Hum Genet. 2015;96:955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitagaki J, Miyauchi S, Asano Y, Imai A, Kawai S, Michikami I, et al. A putative association of a single nucleotide polymorphism in GPR126 with aggressive periodontitis in a Japanese population. PLoS ONE. 2016;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonetti MS, Greenwell H, Kornman KS. Staging and grading of periodontitis: framework and proposal of a new classification and case definition. J Clin Periodontol. 2018;45:S149–61. [DOI] [PubMed] [Google Scholar]
- 26.Pearce DL, Edson JE, Jennelle CS, Walter WD. Evaluation of DNA yield from various tissue and sampling sources for use in single nucleotide polymorphism panels. Sci Rep. 2024;14:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hose L, Schürmann M, Mennebröcker I, Kim R, Busche T, Goon P, et al. Characterization of non-invasive oropharyngeal samples and nucleic acid isolation for molecular diagnostics. Sci Rep. 2024;14:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shungin D, Haworth S, Divaris K, Agler CS, Kamatani Y, Keun Lee M et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat Commun. 2019;10. [DOI] [PMC free article] [PubMed]
- 29.Moriguchi T, Haraguchi K, Ueda N, Okada M, Furuya T, Akiyama T. DREG, a developmentally regulated G protein-coupled receptor containing two conserved proteolytic cleavage sites. Genes Cells. 2004;9:549–60. [DOI] [PubMed] [Google Scholar]
- 30.Stehlik C, Kroismayr R, Dorfleutner A, Binder BR, Lipp J. VIGR - A novel inducible adhesion family G-protein coupled receptor in endothelial cells. FEBS Lett. 2004;569:149–55. [DOI] [PubMed] [Google Scholar]
- 31.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaithilingam RD, Safii SH, Baharuddin NA, Ng CC, Cheong SC, Bartold PM, et al. Moving into a new era of periodontal genetic studies: relevance of large case-control samples using severe phenotypes for genome-wide association studies. J Periodontal Res. 2014;49:683–95. [DOI] [PubMed] [Google Scholar]
- 33.Schwendicke F, Krois J. Precision dentistry—what it is, where it fails (yet), and how to get there. Clin Oral Investig. 2022;26:3395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data that support the findings of the current study are available in the GenBank repository, GenBank accession numbers PQ509770-PQ509851.