Abstract
Microbial communities in hydrothermally active sediments of the Guaymas Basin (Gulf of California, Mexico) were studied by using 16S rRNA sequencing and carbon isotopic analysis of archaeal and bacterial lipids. The Guaymas sediments harbored uncultured euryarchaeota of two distinct phylogenetic lineages within the anaerobic methane oxidation 1 (ANME-1) group, ANME-1a and ANME-1b, and of the ANME-2c lineage within the Methanosarcinales, both previously assigned to the methanotrophic archaea. The archaeal lipids in the Guaymas Basin sediments included archaeol, diagnostic for nonthermophilic euryarchaeota, and sn-2-hydroxyarchaeol, with the latter compound being particularly abundant in cultured members of the Methanosarcinales. The concentrations of these compounds were among the highest observed so far in studies of methane seep environments. The δ-13C values of these lipids (δ-13C = −89 to −58‰) indicate an origin from anaerobic methanotrophic archaea. This molecular-isotopic signature was found not only in samples that yielded predominantly ANME-2 clones but also in samples that yielded exclusively ANME-1 clones. ANME-1 archaea therefore remain strong candidates for mediation of the anaerobic oxidation of methane. Based on 16S rRNA data, the Guaymas sediments harbor phylogenetically diverse bacterial populations, which show considerable overlap with bacterial populations of geothermal habitats and natural or anthropogenic hydrocarbon-rich sites. Consistent with earlier observations, our combined evidence from bacterial phylogeny and molecular-isotopic data indicates an important role of some novel deeply branching bacteria in anaerobic methanotrophy. Anaerobic methane oxidation likely represents a significant and widely occurring process in the trophic ecology of methane-rich hydrothermal vents. This study stresses a high diversity among communities capable of anaerobic oxidation of methane.
Deep-sea hydrothermal vent sites encompass very diverse environments for microbial life, such as black smoker chimney walls and orifices, warm vents and seeps, symbiont-harboring invertebrates, surface-attached microbial mats, and floating particles in vent plumes. Hydrothermally heated sediments characterize the Guaymas Basin hydrothermal vent site in the Gulf of California. The basin has a very high sedimentation rate (1 to 2 mm/year) due to high biological productivity in surface waters and a large terrigenous input. Consequently, the Guaymas Basin vent sites are covered with, on average, 100-m- and up to 500-m-thick layers of organic-rich, diatomaceous sediments (11, 73). Chemical interactions between hydrothermal vent fluids and sediment accelerate diagenetic processes such as metal sulfide precipitation and high-temperature pyrolysis of organic material (43) and create chemical conditions that differ significantly from those at the more common bare lava vent sites. The metal content of the vent fluids is reduced by several orders of magnitude (85). Diffuse venting through the sediments at temperatures up to 200°C releases vast quantities of petroleum hydrocarbons, short-chain organic acids, and ammonia via pyrolysis of complex organic substrates (4, 52, 86). The methane content in the vent fluids is in the range of 12 to 16 mM (270 to 370 ml kg−1 at standard temperature and pressure), which is approximately 2 orders of magnitude higher than those of most bare lava vent sites (87). The carbon isotopic composition (δ-13C) of the methane ranges from −51 to −43‰, suggesting a thermocatalytic origin from sedimentary organic material (87). The high concentration and relatively low isotopic composition of methane differ from those of most other mid-ocean ridge systems, where methane originates from high-temperature reactions involving H2 and CO2, and from mantle outgassing (17).
This metabolic menu of Guaymas vent fluids and sediments supports unusually rich microbial diversity in all three domains of life. A large variety of unusual bacteria, archaea, and eukaryotes have been isolated or described from the Guaymas Basin. The archaea include hyperthermophilic archaeal sulfate reducers of the genus Archaeoglobus (10) and hyperthermophilic methanogens of the genera Methanopyrus and Methanococcus (36, 40). Many bacterial populations that were found at the Guaymas Basin have adapted to the petroleum hydrocarbon-rich environment: for example, anaerobic sulfate-reducing bacteria that oxidize alkanes, aromatic compounds, and fatty acids (69) and aerobic, aromatic hydrocarbon-degrading bacteria with a preference for aromatic carboxylic acids (26). The sulfidic vent fluids and high sulfate reduction rates fuel massive bacterial mats of autotrophic, sulfur-oxidizing, filamentous Beggiatoa spp. (30, 57). The first comparative survey of cultivable eukaryotic protists at Guaymas and other Pacific Ocean vent sites (2) was complemented by a eukaryotic 18S rRNA census of Guaymas that revealed novel, in part deeply branching eukaryotic lineages (21). However, the possibility that methane could sustain extensive microbial communities at Guaymas Basin vent sediments has not yet been investigated.
In this study, we have combined bacterial and archaeal 16S rDNA sequence surveys with structural and isotopic analyses of archaeal and bacterial lipids to constrain structural and metabolic features of the microbial community.
MATERIALS AND METHODS
Sampling sites and characteristics.
Sediment cores were retrieved during dives with the research submersible Alvin (Woods Hole Oceanographic Institution) on a cruise to the Guaymas Basin in April and May 1998. Two sediment cores were investigated in detail. Core A (Alvin dive 3203, 26 April 1998; diameter = 15 cm, length = 35 cm) from the Everest Mound area in the southern Guaymas vent field (27°1′388"N, 111°24′112"W) was covered with an orange Beggiatoa spp. mat, indicating a flow of sulfide from the sediment that sustains these chemolithoautotrophic sulfide oxidizers. The sediment consisted of very soft olive-green ooze in the surface layers and, below a depth of 7 cm, became grayish and developed a petroleum-like sheen. Below 10 cm, gas bubbles were noted after recovery. During Alvin dive 3207 (30 April 1998) at the Orpheus site (27°0′435"N, 111°24′612"W), core C, a 6-inch-diameter core without a Beggiatoa mat, was selected as a contrasting sample to core A. The upper 6 cm consisted of a black layer of liquid petroleum ooze; below 7 cm in depth, the sediment turned olive and became thicker. Gas bubbles in the sediment appeared at a 15-cm depth. In situ temperature profiles of both sediments were determined before coring by using Alvin's thermoprobe; the uppermost 5 cm of each core had a temperature range from cold ambient seawater to moderately hot vent fluids (ca. 2°C to 57°C, core C, and 2°C to 74°C, core A). Both cores came from sediments with high rates of sulfate reduction and organic carbon turnover (A. Weber and B. B. Jørgensen, submitted for publication). The cores were brought to the surface no more than 6 h after sampling and were sliced in 1-cm intervals within an additional 6 h after retrieval. Multiple 2-ml portions of sediment were taken from each centimeter layer and immediately frozen at −80°C for nucleic acid extraction and lipid analysis onshore. A sediment core covered with a Beggiatoa mat, without a temperature profile (core T), was obtained from the Rebecca's Roost area (27°1′131"N, 111°40′687"W) in the southern Guaymas vent field by the remotely operated vehicle JASON. From core T, sample material for phylogenetic analyses was only available from the 2.5- to 5-cm layer. Lipid analyses of an aliquot of this layer revealed a high degree of similarity of the biomarker distribution to the overlying 0- to 2.5-cm layer. Isotope data of individual compounds were not determined for the 2.5- to 5-cm layer.
Nucleic acid isolation.
A bead-beating procedure combined with hot phenol extraction was used to isolate high-molecular-weight DNA and rRNA. The following protocol is a modification of the method used by Stahl and coworkers (74). Sediment (0.5 g) was mixed with 895 μl of lysis buffer (50 mM Tris [pH 8], 25% sucrose), 62.5 μl of 20% (wt/vol) sodium dodecyl sulfate solution, and 0.5 g of baked zirconium beads (0.1-mm diameter). The sample was subjected to 15 s of bead-beating by using a Mini-beadbeater (BioSpec Products, Bartlesville, Okla.). After the bead-beating step, 42 μl of a solution consisting of 20 mg of proteinase K/ml was added, followed by 60 min of incubation at 55°C under constant agitation. The sample (total volume, approximately 1.6 ml) was divided into two equal aliquots, mixed with equal volumes of a phenol-chloroform-isoamyl alcohol mixture (25:24:1 [vol/vol/vol]; pH 7.9), and incubated at 55°C for 10 min. After centrifugation, the aqueous phases (volume, approximately 0.8 ml each) were transferred to two new vials. The two organic phases were mixed with 200 μl of Tris-EDTA buffer each (pH 7.5), incubated at 55°C for 10 min, and centrifuged, and the aqueous extracts were combined with the previous aqueous phases. Both combined aqueous phases were extracted two to three times more with a phenol-chloroform-isoamyl alcohol mixture as described above. The two samples were recombined in a 15-ml Falcon tube (total volume, approximately 4 ml) and mixed with 0.4 ml of a 3 M sodium acetate solution and 8 ml of ethanol. Nucleic acids were precipitated overnight at −20°C and centrifuged at 8,000 rpm in a Sorvall RC2-B centrifuge for 40 min at 4°C in baked 50-ml Corex glass tubes. The pellet was washed with 70% ethanol, dried, taken up in 100 μl of Tris-EDTA (pH 7.5), transferred to a microcentrifuge tube, and stored at −80°C.
16S rRNA gene amplification, cloning, and sequencing.
Near-complete 16S rRNA genes were PCR amplified on a Gene-Amp PCR Cycler 9700 (Perkin-Elmer) by using archaeal primers ARC-8F (5′-TCCGGTTGATCCTGCC-3′) and ARC-1492R (5′-GGCTACCTTGTTACGACTT-3′) and bacterial primers BAC-8F (5′-AGRGTTTGATCCTGGCTCAG-3′) and BAC-1492R (5′-CGGCTACCTTGTTACGACTT-3′). The archaeal PCR primers were checked on the basis of a comprehensive Ribosomal Database Project archaeal alignment to make them as unbiased as possible for use in multitemplate PCR. The 8F archaeal primer was terminated at position 23, thus avoiding ambiguities in archaeal 16S rRNA sequences (positions 24 to 27) that would otherwise form a source of PCR bias (66). This primer was used together with the reverse archaeal primer, 1492R, which also did not contain ambiguities. The bacterial primers were checked in the same way, with a general bacterial alignment of near-complete bacterial 16S sequences that included 3′ and 5′ regions. Each PCR cycle (30 total) consisted of a 1-min denaturation step at 94°C, a 1-min annealing step at 55°C, and a 2-min elongation step at 72°C. PCR products were checked on an agarose gel, the bands were cut out, and PCR products were extracted with the Prep-A-Gene DNA purification kit (Bio-Rad, Hercules, Calif.), following the manufacturer's protocols. The PCR product was resuspended in 10 μl of 10 mM Tris (pH 7.5). The purified PCR products were a-tailed as follows, to improve cloning efficiency: approximately 50 ng of purified PCR product was mixed with 5 μl of 10× PCR buffer [200 mM Tris (pH 8.55), 100 mM (NH4)2SO4, 20 mM MgCl2, 300 mM KCl, 0.1% (wt/vol PCR buffer) gelatin, 0.5% (vol/vol PCR buffer) NP-40], 5 μl of deoxynucleoside triphosphate (2 mM), and 1 U of Promega Taq DNA polymerase in a 50-μl total volume. The mixture was incubated at 72°C for 10 min, extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and centrifuged for 5 min. The aqueous top layer was then transferred to a fresh microcentrifuge tube. The a-tailed PCR product was precipitated with 10% (vol/vol aqueous layer) 3 M sodium acetate and 2 volumes of 100% ethanol, washed with 70% ethanol, and resuspended in 4 μl of distilled water. The PCR products were cloned by using the TOPO XL PCR cloning kit (Invitrogen Corp., Carlsbad, Calif.) and transformation by electroporation according to the manufacturer's specifications.
Sequence data were gathered on a LI-COR 4200L automated sequencing apparatus by using infrared dye-labeled T7F and M13R primers (LI-COR, Lincoln, Nebr.) and a cycle sequencing protocol (Sequitherm; Epicenter Inc., Madison, Wis.) according to the manufacturers' instructions. This system stores the sequence results as image files for subsequent base determination and sequence assembly. The 3′ and 5′ ends of each PCR product were covered by one sequence read each; the central 0.6- to 0.9-kb portion was covered double-stranded. Base calls and PHD quality values were determined by using e.Seq (Build 1.0.52) DNA sequence analysis software from LI-COR. For each individual clone, forward and reverse reads were assembled by using Phrap version 0.960731 (28).
Phylogenetic analysis.
Sequence data were compiled by using ARB software (www.arb-home.de) and aligned with sequences obtained from the GenBank database by using the ARB FastAligner utility. Resulting alignments were manually verified against known secondary structure regions. Representative clones (determined by neighbor-joining clustering patterns) were selected for further analyses. We limited comparisons to 1,353, 1,352, and 1,432 sites that could be unambiguously aligned according to conservation of primary and secondary structures for the nonproteobacterial, proteobacterial, and archaeal data sets, respectively. Alignments and sites included in our analyses are available upon request.
Analyses were performed by using minimum evolution and parsimony methods implemented in PAUP* version 4.08b (76). Heuristic searches under minimum evolution criteria used 1,000 random-addition replicates per data set, each followed by tree bisection-reconnection topological rearrangements. Models for use in minimum evolution analyses of the archaeal, proteobacterial, and nonproteobacterial data sets were evaluated using the likelihood ratio test implemented in Modeltest v.3.0 (67). For all three data sets, all models tested that were simpler than the general time reversible (GTR) + invariant sites (I) + gamma model had a significantly poorer fit to the data for the reference topology (based on a neighbor-joining tree according to Jukes-Cantor). Therefore, minimum evolution distance trees were inferred by using a GTR model with six classes of substitutions, unequal base frequencies, proportion of invariant sites, and the evolutionary rate of the remaining portions of sites differing according to a gamma distribution (with shape parameter α). Model parameters estimated for minimum evolution analyses for the archaeal, proteobacterial, and nonproteobacterial datasets were α = 0.7636, 0.6718, and 0.8071 and I = 0.1902, 0.3858, and 0.1997, respectively. Bootstrapping for minimum evolution and parsimony was done with 1,000 replicates per data set.
Analysis of sedimentary microbial lipids.
Lipid analyses were carried out by following a procedure reported earlier (32, 59). Sediment samples (∼0.6 to 0.7 g dry weight, 22 g for core T, 0- to 2.5-cm layer) were sterilized by ultrasonication (ultrasonic bath) in ∼20 ml of a mixture of dichloromethane and methane (9:1 [vol/vol]) for ∼30 min. After evaporation of the residual solvent, the sediments were oven dried at ∼50°C. Free lipids were extracted from dried and homogenized sediments by using a DIONEX Accelerated Solvent Extraction 200 system at 100°C and 1,000 lb/in2, with dichloromethane (DCM)-methanol (90:10 [vol/vol]) as the solvent. Extracts were separated into four fractions of increasing polarity by using SUPELCO LC-NH2 glass cartridges (500 mg of sorbent) and a sequence of solvent mixtures of increasing polarity (hydrocarbons, 4 ml of n-hexane; ketone-esters, 6 ml of 3:1 [vol/vol] n-hexane-DCM; alcohols, 7 ml of 9:1 [vol/vol] DCM-acetone; carboxylic acids, 8 ml of 2% formic acid in DCM). Individual compounds were quantified and identified by using an HP 6890 gas chromatograph (GC) equipped with a J&W DB-5 capillary column (60-m length, 0.32-mm inner diameter, 0.25-μm film thickness) and coupled to an HP 5973 mass-selective detector and a flame ionization detector. Stable carbon isotopic compositions of individual compounds were determined by using a Finnigan Delta Plus mass spectrometer coupled to an HP 6890 GC and equipped with a CP-Sil 5 column (60-m length, 0.25-mm inner diameter, 0.25-μm film thickness). Column temperatures of both GC systems were programmed to increase from 40°C (1 min, isothermal) to 130°C at a rate of 20°/min and then to 320°C (60 min, isothermal) at a rate of 4°C/min. Alcohols were analyzed for their trimethylsilyl ethers and esters, respectively, after reaction with a N,O-bis(trimethylsilyl)trifluoroacetamide reagent (BSTFA + 1% trimethylchlorosilane) in a 1:1 mixture of DCM and pyridine. Reported stable carbon isotopic values are means of at least two analyses and have been corrected for the presence of carbon atoms added during derivatization. Differences between individual analyses were mostly less than 1‰, with the exception of core T, which was strongly affected by an unresolved complex mixture (UCM) related to hydrothermally generated petroleum. Quantification of individual compounds was based on their flame ionization detector signal relative to that of known amounts of internal standards.
Nucleotide sequence accession numbers.
The Guaymas 16S rRNA gene sequences have the following GenBank accession numbers: archaea, AF419624 to AF419656; proteobacteria, AF420334 to AF420370; all other bacteria, AF419657 to AF419699. Sequences were checked for chimera formation by using the Chimera check program of the Ribosomal Database Project II database; three chimeric bacterial sequences were found and removed from the analysis (51).
RESULTS
Archaeal and bacterial lipids and biomarkers.
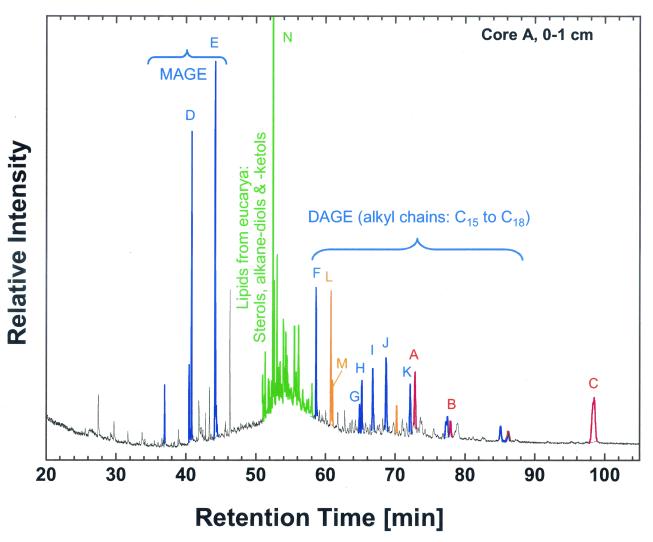
The archaeal lipids archaeol, diagnostic for nonthermophilic members of the Euryarchaeota, and sn-2-hydroxyarchaeol, predominantly occurring in cultured members of the Methanosarcinales, were found in very high abundance in the surface layers of cores A and T (Table 1). To date, observations of similarly high accumulations of these two compounds have been limited to mud volcano sites in the Mediterranean (61). In contrast, cold seep sites from the California and Oregon continental margin exhibited lower concentrations (7, 32), i.e., in a range similar to those observed in core C. The stable carbon isotopic compositions of these compounds (δ-13C, −89 to −58‰) are indicative of an origin from oxidation and assimilation of 13C-depleted methane (Fig. 1). In addition, cores A and T contain very significant amounts of an additional compound of archaeal origin, a biphytanediol with two pentacyclic rings (http://docserver.bis.uni-oldenburg.de/publikationen/dissertation/2001/ruetra01/ruetra01.html) (70) with a 13C depletion similar to those of archaeol and hydroxyarchaeol (Table 1 and Fig. 1).
TABLE 1.
Stable carbon isotopic compositions and concentrations of selected biomarkers from archaea, bacteria, and eukaryotes
| Compounda | Symbol (Fig. 1)c | Source | δ13C/abundance (μg/g dry sediment)
|
|||
|---|---|---|---|---|---|---|
| Core A (layer 0-1 cm) | Core A (layer 1-2 cm) | Core C (layer 0-1 cm)b | Core T (layer 0-2.5 cm)b | |||
| Archaeol | A | Archaea | −58‰/22 | −60‰/20 | −81‰/1.4 | −72‰/18 |
| sn-2-Hydroxyarchaeol | B | Archaea | −70‰/6 | nd/1 | −81‰/1.5 | −89‰/14 |
| Bicyclic diphytanediol | C | Archaea | −68‰/54 | −68‰/18 | Not present | −66‰/31 |
| MAGE-C16 | D | Bacteria | −41‰/26 | −41‰/3 | Not present | UCM/2 |
| MAGE-C18:1 | E | Bacteria | −38‰/34 | −37‰/3 | Not present | UCM/5 |
| DAGE-C30:1 | F | Bacteria | −28‰/21 | −33‰/3 | Not present | UCM/1 |
| DAGE-C16/17 | G | Bacteria | −40‰/7 | −40‰/7 | Not present | −34‰/3 |
| DAGE-C16/17 | H | Bacteria | −45‰/12 | −47‰/10 | Not present | −44‰/3 |
| DAGE-C17/17 | I | Bacteria | −46‰/19 | −51‰/13 | Not present | −53‰/2 |
| DAGE-C17/17 | J | Bacteria | −45‰/23 | −49‰/26 | Not present | −43‰/2 |
| DAGE-C17/18 | K | Bacteria | −44‰/18 | −49‰/27 | Not present | nd/3 |
| Hopan-29-ol | L | Bacteria | −29‰/18 | −32‰/20 | nd/1.4 | −35‰/8 |
| Hopan-22-ol | M | Bacteria | −37‰/7 | −36‰/5 | nd/0.5 | −34‰/6 |
| Cholesterol | N | Eukarya | −21‰/64 | −28‰/73 | −25‰/6 | UCM/17 |
C16 designates the carbon number of the alkyl moiety. Carbon numbers of ether-bound alkyl rests are assigned as follows: C30:1 indicates the total carbon number of the two alkyl groups, in this example with a total of one degree of unsaturation, i.e., a double bond or a ring; C16/17 indicates a hexadecylether moiety bound to the sn-1 glycerol carbon and a heptadecylether moiety at sn-2.
nd, not determined; UCM, not determined due to unresolved complex mixture.
Symbols refer to Fig. 1.
FIG. 1.
Reconstructed ion chromatogram of alcohol fraction from core A sample, 0 to 1 cm. Colors of peaks designate their biological sources: red, methanotrophic archaea; blue, presumed bacterial members of methanotrophic community; orange, aerobic bacteria; green, eucaryotes, e.g., marine algae. Refer to Table 1 for further details.
Both cores A and T contain abundant bacterial lipid biomarkers, namely, monoalkylglycerol ethers (MAGE) and dialkylglycerol ethers (DAGE) (Fig. 1 and Table 1). Related and identical compounds had been observed earlier in sediments hosting active methanotrophic communities (32, 62). Structural analogs of the latter group are known only from the most deeply branching bacteria, Thermodesulfobacterium commune (45) and the Aquificales (37). In both cores, their isotopic compositions (δ-13C values in the range of −50 to −35‰; with the sole exception, that of DAGE-C30:1, at around −30‰) are significantly lower than those of lipids from marine eukaryotic algae (e.g., cholesterol) (Table 1) and bulk organic matter (∼−27‰), a pattern consistent with previous observations from cold methane seeps. In contrast, these compounds were not present in core C.
Archaeal 16S rRNA gene sequence profiles.
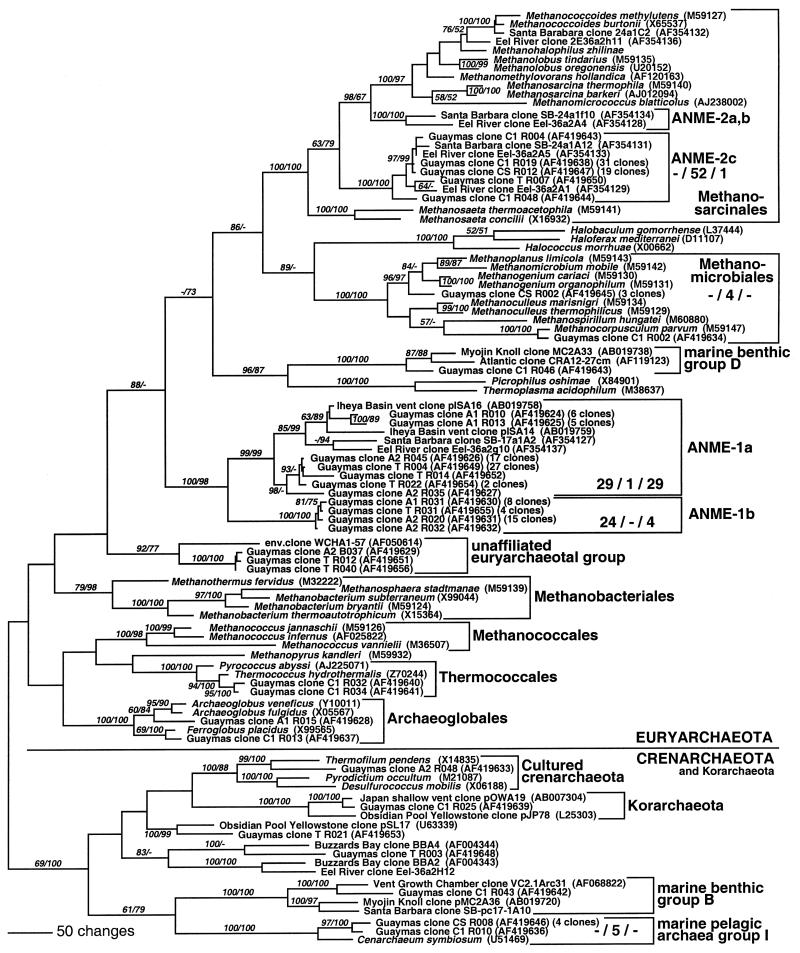
Cores A, T, and C yielded predominantly archaeal 16S rRNA clones of two phylogenetic lineages of uncultured archaea, presumed anaerobic methane-oxidizing archaeal group 1 (ANME-1) and ANME-2 (31, 59). The clones from cores A and T were dominated by members of the ANME-1 cluster, which constituted 95% and 87% of all archaeal clones, respectively. These euryarchaeota were originally suggested as likely mediators of anaerobic methanotrophy (31), but in the past few years, evidence has accumulated mostly for an involvement of the ANME-2 cluster (7, 59, 60). The ANME-1 group clones obtained from the Guaymas Basin showed considerable phylogenetic diversity and formed two major lineages. The ANME-1a group, which corresponds to the previously defined ANME-1 cluster (31, 59), has a sister group, ANME-1b, that is so far represented by sequences from the Guaymas Basin only. Although the two groups are not closely related to each other (83 to 87% sequence similarity between ANME-1a and ANME-1b, compared to >89% within ANME-1a members alone), their sister-group relationship is always supported with 98 to 100% bootstrap support. The ANME-1 archaea have no close cultured relatives; in all analyses, they formed an independent phylogenetic cluster among the Euryarchaeota that could not be subsumed under another archaeal group, for example, the methanogenic lineages of Thermoplasma, Thermococcus, or Archaeoglobus (Fig. 2). In contrast to cores A and T, which yielded mostly ANME-1 and ANME-1b archaeal clones (96 and 87% of all archaeal clones obtained from cores A and T), core C predominantly yielded clones of the ANME-2 group of archaea (Fig. 2). In core C, they constitute 76% (52 clones) of all archaeal clones sequenced. The ANME-2 lineages form distinct branches among the cultured members of the Methanosarcinales, of comparable phylogenetic depth to those of the genera Methanosaeta, Methanosarcina, and Methanolobus (Fig. 2). All Guaymas Basin ANME-2 clones fell into the subgroup ANME-2c of the ANME-2 cluster (59), with mutual sequence similarity greater than 95%. The Guaymas sequences showed lower sequence similarities (87.2 to 89.2%) to the ANME subgroups 2a and 2b, here represented by the Santa Barbara and Eel River sequence clones 24a1f10 and 2E36a2A4. The ancestral node of the ANME-2c subgroup includes not only the ANME-2a and 2b subgroups but also cultured members of the Methanosarcinales, for example, the acetoclastic and methyl-disproportionating genera Methanosarcina and Methanolobus. In other words, the ANME-2 lineages are paraphyletic within the Methanosarcinales (Fig. 2).
FIG. 2.
Distance tree of archaeal clones from Guaymas sediment cores A, C, and T, based on near-complete 16S rDNA sequences. Bootstrap values (in percent) are based on 1,000 replicates each (distance and parsimony) and are shown for branches with more than 50% bootstrap support. The sequence for Methanohalophilus zhilinae was obtained from the ARB sequence server (version May 2001). Crenarchaeotal sequences SB-pc17-1A10 and Eel-36a2H12 were provided by Victoria Orphan. For the major phylogenetic divisions, the numbers of core A, core C, and core T clones are given in that order. For multiple near-identical clones (>99% identity), the number of multiples is given after the GenBank number of a representative clone. Some sequence types occurred in several sediment layers: Guaymas clone A1 R010 was found four times in the 0- to 1-cm layer of core A and two times in the 1- to 2-cm layer of core A. Guaymas clone A2 R045 was found 15 times in the 0- to 1-cm layer of core A and 2 times in the 1- to 2-cm layer of core A. Guaymas clone T R022 was found one time in the 0- to 1-cm layer of core C and one time in core T.
Besides ANME-2 clones within the Methanosarcinales, the only members of other methane-metabolizing families that were found in the Guaymas sediments belonged to the mesophilic or moderately thermophilic family Methanomicrobiales. Three clones, represented by CSR002, were related to the H2-/CO2- and formate-utilizing genera Methanogenium and Methanomicrobium, and the single clone C1R002 was closely related to the species Methanocorpusculum parvum, which uses H2, formate, and secondary alcohols as electron donors (Fig. 2).
Rarely retrieved archaeal clones.
In addition to the ANME groups, diverse archaeal lineages and relatives of hyperthermophilic cultured archaea were found in the Guaymas sediments, each represented by a few or single clones. Four clones were closely related to the hyperthermophilic, sulfur-reducing thermococcales and the sulfate- and metal-reducing archaeoglobales that have both been cultured previously from Guaymas sediments (10, 39). Two crenarchaeotal clones were related to uncultured hyperthermophilic lineages. Clone TR021 was affiliated to clone pSL17 from the hot, sulfidic spring Obsidian Pool in Yellowstone Park (3). A single clone (C1R025) was a member of the Korarchaeota; its closest, nearly identical relative was clone pOWA19 from a shallow marine hydrothermal vent in Japan (78). The temperatures observed in the studied sediments are below those optimal for hyperthermophiles, which explains the rare occurrences of hyperthermophile-related clones.
Five core C clones, four of them from supernatant seawater, belonged to the pelagic marine crenarchaeal group I that includes Cenarchaeum symbiosum. In this tree, they are represented by clones C1R001 and CSR008 (Fig. 2). These archaea most likely have their origin in the water column, their natural habitat. Other archaea occur in a wide variety of sedimentary habitats that includes vent sites but is not limited to them. Core C harbored members (clones C1R043 and C1R046) of euryarchaeotal lineages that were originally described in a study of deep Atlantic nonhydrothermal sediment, namely, the marine benthic groups B and D (84).
The core A clone A2B037, which was obtained with bacterial primers, and the core T clones TR012 and TR040 formed a phylogenetic lineage among the Euryarchaeota that also included the euryarchaeotal clone WCHA1-57 from the methanogenic zone of an oil- and chlorinated solvent-contaminated aquifer (20). The natural petroleum deposits at Guaymas and the aquifer with anthropogenic pollution both harbor members of this euryarchaeotal lineage that at present cannot be subsumed under any other cultured or uncultured archaeal lineage.
Bacterial 16S rRNA gene sequences.
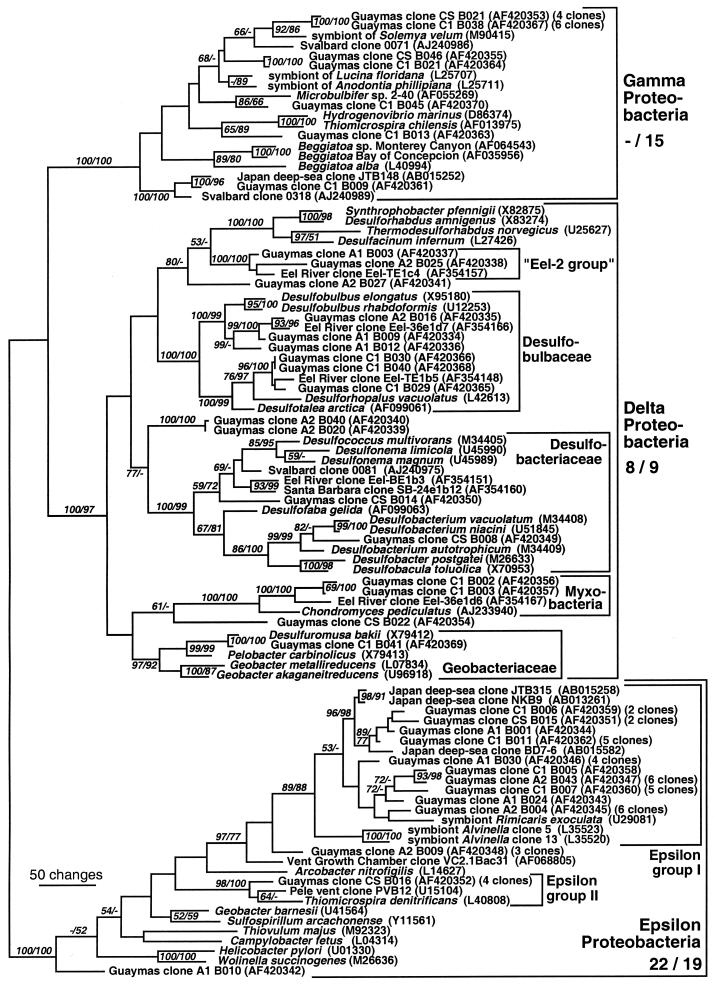
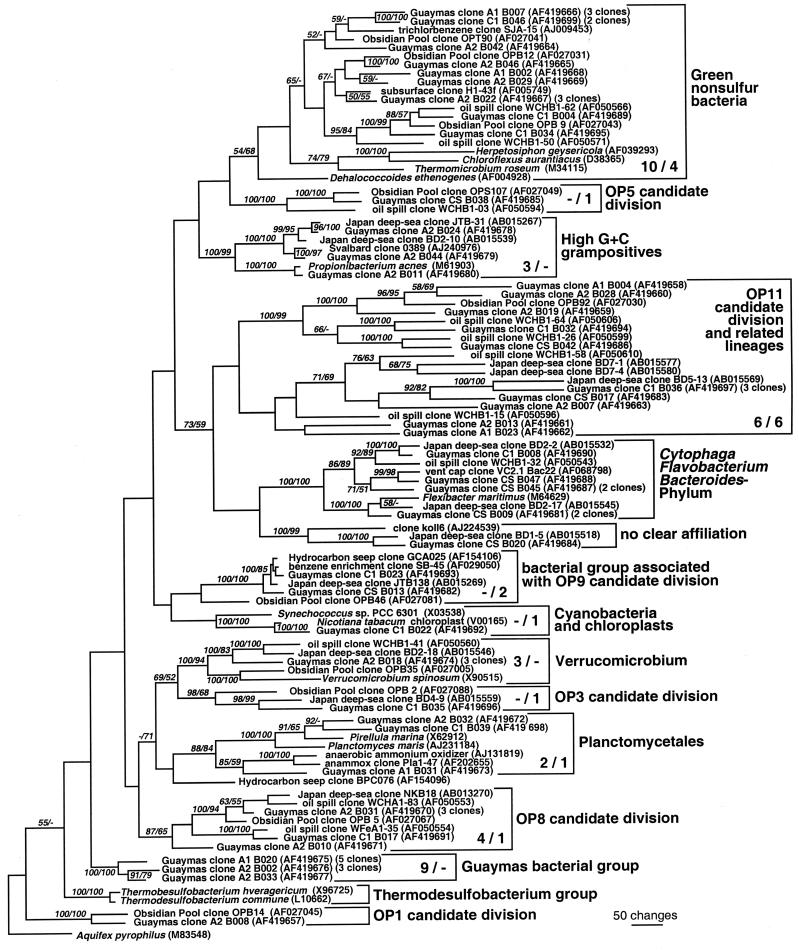
The bacterial clone libraries obtained are phylogenetically highly diversified and include numerous cultured as well as uncultured bacterial lineages (Fig. 3 and 4). In contrast to the archaeal sequences, no single phylogenetic group of bacteria dominated the clone collection. Instead, the four most common bacterial phylotypes that occurred in both cores A and C included members of the epsilon and delta Proteobacteria, the green nonsulfur bacterial phylum, and the candidate division OP11 (Fig. 3 and 4).
FIG. 3.
Distance tree of proteobacterial clones from Guaymas sediment cores A and C, based on near-complete 16S rDNA sequences. Bootstrap values (in percent) are based on 1,000 replicates each (distance and parsimony) and are shown for branches with more than 50% bootstrap support. For the major phylogenetic divisions, the numbers of core A and core C clones are given in that order. For multiple near-identical clones (>99% identity), the number of multiples is given after the GenBank number of a representative clone. Some sequence types occurred in several sediment layers: Guaymas clone A1 B030 was found three times in the 0- to 1-cm layer of core A and one time in the 1- to 2-cm layer of core A, Guaymas clone A2 B043 was found one time in the 0- to 1-cm layer of core A and five times in the 1- to 2-cm layer of core A, and Guaymas clone A2 B004 was found three times in the 0- to 1-cm layer of core A and three times in the 1- to 2-cm layer of core A.
FIG. 4.
Distance tree of bacterial clones, excluding proteobacteria, from Guaymas sediment cores A and C, based on near-complete 16S rDNA sequences. Bootstrap values (in percent) are based on 1,000 replicates each (distance and parsimony) and are shown for branches with more than 50% bootstrap support. For the major phylogenetic divisions, the numbers of core A and core C clones are given in that order. For multiple near-identical clones (>99% identity), the number of multiples is given after the GenBank number of a representative clone. Some sequence types occurred in several sediment layers: Guaymas clone A1 B007 was found two times in the 0- to 1-cm layer and one time in the 1- to 2-cm layer of core A, Guaymas clone A2 B031 was found two times in the 0- to 1-cm layer and one time in the 1- to 2-cm layer of core A, Guaymas clone A1 B020 was found three times in the 0- to 1-cm layer and two times in the 1- to 2-cm layer of core A, and Guaymas clone A2 B002 was found one time in the 0- to 1-cm layer and two times in the 1- to 2-cm layer of core A.
Delta-proteobacterial clones constituted 13% of the bacterial clone libraries of cores A and C (eight and nine clones, respectively); they included members of the propionate-oxidizing family Desulfobulbaceae, members of Myxobacteria, several deeply branching delta-Proteobacteria, and a member of the sulfur-reducing genus Desulfuromusa (Fig. 3). Representatives of the Eel-2 group, an uncultured group of delta-Proteobacteria from Eel River methane seep sediments (59), were also found. In the Eel River as well as the Guaymas Basin sediments, these Eel-2 group delta-proteobacterial clones came from samples that yielded predominantly ANME-1 archaeal clones.
Epsilon-proteobacterial clones constituted the largest segment (28 to 31%) of the bacterial clone library (22 clones in core A and 15 clones in core C). Most epsilon-proteobacterial clones branched with the epsilon 1 group of uncultured epsilon-Proteobacteria (1) from a wide variety of hydrothermal and nonhydrothermal deep-sea sediments (clones NKB8, -9, -10, and -12 in cold seeps of the Nankai Trough [47]; Japanese deep-sea clones BD1-4, BD 2-1, BD2-5, and BD7-6 [48]; and cold seep deep-sea clone JTB315 [49]) and with sulfide-oxidizing exosymbionts of the vent shrimp Rimicaris exoculata (65) and the hydrothermal vent annelid Alvinella pompejana (12). A phylogenetically distinct group of epsilon-Proteobacteria (epsilon group II) (1) contained four Guaymas clones, the nitrate-reducing sulfur oxidizer Thiomicrospira denitrificans, and several clones from Pele's vents near Hawaii (55).
The green nonsulfur bacterial phylum accounted for 6 to 14% of the clones in cores A and C. This bacterial phylum has been named for its anaerobic and thermophilic cultured representatives, the green nonsulfur phototrophic bacteria from terrestrial hot springs, such as Chloroflexus aurantiacus and Herpetosiphon geysericola. The Guaymas clones branched among the uncultivated members of this phylum, clones from Obsidian Pool (38), from subsurface palaeosol (15), from a methanogenic aquifer contaminated with jet fuel waste and aromatic and chlorinated hydrocarbons (20), and from enrichments growing on chlorinated aromatic compounds (89).
Members of the uncultured candidate division OP11 and OP11-affiliated lineages constituted 9% of the clones from cores A and C. The OP11 division was found originally in Obsidian Pool (38), in the methanogenic zone of a jet fuel-contaminated aquifer (20), and in cold deep-sea sediments (48); cultured representatives are not known at present.
Not all bacterial phyla were equally represented in the clone libraries of cores A and C. In particular, core C harbored two specific bacterial populations that were entirely missing from the core A clone library, gamma-Proteobacteria (22%) and members of the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum (9%). Most gamma-proteobacterial clones were related to sulfur-oxidizing bacterial endosymbionts of bivalves at hydrothermal vents and sulfidic mud flats (13). The clones of the CFB phylum, ubiquitous degraders of carbohydrate polymers in the marine environment, were mostly obtained from the supernatant water portion of core C, consistent with their aerobic physiology and their abundance in the marine water column (18).
Core A yielded several clones that belonged to a deeply branching bacterial phylotype (13%), which could not be subsumed under another bacterial group. Depending on the method of phylogenetic analysis, a marginal affiliation with the thermophilic sulfate reducers of the genus Thermodesulfobacterium (uncorrected sequence similarity to Thermodesulfobacterium, ca. 85%) was obtained, but a sister-group relationship, suggested by preliminary ARB parsimony trees, could not be substantiated by bootstrap. No related sequences were available in GenBank. So far, this cluster of sequences has been obtained from Guaymas sediments only (Fig. 4).
Clones affiliated to other bacterial lineages were retrieved in small members, including members of the Verrucomicrobium lineage and Planctomycetales, the high-G+C gram-positive bacteria, the Chloroplasts, and the uncultured OP5, OP3, and OP1 candidate divisions. Four clones from core A and one clone from core C fall into the OP8 candidate subdivision; their closest relatives included clones from deep-sea cold seep sediments (the Nankai Trough clone NKB18 [47]), Obsidian Pool (38), and the jet-fuel-contaminated aquifer (20).
Two clones from core C formed a well-supported cluster with clones SB-45 (included here) and SB-15, both from a benzene-mineralizing, sulfate-reducing mixed culture (64); with clones JTB138 (included here) and JTB243 from a deep-sea cold seep area in the Japan Trench (49); and with clone GCA025 (58) from hydrocarbon seep sediment. Minimum evolution trees suggested an affiliation of this group with a representative clone of the candidate division OP9, clone OPB46, but this grouping remains so far unsupported by bootstrap analysis (Fig. 4).
DISCUSSION
Prokaryotic lipids and their isotopic composition as evidence for anaerobic methanotrophy.
Stable carbon isotopic signatures of diagnostic prokaryotic membrane lipid products indicate the presence and activity of anaerobic methanotrophic consortia in hydrothermally active Guaymas sediments. Moreover, the high abundance of prokaryotic biomarkers with distinctly low carbon isotopic compositions (Table 1) suggests that methanotrophy is an important mode of microbial carbon assimilation in these sediments. It has to be noted that these biomarkers likely are degradation products of their biologically produced polar lipid counterparts that constitute the cell membrane. Although in the strictest sense these lipid biomarkers are presumed to be more stable than the 16S rRNA SSU sequence biomarkers to which they were compared, a series of studies suggests that these and other isotopically depleted archaeal lipids are particularly abundant in environments that host active anaerobic methanotrophic communities (7, 31, 32, 33, 59, 60, 61, 72, 80). Complementary to the lipid biomarker isotopic signatures, 16S rRNA sequences have identified the uncultured archaeal ANME-1 and ANME-2 lineages in the Guaymas sediments that are characteristic for methane-oxidizing microbial communities; the sequence data sets are broadly congruent with the biomarker evidence. However, substantial differences in both the distribution and isotopic compositions of prokaryotic lipids and in the composition of 16S rRNA clone libraries were observed between the cores.
The membrane lipid products of the Guaymas Basin are derived from present and active microbial populations, for several reasons. In surface sediments of Guaymas Basin, the concentration of the analyzed glycerol ethers diminished rapidly; in core T, their concentration was near the detection limit below a depth of 2.5 cm. Unresolved organic compounds that could not be resolved by gas chromatography predominated in deeper sediment layers. Sedimentary conditions do not favor long-term burial for neutral lipids; they are limited to the surface layers and thus show the same vertical distribution as phospholipid ester-linked fatty acids that occur only in the upper few centimeters of the Guaymas sediments (29). These observations are consistent with very high microbial activity in these sediment surface layers (5, 86a) and with the complete oxidation of fatty acids that has been demonstrated for pure cultures of sulfate-reducing bacteria from the Guaymas Basin (69). Therefore, it is unlikely that the neutral lipids in the Guaymas sediment surface layer reflect fossil remnants of past microbial activity. The alternative, import of lipids from the aerobic water column, is also extremely unlikely. Producers of archaeol and hydroxyarchaeol commonly inhabit anoxic sediments or anoxic, methane-rich water columns in anoxic basins such as the Black Sea (72). Compounds related to the bicyclic biphytanediol in Guaymas occur in oceanic water columns and sediments, but they have isotopic compositions around −20‰ (71) and are derived from cosmopolitan, cold-water planktonic archaea (19, 34).
The occurrence of isotopically highly fractionated archaeol and sn-2-hydroxyarchaeol in the Guaymas sediments is strong evidence for anaerobic methane oxidation by archaea. The isotopic composition of these archaeal compounds in the Guaymas sediments is compatible with an incorporation of carbon derived from isotopically depleted methane. Archaeol and sn-2-hydroxyarchaeol have been found in a variety of methane-rich, sedimentary environments with isotopic compositions from −133 to −41‰ (33). Such low isotopic compositions indicate a partial to exclusive biosynthesis from methane-derived carbon (31, 33, 59, 61, 79). The relatively broad range of isotopic compositions is related to differences in isotopic composition of methane and presumably to other poorly constrained environmental and physiological factors (33). Another biomarker of the Guaymas sediments that is conspicuously tied to anaerobic methane oxidation is a biphytanediol with two pentacyclic rings with an isotopic signature similar to those of archaeol and sn-2-hydroxyarchaeol (Table 1). This and other biphytanyl derivatives were originally only found in cultured, extremely thermophilic crenarchaeota but more recently have also been identified in planktonic coldwater archaea of the marine water column (34), specifically in group I cold-water crenarchaeota related to Crenarchaeum symbiosum (19). Also, biphytanyl derivatives occur in sediments at methane seeps with isotopic signatures indicative of methane assimilation (61, 71). Notably, the biphytanyl derivative is absent in the ANME-2-dominated core C.
The ANME-1 archaea as candidate for anaerobic methane oxidation.
The molecular, isotopic, and phylogenetic evidence in this study suggests that ANME-1 archaea are a source of these 13C-depleted lipids, as implied by earlier observations of ANME-1 archaea at sites of active anaerobic methanotrophy (31, 79). Specifically, the Guaymas data set argues for an active role of ANME-1 archaea in anaerobic methane oxidation, since the ANME-I clones were obtained from Guaymas sediment samples that consistently showed archaeal lipid signatures for methane oxidation. Cores A and T have yielded exclusively ANME-1 clones, combined with abundant, 13C-depleted sn-2-hydroxyarchaeol, archaeol, and biphytanediol.
An important argument for the involvement of ANME-1 archaea in anaerobic methane oxidation is their consistent occurrence pattern in methane-rich sediments and cold methane seeps. These sites include methane seeps in Eel River Basin (31), Santa Barbara Basin (59), the methane oxidation zone in near-shore sediments in Aarhus Bay, Denmark (80), and hydrothermal sediments in the Iheya Basin in the Okinawa Trough, Japan (77). Recent observations by Thomsen et al. (80) lend additional support for the involvement of ANME-1 archaea in anaerobic methanotrophy, showing a predominance of ANME-1 archaea in the sulfate reduction-methanogenesis transition layer together with unusual dissimilatory sulfite reductase genes. These findings are consistent with a coupling of anaerobic methanotrophy with sulfate reduction (35, 82).
In the Guaymas samples, ANME-1 archaea were found associated with 13C-depleted cyclic biphytanyldiol biomarkers. The cyclic, 13C-depleted biphytanediol, commonly considered characteristic for cold-water crenarchaeota (e.g., see reference 71), occurred only in cores A and T, which yielded ANME-1 clones, whereas it was absent in the ANME-2-dominated core C. The association of biphytanediol derivatives with ANME-1 archaea could explain previous observations of 13C-depleted, cyclic C40 biphytanyl derivatives in Mediterranean cold seep sediments (61) (no phylogenetic data shown) and in Black Sea sediments (79) but not in others such as ANME-2 dominated sediments at methane seeps in Santa Barbara Basin and Eel River Basin (32, 59). Biphytanyl derivatives found at those seep sites have isotopic compositions (∼−20‰) indicative of an origin from archaeal plankton. The sequence data and the temperature range of the surface samples do not suggest a high abundance of hyperthermophilic crenarchaeota, a possible alternative source of cyclic biphytanediols, in the Guaymas sediment surface samples. A precise quantification of both ANME groups in environmental samples would clarify the question of whether either ANME group could be solely responsible for anaerobic methane oxidation when the other group is absent.
ANME-2 archaea in Guaymas.
Previous studies have left no doubt that archaea affiliated with the ANME-2 lineage, together with sulfate-reducing bacteria affiliated with the Desulfosarcinales, oxidize methane in several sedimentary environments (7, 59, 60). Sequences of the ANME-2 group were always found in sampling sites that either showed anaerobic methane oxidation or were compatible with this process, such as gas hydrates in the Gulf of Mexico (46) and at cold methane seeps in the Eel River Basin (the Eel River clones BA2H11 and TA1a4 [31] and other close affiliates [59]) and in salt marsh sediments in England (clones 2MT7 and 2C83) (56), sediments of the Santa Barbara Basin (59), and methane hydrate sediments at the Cascadia Margin (7).
With fluorescent in situ hybridization (FISH) of methane-laden sediments, it was shown that ANME-2 archaea occurred in syntrophic association with sulfate-reducing bacteria of the Desulfosarcina lineage (7, 59). The stable carbon isotopic signatures of these syntrophic bacterial-archaeal associations, revealed by a new combination of FISH and secondary ion mass spectrometry, clearly indicated isotopically depleted methane as the carbon substrate of these archaea and their associated sulfate reducers (60). In the Guaymas samples, ANME-2 clones have been obtained only from core C, together with highly 13C-depleted archaeol and sn-2-hydroxyarchaeol (Table 1). Interestingly, only clones of the ANME-2c subgroup have been obtained from Guaymas; clones of the ANME-2a and 2b subgroups are missing. The existing phylogenetic diversity among ANME-2 lineages suggests some degree of physiological and biochemical divergence, similar to the divergence among the cultured lineages within members of the Methanosarcinales lineage that contain acetoclastic and methyl-disproportionating lineages (8). Ecophysiological differences and distinct occurrence patterns can set the different ANME-2 subgroups apart from each other; a particular habitat or methane-oxidizing microbial community may harbor only one of the ANME-2 groups but not all of them simultaneously.
Diverse archaea at sites of anaerobic methanotrophy.
Many archaea exist in the clone libraries with no evidence for their involvement in anaerobic methanotrophy. Group B and D benthic archaea were found in the Guaymas sediments; they are widespread in methane-oxidizing sediments and vents but are not limited to these habitats. Group D benthic archaea, a sister group of the Thermoplasmales, include Japanese hydrothermal vent clones pMC2A203 and pMC2A33, from a black smoker chimney at the Myojin Knoll on the Izu-Ogasawara arc (78); the Eel River methane seep clones TA1f2, TA1e6, and TA1c9 (31); and clones ANME1, ANME7, ANME17, ANME19, and ANME23 from Baltic sediments (80). Orphan and coworkers noted the wide environmental distribution of this group, the “Thermoplasmales relatives,” in nonhydrothermal marine sediments as well as methane seeps (59). Similarly, archaea of the marine benthic group B have been found at Japanese hydrothermal vents (clone pMC2A36) (77), in hydrothermal vent in situ growth chambers at the Mid-Atlantic Ridge (clone VC2.1Arc31) (68), in sediments of the Santa Barbara Basin (clone sbpc17-1A) (59), and in near-shore and deep Atlantic sediments (83, 84).
Sulfate-reducing bacteria involved in anaerobic methanotrophy.
The bacterial isotopic signatures of the Guaymas sediment samples strongly argue for an involvement of bacteria in anaerobic methane oxidation. Isotopically light, 13C-depleted bacterial MAGE and DAGE lipids with a presumed origin from unknown methane-consuming bacterial syntrophs (32) cooccurred with ANME-1 archaeal clones in cores A and T (Table 1), in contrast to the Eel River sediments, where they mostly coincided with ANME-2 clones (59). These compounds had been observed earlier in sediments at methane seeps in the Eel River and Santa Barbara basins (32, 59) and at mud volcanoes in the Mediterranean Sea (61), where they have been interpreted as being associated with bacterial syntrophic partners of methanotrophic archaea. Their δ-13C signature of mostly −50 to −35‰ is intermediate between the isotopic signatures of archaeal methane-derived biomarkers and eukaryotic photosynthetic biomasses, suggesting that these bacteria assimilate carbon of mixed origin. In particular, the dialkyl ethers are not known from proteobacterial sulfate-reducing bacteria, which have been previously identified as syntrophic symbionts within methane-oxidizing consortia at cold gas hydrate sites (7). The monoalkyl ethers could have a more general bacterial source as products of the relatively widespread alkyl-acyl glycerol derivatives with mixed ether- and ester-linked alkyl moieties (27). Notably, a simple distribution of monoalkyl ethers with C16 and C18 alkyl moieties has been observed recently in members of Desulfosarcinales (http://docserver.bis.uni-oldenburg.de/publikationen/dissertation/2001/ruetra01/ruetra01.html) (70).
As long as bacteria are capable of syntrophic interaction with methane-oxidizing archaea and of consuming reaction products of anaerobic methane oxidation, syntrophic candidates are not necessarily limited to classical delta-proteobacterial sulfate-reducing bacteria. The highly diverse members of the uncultured bacterial phyla that were detected in cores A and C could include alternative sulfate-reducing bacteria that could fill this niche. The bacterial sequence profiles in samples from the Guaymas Basin show some broad overlap with those of other methane-rich sediments and seeps. A comparison of different methane-rich sediments and seeps has shown that gamma-, delta-, and epsilon-Proteobacteria, members of the CFB phylum, members of the gram-positive, fermentative genus Propionigenium, and uncultured members of the OP9 candidate subdivision and of the green nonsulfur phylum were found in at least half of these environments (59). Since anaerobic methane oxidation is coupled to sulfate reduction (82), it is possible that members of uncultured bacterial lineages that cooccur with anaerobic methane-oxidizing archaea could be novel sulfate reducers. Since DAGE lipids have so far been found in phylogenetically deeply branching bacteria, the new, deeply branching bacterial phylotype in core A could represent a candidate group (represented by clones A1B020, A2B002, and A2B033) (Fig. 4). This phylotype is present in core A but absent in core C and thus matches the distribution of the 13C-depleted bacterial DAGE lipids.
Interestingly, an autonomous character for some methanotrophic archaea—at least under some specific conditions—cannot be excluded. Clumps of ANME-2 archaea have been found repeatedly by FISH analysis which appear not to form consortia with any bacterial syntrophs (60). A systematic survey of ANME archaea and their syntrophs in natural environments, optimally by FISH-secondary ion mass spectrometry, is needed to clarify which bacterial syntrophs are required for anaerobic methane oxidation and to what extents (V. J. Orphan, C. H. House, K.-U. Hinrichs, K. D. McKeegan, C. Paull, W. Ussler, and E. F. DeLong, Eos. Trans. AGU Fall Meet. Suppl. 82[47], abstr. B12B-0127, 2001).
Although the question of their role as methane-oxidizing syntrophs remains open, sulfate-reducing bacteria are highly active in the Guaymas sediments. The sequencing survey identified mesophilic sulfate-reducing bacteria (mostly members of the mesophilic Desulfobulbaceae) as well as hyperthermophilic sulfate-reducing archaea (members of Archaeoglobales) in the Guaymas surface sediments (Fig. 2 and 3). Sulfate reduction rate measurements in Guaymas sediments have consistently shown high activities for mesophilic, thermophilic, and hyperthermophilic sulfate reducers (22, 41, 42, 86a). Cores taken in the vicinity of core A on the same dive (no. 3203) showed high sulfate reduction maxima of 300 to 2,000 nmol cm−3 day−1 for the temperature range of 12 to 35°C near the sediment surface, with the highest rate invariably occurring in the uppermost-centimeter layer (86a). Also, thermophilic sulfate-reducing activity at 70 and 80°C peaked in this layer, with a second peak for 70°C between the 5- and 10-cm depths (86a). Carbon substrates for sulfate reduction are available in the Guaymas sediments. Propionate, the signature substrate of the Desulfobulbaceae, and acetate, used by many sulfate-reducing bacteria, including members of the Archaeoglobales (88), are abundant in the Guaymas sediments. Their concentrations decrease towards the sediment surface, where sulfate-reducing bacteria are most active (52).
Bacterial diversity in the Guaymas sediments.
Numerous bacterial lineages in the Guaymas sediments were not linked to anaerobic methane oxidation. Several phylogenetic groups have been typically recovered from sulfidic, oxic-anoxic interfaces. The epsilon-Proteobacteria of epsilon group I (1) have been found in a wide variety of microoxic or anoxic aquatic habitats, including a sulfidic cave river with white filamentous microbial mats (1), the anoxic portion of the chemocline of the Cariaco Basin (50), and anoxic or microoxic deltaic muds in the Gulf of Papua (81). Analogous to the epsilon-Proteobacteria, gamma-proteobacterial clones in Guaymas that are related to sulfur-oxidizing bivalve symbionts (13) could be involved in sulfur oxidation.
Clones of the green nonsulfur bacterial phylum in Guaymas Basin have their closest relatives in geothermal springs, subsurface soil, petroleum hydrocarbon-rich sites, and enrichment cultures on aromatic hydrocarbons (15, 20, 38, 89). The only cultured representative of the nonphotosynthetic members of the green nonsulfur phylum, the anaerobic dechlorinating bacterium “Dehalococcoides dehalogenes,” can grow by oxidation of hydrogen and concomitant dechlorination of tetrachlorethene to ethene; mixed microbial extracts are also required for growth (53). These habitat preferences of nonphotosynthetic green-nonsulfur bacteria are compatible with the geochemical features of the Guaymas hydrothermal sediments. In this natural reactor, organic biomass undergoes pyrolysis and thermal alteration to a wide variety of petroleum hydrocarbons, including unbranched alkanes, cycloalkanes, triterpanes, steranes and diasteranes, and aromatic hydrocarbons (4).
Similar to those of the green nonsulfur bacteria, the habitat characteristics of members of the OP11 candidate division (20, 38, 48) suggest an anaerobic phenotype, with thermophilic as well as nonthermophilic representatives. OP11 bacteria also occur in cold, nonhydrothermal deep-sea sediments without obvious petroleum hydrocarbon contamination, where green nonsulfur bacteria have not been found (for example, clones BD2-3, BD5-13, BD7-1 and BD7-4) (48). No cultured representative of this group is known at present.
Like the clones of the nonphotosynthetic green nonsulfur bacteria and OP11 candidate division, members of the OP8 cluster and the OP9-affiliated clusters that occurred at Guaymas were generally found at cold seeps and in hydrocarbon-rich environments and enrichments (49, 58, 64). Although their environmental distribution suggests an anaerobic metabolism and involvement in degradation of aromatic hydrocarbons and petroleum compounds, the physiology and environmental activity of these diverse bacterial groups remain to be determined by pure culture studies.
Alternative causes of carbon isotopic depletions.
The question arises whether low δ-13C values in prokaryotic lipids could also be produced by other pathways than methane oxidation, for example, by autotrophic archaeal methanogenesis. δ-13C values in the Guaymas sediments ranged from −13.3 to −8.5‰ for CO2 and −14 to −9.6‰ for calcite (63). Data on Δδ-13C between carbon source and biomass exist for a few autotrophic methanogenic species in pure culture and allow estimation of projected δ-13C values of lipids in the Guaymas environment. The biomass of the obligate autotroph Methanobacterium thermoautotrophicum was depleted in 13C relative to CO2 by 24‰ (24). The Δδ-13C towards lipids should be higher, as suggested by results with the facultative autotroph Methanosarcina barkeri. Here, cell biomass was depleted by ca. 20‰ after growth on trimethylamine, whereas lipid phytanyl chains showed a larger Δδ-13C of −37‰ (75). Assuming a Δδ-13C of −40‰ between Guaymas CO2 and lipid phytanyl chains of autotrophic methanogens, projected δ-13C values for lipids in the sediment would reach −50 to −55‰. Actual minimum values in archaeal lipids, however, reach −89‰ (Table 1). For other significant chemolithoautotrophic CO2 assimilation pathways, namely, the Calvin cycle and the reverse citric acid cycle, the Δδ-13C values between CO2 and cell biomass amount to −25‰ and −8 to −12‰, respectively, leading to smaller fractionation effects (25). As in other environments where archaeal lipid δ-13C values reach minima of ∼130‰ (33), it is inconceivable those in the Guaymas Basin are derived from autotrophic methanogens or other autotrophic archaea. The relatively high (in comparison to other sites) δ values of the archaeal lipids likely result from the relatively high δ value of the substrate methane in Guaymas Basin sediments (87).
Anaerobic methane oxidation and trophic ecology of vent sites.
Anaerobic oxidation of methane provides a notable source of energy and biomass at hydrothermal vents and cold hydrocarbon seeps, in addition to the better known oxidation of sulfide by free-living and symbiotic bacteria and the aerobic oxidation of methane by symbiotic bacteria of mussels and pogonophorans (9). Anaerobic oxidation of methane is a significant process in coastal marine sediments and in particular at cold methane seeps (33). Methane seeps that harbored conspicuous populations of vent animals in the Oregon subduction zone showed anaerobic oxidation of methane (44); these findings implied that anaerobic methane oxidation should be considered as part of the trophic ecology of vent ecosystems. Based on results at a cold methane seep with dense giant clam communities in Sagami Bay, Japan, Mazusawa and coworkers (54) suggested that a mixture of methane of biogenic origin (δ-13C = −85 to −60‰) and thermogenic origin (δ-13C = −50 to −35‰) is oxidized in the sediment column beneath the cold seep clam community (Calyptogena soyoae). The resulting dissolved inorganic carbon of the pore water with an isotopic composition of −45 to −40‰ is incorporated into the soft tissue of the symbiont-harboring clams via sulfur-oxidizing bacterial symbionts. The clam tissue has a δ-13C value of around −35‰ (54). This 13C depletion is at the upper margin of isotopic signatures in animals with sulfur-oxidizing bacterial symbionts (including calyptogena) that fix carbon via the Calvin-Benson-Bassham cycle, also named the −30‰ group for the typical isotopic signature (16). A comparison of δ-13C values in bivalve mollusks from different sites showed generally lighter 13C signatures (−40 to −35‰) for vent bivalves at methane-rich cold seeps and hydrocarbon seeps than for bivalves from sulfide-dominated hydrothermal vent sites (23), suggesting a contribution of sedimentary methane oxidation to the CO2 pool that is subsequently incorporated into bivalve biomass via their autotrophic, sulfur-oxidizing symbionts. Aerobic, methylotrophic symbionts of mytilid clams and pogonophorans produce distinct, significantly lighter isotopic signatures in the tissue of their host animals, in the range of approximately −80‰ to −40‰ (14). Interestingly, contribution of CO2 from methane oxidation has been observed in sulfide-oxidizing, autotrophic Beggiatoa spp. from methane seeps, which showed unusually low isotopic signatures (approximately −50‰) in their biomass (Orphan et al., Eos. Trans. AGU Fall Meet. Suppl. 82[47], 2001).
The highly diverse bacterial and archaeal communities of the Guaymas basin sediments suggest a wide diversity of microbial physiologies and an untapped potential for novel microorganisms. The uncultured OP subdivision bacteria, the green nonsulfur bacteria, several proteobacterial lineages, and the ANME archaea lineages in Guaymas have evolutionary cousins in natural and anthropogenic hydrocarbon-rich sites, methane seeps and sediments, geothermal environments, and sulfide gradients. The Guaymas hydrothermal vent sites combine features of these habitats, which is reflected in their complex, multifaceted microbial communities. The enrichment and isolation of microbial key species, and the concomitant analysis of large genomic fragments from environmental DNA (6), will lead towards a better understanding of metabolic potential and activities of these microbial ecosystems.
Acknowledgments
This study was supported by the NSF Life in Extreme Environments program (grant OCE 9714195 to A.T. and H.W.J.), and by the NASA Astrobiology Institutes (Environmental Genomes; A.T., K.H., V.E., D.K., A.D.V.G., and M.L.S.; Subsurface Biospheres; A.T. and K.H.).
We thank Carl Wirsen and Stephen Molyneaux for their steady hand in organizing and preparing this cruise (no. 1997-116) as well as for their invaluable help at sea. This cruise greatly benefited from the good teamwork and good spirits of all cruise participants. Russell Cuhel collected additional sample material in Guaymas immediately before our cruise leg. We thank Victoria Orphan for providing unpublished sequence data and for sharing early versions of her work. We thank the crew and pilots of RV Atlantis and Research Submersible Alvin for their expert handling of our dives and equipment at the Guaymas Basin and at the 21°N EPR Vent sites and the Mexican government for permission to carry out research in Mexican territorial waters.
Footnotes
This is WHOI contribution 10624.
REFERENCES
- 1.Angert, E. R., D. E. Northup, A.-L. Reysenbach, A. S. Peek, B. M. Goebel, and N. R. Pace. 1998. Molecular phylogenetic analysis of a bacterial community in Sulphur River, Parker Cave, Kentucky. Am. Mineralogist 83:1583-1592. [Google Scholar]
- 2.Atkins, M. S., A. P. Teske, and O. R. Anderson. 2000. A survey of flagellate diversity at four deep-sea hydrothermal vents in the Eastern Pacific Ocean using structural and molecular approaches. J. Eukaryot. Microbiol. 47:400-411. [DOI] [PubMed] [Google Scholar]
- 3.Barns, S. M., C. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazylinski, D. A., J. W. Farrington, and H. W. Jannasch. 1988. Hydrocarbons in surface sediments from a Guaymas Basin hydrothermal vent site. Org. Geochem. 12:547-558. [Google Scholar]
- 5.Bazylinski, D. A., C. O. Wirsen, and H. W. Jannasch. 1989. Microbial utilization of naturally occurring hydrocarbons at the Guaymas Basin hydrothermal vent site. Appl. Environ. Microbiol. 55:2832-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of photography in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 7.Boetius, A., K. Ravenschlag, C. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 8.Boone, D. R., W. B. Whitman, and P. Rouvière. 1993. Diversity and taxonomy of methanogens, p. 35-80. In J. G. Ferry (ed.), Methanogenesis. Chapman and Hall, New York, N.Y.
- 9.Brooks, J. M., M. C. Kennicutt II, C. R. Fisher, S. A. Macko, K. Cole, J. J. Childress, R. R. Bidigare, and R. D. Vetter. 1987. Deep-sea hydrocarbon seep communities: evidence for energy and nutritional carbon sources. Science 238:1138-1142. [DOI] [PubMed] [Google Scholar]
- 10.Burggraf, S., H. W. Jannasch, B. Nicolaus, and K. O. Stetter. 1990. Archaeoglobus profundus sp. nov. represents a new species within the sulfate-reducing archaebacteria. Syst. Appl. Microbiol. 13:24-28. [Google Scholar]
- 11.Calvert, S. E. 1966. Origin of diatom-rich varved sediments from the Gulf of California. J. Geol. 76:546-565. [Google Scholar]
- 12.Cary, S. C., M. T. Cottrell, J. L. Stein, F. Camacho, and D. Desbruyeres. 1997. Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl. Environ. Microbiol. 63:1124-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavanaugh, C. M. 1994. Microbial symbiosis: patterns of diversity in the marine environment. Am. Zool. 34:79-89. [Google Scholar]
- 14.Cavanaugh, C. M. 1996. Methanotroph-invertebrate symbioses in the marine environment: ultrastructural, biochemical, and molecular studies, p. 315-328. In M. E. Lidstrom and R. F. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishing, Dordrecht, The Netherlands.
- 15.Chandler, D. P., F. J. Brockman, T. J. Bailey, and J. K. Fredrickson. 1998. Phylogenetic diversity of Archaea and Bacteria in a deep subsurface palaeosol. Microb. Ecol. 36:37-50. [DOI] [PubMed] [Google Scholar]
- 16.Childress, J. J., and C. R. Fisher. 1992. The biology of hydrothermal vent animals: physiology, biochemistry, and autotrophic symbioses. Oceanogr. Mar. Biol. Annu. Rev. 30:337-441. [Google Scholar]
- 17.Claypool, G. E., and K. A. Kvenvolden. 1983. Methane and other hydrocarbon gases in marine sediment. Annu. Rev. Earth Planet. Sci. 11:299-327. [Google Scholar]
- 18.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delong, D. F., L. L. King, R. Massana, H. Cittone, A. Murray, C. Schleper, and S. G. Wakeham. 1998. Dibiphytanyl ether lipids in nonthermophilic crenarchaeotes. Appl. Environ. Microbiol. 64:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dojka, M. A., P. Hugenholtz, S. H. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgcomb, V., D. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 22.Elsgaard, L., M. F. Isaksen, B. B. Jørgensen, A.-M. Alayse, and H. W. Jannasch. 1994. Microbial sulfate reduction in deep-sea sediments at the Guaymas basin hydrothermal vent area: influence of temperature and substrates. Geochim. Cosmochim. Acta 58:3335-3343. [Google Scholar]
- 23.Fiala-Medioni, A., J. Boulegue, S. Ohta, H. Felbeck, and A. Mariotti. 1993. Source of energy sustaining the Calyptogena populations from deep trenches in subduction zones off Japan. Deep-Sea Res. I 40:1241-1258. [Google Scholar]
- 24.Fuchs, G., R. Thauer, H. Ziegler, and W. Stichler. 1979. Carbon isotope fractionation by Methanobacterium thermoautotrophicum. Arch. Microbiol. 120:135-139. [Google Scholar]
- 25.Fuchs, G. 1989. Alternative pathways of autotrophic CO2 fixation, p. 365-382. In H. G. Schlegel and B. Bowien (ed.), Autotrophic bacteria. Springer-Verlag, KG, Berlin, Germany.
- 26.Goetz, F. E., and H. W. Jannasch. 1993. Aromatic hydrocarbon-degrading bacteria in the petroleum-rich sediments of the Guaymas basin hydrothermal vent site: preference for aromatic carboxylic acids. Geomicrobiol. J. 11:1-18. [Google Scholar]
- 27.Goldfine, H., and T. A. Langworthy. 1988. A growing interest in bacterial ether lipids. Trends Biochem. Sci. 13:217-221. [DOI] [PubMed] [Google Scholar]
- 28.Green, P. 1996. Phrap sequence assembly program. Version 0.96073.1. University of Washington, Seattle.
- 29.Guezennec, J. G., J. Dussauze, M. Bian, F. Rocchiccioli, D. Ringelberg, D. B. Hedrick, and D. C. White. 1996. Bacterial community structure from Guaymas Basin, Gulf of California, as determined by analysis of phospholipid ester-linked fatty acids. J. Mar. Biotechnol. 4:165-175. [Google Scholar]
- 30.Gundersen, J. K., B. B. Jørgensen, E. Larsen, and H. W. Jannasch. 1992. Mats of giant sulphur bacteria on deep-sea sediments due to fluctuating hydrothermal flow. Nature 360:454-455. [Google Scholar]
- 31.Hinrichs, K.-U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 32.Hinrichs, K.-U., R. E. Summons, V. J. Orphan, S. P. Sylva, and J. M. Hayes. 2000. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 31:1685-1701. [Google Scholar]
- 33.Hinrichs, K.-U., and A. Boetius. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry. In G. Wefer, D. Billet, D. Hebbeln, B. B. Jørgensen, M. Schlüter, and T. van Weering (ed.), Ocean margin systems, in press. Springer-Verlag, Berlin, Germany.
- 34.Hoefs, M. J. L., S. Schouten, J. W. de Leeuw, L. L. King, S. G. Wakeham, and J. S. Sinninghe Damsté. 1997. Ether lipids of planktonic archaea in the marine water column. Appl. Environ. Microbiol. 63:3090-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem. Cycles 8:451-463. [Google Scholar]
- 36.Huber, R., M. Kurr, H. W. Jannasch, and K. O. Stetter. 1989. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110°C. Nature 342:833-834. [Google Scholar]
- 37.Huber, R., T. Wilharm, D. Huber, A. Trincone, S. Burggraf, H. Koenig, R. Rachel, I. Rockinger, H. Fricke, and K. O. Stetter. 1992. Aquifex pyrophilus gen. nov. sp. nov. represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst. Appl. Microbiol. 15:340-351. [Google Scholar]
- 38.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jannasch, H. W., C. O. Wirsen, S. J. Molyneaux, and T. A. Langworthy. 1992. Comparative physiological studies on hyperthermophilic archaea isolated from deep-sea hot vents with special emphasis on Pyrococcus strain GB-D. Appl. Environ. Microbiol. 58:3472-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones, W. J., J. A. Leigh, F. Mayer, C. R. Woese, and R. S. Wolfe. 1983. Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 136:254-261. [Google Scholar]
- 41.Jørgensen, B. B., L. X. Zawacki, and H. W. Jannasch. 1990. Thermophilic bacterial sulfate reduction in deep-sea sediments at the Guaymas Basin hydrothermal vents (Gulf of California). Deep-Sea Res. I 37:695-710. [Google Scholar]
- 42.Jørgensen, B. B., M. F. Isaksen, and H. W. Jannasch. 1992. Bacterial sulfate reduction above 100°C in deep-sea hydrothermal vent systems. Science 258:1756-1757. [DOI] [PubMed] [Google Scholar]
- 43.Kawka, O. E., and B. R. T. Simoneit. 1987. Survey of hydrothermally-generated petroleums from the Guaymas Basin spreading center. Org. Geochem. 11:311-328. [Google Scholar]
- 44.Kulm, L. D., E. Suess, J. C. Moore, B. Carson, B. T. Lewis, S. D. Ritger, D. C. Kadko, T. M. Thornburg, R. W. Embley, W. D. Rugh, G. J. Massoth, M. G. Langseth, G. R. Cochrane, and R. L. Scamman. 1986. Oregon subduction zone: venting, fauna, and carbonates. Science 231:561-566. [DOI] [PubMed] [Google Scholar]
- 45.Langworthy, T. A., G. Holzer, J. G. Zeikus, and T. G. Tornabene. 1983. Iso- and anteiso-branched glycerol diethers of thermophilic anaerobic Thermodesulfobacterium commune. Syst. Appl. Microbiol. 4:1-17. [DOI] [PubMed] [Google Scholar]
- 46.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, L., J. Guenzennec, P. Nichols, P. Henry, M. Yanagibayashi, and C. Kato. 1999. Microbial diversity in Nankai Trough sediments at a depth of 3,843 m. J. Oceanogr. 55:635-642. [Google Scholar]
- 48.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 49.Li, L., C. Kato, and K. Horikoshi. 1999. Microbial diversity in sediments collected from the deepest cold-seep area, the Japan trench. Mar. Biotechnol. 1:391-400. [DOI] [PubMed] [Google Scholar]
- 50.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martens, C. S. 1990. Generation of short chain organic acid anions in hydrothermally altered sediments of the Guaymas Basin, Gulf of California. Appl. Geochem. 5:71-76. [Google Scholar]
- 53.Maymo-Gatell, X., Y. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 54.Mazusawa, T., N. Handa, H. Kitagawa, and M. Kusakabe. 1992. Sulfate reduction using methane in sediments beneath a bathyal “cold seep” giant clam community off Hatsushima Island, Sagami Bay, Japan. Earth Planet. Sci. Lett. 110:39-50. [Google Scholar]
- 55.Moyer, C. L., F. C. Dobbs, and D. M. Karl. 1995. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munson, M. A., D. B. Nedwell, and T. M. Embley. 1997. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl. Environ. Microbiol. 63:4729-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson, D. C., C. O. Wirsen, and H. W. Jannasch. 1989. Characterization of large autotrophic Beggiatoa abundant at hydrothermal vents of the Guaymas Basin. Appl. Environ. Microbiol. 55:2909-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Neill, K. R. 1998. Molecular microbial ecology of the Gulf of Mexico hydrocarbon seep sediment. Ph.D. thesis. University of Maryland, College Park.
- 59.Orphan, V. J., K.-U. Hinrichs, C. K. Paull, L. T. Taylor, S. Sylva, and E. F. Delong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 61.Pancost, R. D., S. J. Sinninghe Damsté, S. D. Lint, M. J. E. C. van der Maarel, J. C. Gottschal, and Shipboard Scientific Party. 2000. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic bacteria and archaea. Appl. Environ. Microbiol. 66:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pancost, R. D., I. Bouloubassi, G. Aloisi, J. S. S. Damste, and Medinaut Shipboard Scientific Party. 2001. Three series of non-isoprenoidal dialkyl glycerol ethers in cold-seep carbonate crusts. Org. Geochem. 32:695-707. [Google Scholar]
- 63.Peter, J. M., and W. C. Shanks. 1992. Sulfur, carbon, and oxygen isotope variations in submarine hydrothermal deposits of Guaymas Basin, Gulf of California, USA. Geochim. Cosmochim. Acta 56:2025-2040. [Google Scholar]
- 64.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 65.Polz, M. F., and C. M. Cavanaugh. 1995. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl. Acad. Sci. USA 92:7232-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 68.Reysenbach, A.-L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rueter, P., R. Rabus, H. Wilkes, F. Aeckersberg, F. A. Rainey, H. W. Jannasch, and F. Widdel. 1994. Anaerobic oxidation of hydrocarbons in crude oil by new types of sulphate-reducing bacteria. Nature 372:455-458. [DOI] [PubMed] [Google Scholar]
- 70.Rütters, H. 2001. Tracing viable bacteria in Wadden Sea sediments using phospholipid analysis. Ph.D. thesis. Oldenburg University, Oldenburg, Germany.
- 71.Schouten, S., M. J. L. Hoefs, M. P. Koopmans, H.-J. Bosch, and J. S. Sinninghe Damsté. 1998. Structural characterization, occurrence and fate of archaeal ether-bound acyclic and cyclic biphytanes and corresponding diols in sediments. Org. Geochem. 29:1305-1319. [Google Scholar]
- 72.Schouten, S., S. G. Wakeham, and J. S. Sinninghe Damsté. 2001. Evidence for anaerobic methane oxidation by archaea in euxinic waters of the Black Sea. Org. Geochem. 32:1277-1281. [Google Scholar]
- 73.Schrader, H. 1982. Diatom biostratigraphy and laminated diatomaceous sediments from the Gulf of California, p. 973-981. In J. R. Curray (ed.), Initial reports of the Deep Sea Drilling Project, Leg 64. U.S. Government Printing Office, Washington, D.C.
- 74.Stahl, D. A, B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Summons, R. E., P. D. Franzmann, and P. D. Nichols. 1999. Carbon isotopic fractionation associated with methylotrophic methanogenesis. Org. Geochem. 28:465-475. [Google Scholar]
- 76.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, Massachusetts.
- 77.Takai, K., and K. Horokoshi. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takai, K., and Y. Sako. 1999. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol. Ecol. 28:177-188. [Google Scholar]
- 79.Thiel, V., J. Peckmann, H. H. Richnow, U. Luth, J. Reitner, and W. Michaelis. 2001. Molecular signals for anaerobic methane oxidation in Black Sea seep carbonates and a microbial mat. Mar. Chem. 73:97-112. [Google Scholar]
- 80.Thomson, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Todorov, J. R., A. Y. Chistoserdov, and J. Aller. 2000. Molecular analysis of microbial communities in mobile deltaic muds of Southeastern Papua New Guinea. FEMS Microbiol. Ecol. 33:147-155. [DOI] [PubMed] [Google Scholar]
- 82.Valentine, D. L., and W. S. Reeburgh. 2000. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2:477-484. [DOI] [PubMed] [Google Scholar]
- 83.Vetriani, C., A.-L. Reysenbach, and J. Doré. 1998. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol. Lett. 161:83-88. [DOI] [PubMed] [Google Scholar]
- 84.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Von Damm, K. L. 1990. Seafloor hydrothermal activity: black smoker chemistry and chimneys. Annu. Rev. Earth Planet. Sci. 18:173-204. [Google Scholar]
- 86.Von Damm, K. L., J. M. Edmond, C. I. Measures, and B. Grant. 1985. Chemistry of submarine hydrothermal solutions at Guaymas Basin, Gulf of California. Geochim. Cosmochim. Acta 49:2221-2237. [Google Scholar]
- 86a.Weber, A., and B. B. Joergensen. Bacterial sulfate reduction in hydrothermal sediments of the Guaymas Basin, Gulf of California, Mexico. Deep-Sea Res., in press.
- 87.Welhan, J. A. 1988. Origins of methane in hydrothermal systems. Chem. Geol. 71:183-198. [Google Scholar]
- 88.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 89.Wintzingerode, F. V., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]