Abstract
Iroquois transcription factors regulate diverse aspects of developmental patterning in all metazoans. Despite their widespread importance, the direct targets of the Iroquois are poorly understood. Here, we use in vitro site selection to define the DNA-binding preference of the Drosophila Iroquois Mirror. We use electrophoretic mobility shift assays to determine the critical nucleotides for Mirror binding and to show that this site is recognized by other Drosophila Iroquois transcription factors. This site also is recognized by vertebrate Iroquois transcription factors. Transgenic analysis demonstrates that Drosophila Iroquois proteins recognize this site in vivo to mediate transcriptional repression. We further show that Iroquois transcription factors form homodimers and heterodimers, suggesting that combinatorial binding may contribute to gene regulation by this family.
Keywords: development, Drosophila, Notch, planar polarity, fringe
Iroquois transcription factors (Iro/Irx) are found in all multicellular organisms, from sponges (1) to man, and have essential roles in controlling many aspects of developmental patterning (2). The Iro were first identified in Drosophila, where there are three closely related proteins: Mirror (Mirr), Araucan (Ara), and Caupolican (Caup) (3, 4). The three Drosophila Iro are located in a gene cluster called the Iro-complex. ara and caup are highly related and genetically redundant and share cis-regulatory regions. mirr encodes a more divergent protein, which is expressed in different tissues from the other Iro, and is not redundant with ara and caup. In mammals there are six Irx genes (Irx1--6), which are found in two clusters of three genes each (4, 5). All Iro/Irx proteins share very high homology in the homeodomain (HD); however, there is no direct relationship between any one Drosophila Iro and any vertebrate Irx. Iro genes have been shown to be essential for diverse processes, such as determination of ventricular identity in the heart (6), neural determination in the embryo (7, 8), formation of the organizer during gastrulation (9), establishment of planar polarity in the eye (4, 10-13), follicle cell organization in the ovaries (14), and axonal path-finding in the CNS (15). However, despite extensive work demonstrating the importance of Iro transcription factors in many developmental processes, it is unclear in any system which genes are directly controlled by the Iro.
All Iro family members share a highly conserved HD with little sequence similarity outside the HD (Fig. 1A). Unlike classic Hox transcription factors, which have a characteristic 60-aa HD, Iro family members have a 63-aa HD with a 3-aa loop extension (TALE), which places the Iro into the TALE family of transcription factors (16). Outside the HD, all Iro have a 9-aa region of homology called the Iro box (2, 4, 16).
Fig. 1.
The Iro DNA-binding domain is unique, highly conserved, and recognizes a previously undescribed motif. (A) Alignment of fly and vertebrate Iro HDs. PBX (top) and classic HOX (bottom) HDs are shown for comparison. Pink boxes indicate α-helices in the PBX and Iro families. Blue boxes indicate α-helices in the Antp HD. Note conservation within the DNA binding helix (*) in all Iro. (B) EMSA with samples from the second, third, and fourth rounds of the FLAG IP site selection. (C) Information Content Diagram showing the degree of conservation at each position of the motif. The height of each column corresponds to the frequency of each base at each position (A, red; C, blue; G, yellow; T, green). If no base is significantly enriched, the column is black. (D) Sequences from the site-selection experiments aligned to highlight the consensus motif.
Given their unique, atypical HD, it is surprising that the Iro DNA binding site has been suggested to be a classic HOX binding motif (TAAT) (3). The initial characterization of Iro used DNase I footprinting to demonstrate that Ara binds in vitro to a cis-regulatory element of achaete-scute (ac-sc). Based on these data, it was proposed that the Iro-complex proteins directly activate expression of ac-sc, providing the best evidence for a direct Iro target in any system. However, subsequent data (2) have indicated that ac-sc is not a direct Iro target and brings into question the relevance of this site.
The Drosophila Iro-complex gene mirr has been shown to be essential for regulating dorsal-ventral planar polarity in the eye (4, 11, 13) and positional identity in the ovary (14, 17). In both tissues, mirr functions by repressing expression of fringe (fng). fng encodes a glycosyltransferase that regulates Notch activity (reviewed in ref. 18). The regulation of fng expression is essential for mirr's role in regulating planar polarity in the eye and follicle cell organization in the ovaries. ara and caup also can regulate fng expression (10, 12); however, without knowing the Iro binding site (IBS), it is difficult to determine whether the repression of fng by the Iro is direct.
Here, we use DNA-binding site selection experiments to show that Iro proteins have distinct DNA-binding specificities, which differ from a classic HOX consensus. We find that this specificity is conferred not solely through the HD but also by sequences outside this region. We show that this binding site is recognized by Drosophila and vertebrate members of the Iro family. We present evidence that this site is functional in vivo and that the Iro bind to this site to repress fng expression.
Materials and Methods
Binding-Site Selection. Selection of DNA sequences was carried out according to ref. 20 by using the degenerate oligonucleotide R76 (for details of oligonucleotides and summary of all EMSA results, see Supporting Materials and Methods and Table 1, which are published as supporting information on the PNAS web site). Mirr and FLAG-Mirr proteins were synthesized in vitro by using the coupled transcription/translation rabbit reticulocyte lysate system (Promega).
Fly Strains. yw (y,w), hs-Mirr (w; PKB-mirror), ry (cn;ry), Fng-Gal4 (w; P{w+mW.hs = GawB} fng/TM3), UAS-Mirr (w1118; pUAST12), mirr-lacZ (F7).
In Vivo Reporter Assays. Oligonucleotides with four repeats of the consensus (PAL) and the mutated motif (MUT) were cloned into the pGbe-lacZ vector (19), which carries three copies of the binding element for the Grainyhead transcriptional activator cloned upstream of LacZ. Constructs were injected in cn,ry embryos to generate Pry+ transformants. Discs were stained by using standard procedures (4).
Luciferase Reporter Assays. A 502-bp region flanking the Fng-IBS, was PCR amplified from Drosophila genomic DNA and inserted into the pGL3-Promoter vector (Promega). Point mutations in the IBS were created by using the QuikChange kit (Stratagene) Double-stranded RNA was produced by using the Megascript kit (Ambion, Austin, TX). S2 cells were transfected in triplicate by using Effectene (Qiagen, Hilden, Germany). Luciferase assays were conducted 2 days later according to manufacturer's instructions (Promega).
Results
To determine the Iro DNA binding preference, we used an unbiased in vitro selection assay (20). We immunoprecipitated (IP) from a pool of 32P-labeled random oligonucleotides DNA preferentially bound by the Iro protein, Mirr. The DNA was recovered and subjected to further rounds of selection. Auto-radiography of an EMSA revealed bands that appeared with increased intensity after each round as a result of the enrichment for sequences bound by Mirr. We performed the site-selection experiments in two different ways using in vitro translated Mirr. The first site selection used an untagged Mirr protein and an anti-Mirr antibody (Ab) for the IP. The second approach was to generate a FLAG-tagged Mirr and use anti-FLAG Ab-conjugated beads for the IP. In both experiments we detected an enrichment of Mirr binding sequences by the third round of selection (Fig. 1B and data not shown). The specificity of the enriched DNA for binding Mirr was confirmed by a supershift of the complex by the addition of Abs against the FLAG tag (Fig. 1B) or against Mirr (data not shown). We sequenced 35 clones derived from the fourth round of the Mirr IP and 46 clones from the fourth round of the FLAG IP. Visual inspection of these sequences revealed an enrichment of ACA and TGT sequences (Fig. 1C, S1), with a preponderance of palindromic sequences of ACAnnTGT. There were very few examples of the putative IBS TAAT (3) in the sequences.
The Mirr Preferred Binding Site Is AAAACACGTGTTAA. To further analyze this pool of sequences, we used meme analysis (http://meme.sdsc.edu/meme). meme identifies conserved motifs in a group of related sequences (21), using statistical modeling to choose the best width, number of occurrences, and distribution of motifs in the input sequences. Sequences from both experiments were combined and submitted to the meme analysis tool. The meme identified motif was [A/G],A,[A/T],[A/T]-ACA-[C/T],[G/A]-TGT-[T/A],A,[A/T] (e = 3.6e-062; bold type signifies strong bias) (Fig. 1 C and D). Another way of analyzing the results of the site selection is the use of a positional weight matrix (PWM). The PWM for the sequences of the two site-selection experiments is shown in Fig. 2. From both the MEME and the PWM analyses, it is evident that the in vitro site selection has identified a Mirr consensus motif [A/G]AAAACACGTGTTAA.
Fig. 2.
Positional weight matrix for identification of a consensus Mirr motif from site selections 1 and 2. Each element in the matrix represents how many times nucleotide i was found in position j of the alignment (ref. 25; http://trantor.bioc.columbia.edu/Target_Explorer). Frequencies of prevalent bases at each position are shown in bold. At the bottom of the figure is the consensus motif.
We used EMSA to test whether Mirr specifically binds this motif (AAAAACACGTGTTAA). Addition of FLAG-tagged Mirr resulted in a mobility shift, demonstrating that Mirr binds this sequence in vitro. Addition of Abs to the FLAG tag resulted in a supershift of the Mirr complex, confirming the specificity of the binding (see Fig. 8, which is published as supporting information on the PNAS web site).
ACAnnTGT Is a Minimal Mirr Binding Site. To determine which nucleotides are essential for Mirr binding, we tested 32P-labeled oligonucleotides with point mutations in EMSA. Analysis of the sequences derived from both experiments indicated a common core sequence of ACACGTGT, with an A/T-rich region on either side of the palindrome (Fig. 1D). We tested whether the A/T-rich flanking region was essential by EMSA and found that Mirr can bind ACACGTGT in the context of several different flanking sequences. (Fig. 3A, S2). The complex can be supershifted by the addition of Abs to Mirr (data not shown) or to the FLAG tag (Fig. 3A), demonstrating that ACACGTGT is sufficient for specific Mirr binding.
Fig. 3.
Characterization of Mirr binding specificities. (A) Mirr binds the ACAnnTGT motif irrespective of the flanking sequences. (B) The two nucleotides separating the two halves of the palindrome act as a spacer. Changing the sequence from CG to TA does not compromise binding. (C) Spacer length has a dramatic impact on binding. (D) Increasing the length of the spacer to 4 or 6 nt results in further reduction in affinity. (E) Point mutations changing the ACAnnTGT core to AtAnnTaT abolish Mirr binding. (F) Replacing the inverted with direct repeats also abolishes Mirr binding. Arrows indicate EMSA shift; arrowheads indicate supershift.
The site-selection experiments suggested a preference for CG in the two central positions; however, Mirr can bind to oligos containing AT sequences in these positions (Fig. 3B), suggesting that these nucleotides may act as spacers. The length of the spacer is crucial for high-affinity binding. Sequences with a 2-nucleotide (nt) spacer (ACAnnTGT) bind much more efficiently than those with a 1-, 3-, 4-, or 6-nt spacer (Fig. 3 C and D). Palindromes with no spacer (ACATGT) or palindromes spaced by >6 nt do not show detectable binding (S2). Single-nucleotide mutations within each half of the palindrome (ACAnnTGT → AtAnnTaT) result in loss of Mirr binding (Fig. 3E). Changing the binding site from inverted repeats to direct repeats (ACAnnTGT → ACAnnACA) also abolished binding (Fig. 3F, S2). Competition experiments indicate that Mirr binds more efficiently to AAAAACACGTGTTAA (data not shown), suggesting that the ACAnnTGT is a minimal recognition sequence.
Mirr Does Not Recognize a HOX Sequence in EMSA. The Mirr binding site bears little resemblance to the HOX binding site (TAAT consensus) previously suggested to be an IBS (3). We tested whether Mirr could bind a classic HOX site using a number of oligos bearing a classic HOX consensus sequence. The goosecoid DE enhancer, which contains two classic HOX binding sites (22) showed no detectable interaction with Mirr in EMSA. (see Fig. 9, which is published as supporting information on the PNAS web site). We also failed to find any significant interaction several enhancer elements containing TAAT sites, such as the P3 enhancer from the paired promoter (ref. 23; Fig. 9).
Iro binding to a TAAT site had been suggested because a HD-containing fragment of Ara protected a region of the L3 enhancer that included a TAAT site in footprinting assays (3). We tested whether Mirr binds the L3 enhancer and found that Mirr bound very weakly to this sequence (Fig. 4A). Because we could not detect any binding of Mirr to a classic HOX site (S1), we carefully examined the protected region (3) and found that it contained an ACA sequence, corresponding to one-half of the minimum Mirr site. In EMSA Mirr binds the L3 enhancer weakly (S1): Interestingly, mutation of this ACA to AtA results in loss of Mirr binding to the L3 enhancer (S1).
Fig. 4.
The Mirr-binding site is a general Iro site. (A) Full-length Mirr binds with higher affinity to the ACAnnTGT motif than to the L3 element. (B) Binding of Mirr to ACAnnTGT is competed out by increasing amounts of unlabeled oligos carrying the same motif but not by oligos carrying the mutated motif or the L3 element. (C) Mirr-HD binds ACAnnTGT and the L3 element with similar affinity. (D) Ara binds ACAnnTGT and shows the same requirement for a 2-nt spacer as Mirr. (E) Mouse Irx4 binds the ACAnnTGT motif. (F) Competition assays show that unlabeled ACAnnTGT, but not AtAnnTaT, oligos reduce Ara binding to the labeled ACAnnTGT probe. Arrows indicate shift; arrowheads indicate supershift.
Competition experiments showed that binding of Mirr to the ACAnnTGT sequence is competed out by increasing amounts of unlabeled oligos of the same sequence. Addition of the same amounts of mutant oligonucleotides (AtAnnTaT) or of the L3 enhancer (TAAT) does not affect binding to the labeled probe, demonstrating that the ACAnnTGT is the preferred Mirr binding site (Fig. 4B).
Full Binding Specificity Requires a Full-Length Iro Protein. The experiments that implicated a TAAT sequence had been performed using a HD containing fragment of Ara, rather than the full-length protein (3). We reasoned that a HD alone construct might have reduced specificity. We tested this hypothesis by generating a FLAG-tagged Mirr HD-only construct, containing only the 63-aa HD of Mirr, and examined binding on the L3 enhancer. Strikingly, we found that the Mirr HD construct binds equally well to both the L3 enhancer and to the ACAnnTGT site (Fig. 4C). Binding is specific, because the complex can be supershifted by the addition of FLAG Abs. Mirr HD fails to bind an L3 enhancer that carries a point mutation within the half-site (S1), indicating it is recognizing the ACA motif. The Mirr HD construct fails to bind a classic HOX site (data not shown) suggesting that some specificity is retained. We also generated FLAG-tagged Ara, and tested it on the L3 enhancer. Full-length Ara had only very weak binding (data not shown), and only a HD fragment of Ara shows strong binding (3). Together these data suggest that binding to the L3 enhancer is only a property of truncated Iro proteins.
Vertebrate and Invertebrate Iro Proteins Bind the ACAnnTGT Site. Because the Iro HD is highly conserved (Fig. 1 A), we reasoned that ACAnnTGT might be a universal IBS. We used the FLAG-tagged full-length Ara and generated mammalian FLAG-tagged Irx4 in EMSA and found that both proteins bind ACAnnTGT (Fig. 4 D and E). For both Ara and Irx4, binding to the ACAnnTGT site is weaker than with Mirr, but all Iro showed much stronger binding to the ACAnnTGT site than to the L3 enhancer and had no detectable binding to a TAAT site (data not shown). Binding specificity was verified by a supershift of the complex (Fig. 4 D and E). Binding was specifically competed by ACAnnTGT, but not by incubation with mutated oligonucleotides (Fig. 4F). Neither Ara nor Irx4 bind a mutated Mirr-binding site (data not shown). Together, these data indicate that Iro proteins recognize ACAnnTGT. We therefore refer to this sequence as an IBS.
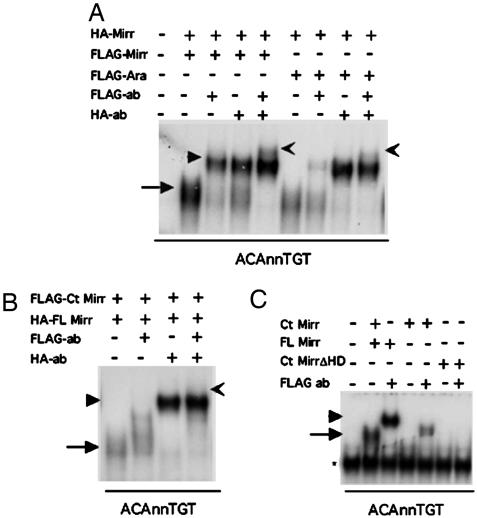
Iro Transcription Factors Form Homodimers and Heterodimers. The palindromic nature of the IBS suggests that Iro proteins might form dimers on DNA. Dimer formation also is supported by the observation that a 2-nt spacer is essential for high-affinity binding (Figs. 3C and 4D). To directly test whether Iro proteins form dimers, we created a hemagglutinin (HA)-tagged version of Mirr and cotranslated it with FLAG-tagged Mirr. Addition of Abs to either the HA-tag or the FLAG-tag supershifted the complex. Addition of both Abs produced a super-supershift, indicating that both FLAG- and HA-tagged Mirr were present in the DNA-protein complex (Fig. 5A).
Fig. 5.
Iro proteins form homodimers and heterodimers in vitro. (A) When FLAG-tagged and HA-tagged Mirr are cotranslated and mixed with labeled ACAnnTGT probe, a DNA-protein shift is formed (arrow). Addition of each of the Abs against the two different tags results in a supershift (arrowheads). Addition of both Abs in the same reaction results in a super-supershift (notched arrow), indicating that the complexes contain both FLAG and the HA-tagged Mirr. The same is shown for HA-Mirr and FLAG Ara indicating that Mirr can form heterodimers with other Iro proteins in vitro. (B) When an N-terminal deletion construct is cotranslated with Mirr and mixed with the ACAnnTGT probe, no super-supershift is detected, suggesting that sequences in the N-terminal region are important for dimerization. (C) The N-terminal deleted Mirr binds DNA weakly, and a complex can only be detected upon addition of the Ab.
A FLAG-tagged Mirr construct that lacks the N-terminal domain but includes the HD binds weakly and fails to show a super-supershift when mixed with full-length Mirr (Fig. 5 B and C), suggesting that the N-terminal domain is important for dimer formation. Incubation of FLAG-tagged Ara and HA-tagged Mirr also resulted in a super-supershift, indicating that Mirr can form heterodimers with Ara on the IBS (Fig. 5A). Pull-down experiments indicate that Mirr can form dimers independent of the presence of DNA (data not shown). The formation of Iro homodimers and heterodimers, together with the differences in strength of binding of the different Iro for the IBS (Fig. 4 A, D, and E), suggests that the overlapping expression patterns of the Iro seen in both vertebrates and invertebrates (reviewed in ref. 2) may result in a combinatorial control of gene expression by the Iro.
The IBS Functions in Vivo to Mediate Transcriptional Repression. The EMSA data showed that Mirr binds specifically and with high affinity to the IBS. However, these data do not demonstrate that this site has in vivo relevance. To directly test whether the IBS is functional in vivo, we generated transgenic flies carrying four repeats of the IBS in a β-gal reporter construct. Because mirr functions in vivo to repress expression of fng, we placed these IBSs in a vector that carries binding sites for the transcriptional activator Grainyhead (GBEs) (Fig. 6A). The GBEs provide a general transcriptional activation, allowing us to detect a predicted transcriptional repression in response to Mirr.
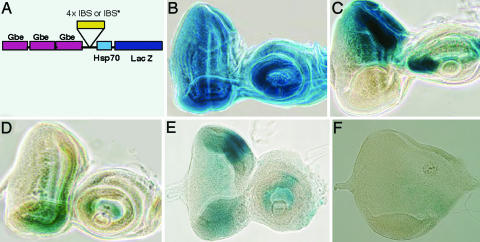
Fig. 6.
IBS mediates transcriptional repression in vivo. (A) Schematic of the construct used to generate the in vivo reporter lines. Gbe, Grainyhead binding element. (B) X-gal staining of a Gbe-lacZ control line: Grainyhead is ubiquitously expressed in the eye disk. (C) X-gal staining of a mirr-lacZ disk (F7) shows the expression pattern of mirr. Mirr is expressed at high levels at the dorsal part of the eye disk. (D) X-gal staining of a Gbe-IBS-lacZ line showing strong repression of the β-gal pattern in the dorsal domain, where Mirr is expressed. (E) Gbe-IBS*-lacZ. Mutating the IBS results in a loss of the dorsal repression of β-gal (compare C). (F) Gbe-IBS-lacZ; UAS mirr. Ectopic expression of mirr using a ventral fngGal4 driver results in repression of β-gal and morphological defects. The antennal disk was absent in all larvae overexpressing mirr ventrally, as a result of ectopic Mirr expression.
Mirr and the other Iro are expressed in the dorsal half of the eye (Fig. 6C) where they act to repress fng, resulting in the ventral fng expression (10-13). Inclusion of four IBSs in the LacZ reporter results in repression of β-gal expression in the dorsal region, where Mirr is expressed (Fig. 6D). In contrast, the parental reporter vector has ubiquitous β-gal expression (Fig. 6B). As an additional control, we also generated transgenic lines carrying four repeats of a mutated version of the IBS (AtAnnTaT). Importantly, mutations of the IBS that inhibit Mirr binding in vitro (Fig. 3B) result in loss of dorsal-specific repression (Fig. 6E). Both the Mirr binding and the mutated constructs have a reduced overall level of β-gal activity, compared with the parental vector. However, the specific dorsal repression of β-gal is only seen in constructs carrying repeats that can bind Mirr in vitro.
To directly test whether Mirr binding leads to repression of β-gal, we examined the effects of Mirr overexpression on the IBS reporter. We used the Gal4 system to ectopically express Mirr in the ventral half of the eye. Significantly, overexpression of Mirr results in a dramatic reduction in β-gal expression (Fig. 6F), indicating that Mirr can recognize the IBS to mediate transcriptional repression in vivo.
Mirr Represses Transcription from the fng Enhancer. To determine whether Mirr also acts as a repressor on endogenous enhancers, we examined the fng genomic region. There is an IBS located 193 bp downstream of fng. EMSA analysis confirmed that this IBS binds Mirr (data not shown). We cloned the 500-bp downstream region containing the fng-IBS into a luciferase reporter vector and examined the effects of altering Mirr levels on reporter expression. S2 cells contain endogenous mirr mRNA (data not shown) and protein (Fig. 7). Depletion of mirr by RNAi leads to significant increases in reporter activity, indicating that physiological levels of Mirr repress transcription. Mutation of the fng-IBS increases reporter activity to the same level as removal of Mirr, consistent with Mirr repressing transcription through the IBS. Depletion of an irrelevant protein (GFP) had no effect on Mirr levels or reporter activity. Together, these data strongly support the IBS as a biologically relevant Mirr-binding site and suggest that the Iro directly repress fng expression in vivo.
Fig. 7.
fng 3′ enhancer RNAi luciferase assay. Mirr mediates transcriptional repression on the fng enhancer. Blue indicates luciferase activity in S2 cells transfected with a pGL3 reporter containing the fng-IBS, treated with dsRNA to GFP. Magenta indicates luciferase units from S2 cells transfected with the same construct but treated with dsRNA to mirr. Yellow indicates luciferase activity from S2 cells with a point mutation in the fng-IBS, treated with RNAi to GFP. Light blue indicates luciferase activity from S2 cells containing the mutated enhancer, treated with RNAi to mirr. (Inset) Western blot from S2 cells treated with dsRNA to either GFP (Left) or Mirr (Right) shows dramatic reduction of Mirr protein levels specifically in response to Mirr RNAi.
Discussion
The Iro are highly conserved, key regulators of diverse developmental processes. Despite the widespread importance of the Iro family, little is known about their binding specificities, and there are no confirmed direct targets. Here, we have identified a previously undescribed binding site for the Drosophila Iro, Mirr, using an in vitro selection assay. We verified the specificity of this binding site in vitro, using mutational analysis and competition studies. Importantly, we demonstrate that this site is functional in vivo in Drosophila imaginal disks, where it can mediate transcriptional repression. We also show that Mirr can repress transcription through the endogenous fng enhancer. Significantly, other Iro from flies and vertebrates recognize the IBS. The finding that Iro proteins bind the IBS is an important step in identifying in vivo targets of this important regulatory family in vertebrates and invertebrates.
The in vitro site selection assay identified AAAACACGTGTTAA as a putative Mirr-binding site. We validated this site in EMSA, demonstrating that Mirr can recognize this site in vitro. By testing the ability of mutated oligonucleotides to bind to Mirr in EMSA, we refined the minimal binding site to a small palindromic sequence ACAnnTGT.
The palindromic nature of the IBS suggested that Mirr binds DNA as a homodimer, with each molecule binding half of the palindrome. We found that increasing spacer length dramatically interfered with Mirr binding, providing support for the idea that Mirr binds the IBS as a dimer. To directly test whether Mirr and the other Iro form dimers in vitro, we used EMSA as well as pull-down assays on in vitro translated proteins. These experiments confirmed that Iro transcription factors can form heterodimers and homodimers in vitro and suggest that they may form dimers in vivo.
What might be the functional significance of dimer formation by the Iro? A hallmark of the Iro family of transcription factors is that different family members are often expressed in overlapping patterns in many tissues (2). Although all of the Iro bind the IBS, they appear to differ in their strength of binding to this site. Therefore, the partially overlapping expression patterns may provide a mechanism to modulate binding to the IBS and, hence, gene regulation. Together, these results suggest that combinatorial DNA binding mediated by Iro heterodimers may contribute to gene regulation.
It is striking that there is little conservation of Iro transcription factors outside the well-conserved HD. This finding suggests that different Iro proteins may have different protein-binding partners and, hence, interact with different signal transduction pathways or corepressors/coactivators to control gene expression. Formation of Iro heterodimers thus may allow for integration of different pathways on target gene enhancers.
Multiple copies of the IBS can be recognized by Mirr in vivo to mediate transcriptional repression in the eye imaginal disk. This finding indicates that Mirr acts as a transcriptional repressor, consistent with the known ability of Mirr to repress fng. Examination of the fng genomic region revealed an IBS immediately downstream of fng, consistent with a direct role of mirr in regulating fng expression. Mirr can bind the fng IBS in EMSA, supporting the proposal that Mirr directly represses fng expression. Reporter assays in Drosophila tissue culture demonstrated that Mirr can repress transcription through the fng downstream enhancer and that this repression was lost when the Mirr binding site was mutated. Together these data suggest that Mirr directly represses fng expression in vivo.
The derepression of reporter activity upon depletion of S2 cells for Mirr, although reproducible and significant, was modest. S2 cells also express mRNA for another Iro, caup (G.C., unpublished data), which is likely to be redundant on the fng enhancer.
Although our data indicate that Mirr acts as a transcriptional repressor in the eye, we cannot exclude the possibility that Mirr may act as a transcriptional activator in other contexts. Recent work has shown that phosphorylation can convert the vertebrate Iro, IRX2, from a transcriptional repressor to an activator (24), suggesting that regulation of transcription by Mirr also may be context dependent. A major goal for the future is to understand how Iro transcription factors regulate gene expression to direct the development of complex structures.
Supplementary Material
Acknowledgments
We thank S. Bray (University of Cambridge, Cambridge, U.K.), V. Christoffels (University of Amsterdam, Amsterdam), I. Dawid (National Institutes of Health, Bethesda), J Gomez-Skarmeta (University of Madrid, Madrid), and J. Modolell (University of Madrid) for DNA constructs and flies. This work was supported by Cancer Research UK.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HA, hemagglutinin; HD, homeodomain; Iro, Iroquois; IBS, Iro binding site; IP, immunoprecipitated.
References
- 1.Perovic, S., Schroder, H., Sudek, S., Grebenjuk, V., Stifanic, M., Muller, M. & Muller, W. (2003) Evol. Dev. 5, 240-250. [DOI] [PubMed] [Google Scholar]
- 2.Cavodeassi, F., Modolell, J. & Gomez-Skarmeta, J. L. (2001) Development (Cambridge, U.K.) 128, 2847-2855. [DOI] [PubMed] [Google Scholar]
- 3.Gomez-Skarmeta, J. L., Diez del Corral, R., de la Calle-Mustienes, E., Ferre-Marco, D. & Modolell, J. (1996) Cell 85, 95-105. [DOI] [PubMed] [Google Scholar]
- 4.McNeill, H., Yang, C., Ungos, J. & Simon, M. (1997) Genes Dev. 11, 1073-1082. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Skarmeta, J. L. & Modolell, J. (2002) Curr. Opin. Genet. Dev. 12, 403-408. [DOI] [PubMed] [Google Scholar]
- 6.Bao, Z., Bruneau, B., Seidman, J., Seidman, C. & Cepko, C. (1999) Science 283, 1161-1164. [DOI] [PubMed] [Google Scholar]
- 7.Bellefroid, E., Kobbe, A., Gruss, P., Gurdon, J. & Papalopulu, N. (1998) EMBO J. 17, 191-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Skarmeta, J. L., Glavic, A., de la Calle-Mustienes, E., Modolell, J. & Mayor, R. (1998) EMBO J. 17, 181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudoh, T. & Dawid, I. B. (2001) Proc. Natl. Acad. Sci. USA 98, 7852-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez, M. & de Celis, J. (1998) Nature 396, 276-278. [DOI] [PubMed] [Google Scholar]
- 11.Cho, K. & Choi, K. (1998) Nature 396, 272-276. [DOI] [PubMed] [Google Scholar]
- 12.Cavodeassi, F., Diez Del Corral, R., Campuzano, S. & Dominguez, M. (1999) Development (Cambridge, U.K.) 126, 4933-4942. [DOI] [PubMed] [Google Scholar]
- 13.Yang, C., Simon, M. & McNeill, H. (1999) Development (Cambridge, U.K.) 126, 5857-5866. [DOI] [PubMed] [Google Scholar]
- 14.Jordan, K., Clegg, N., Morimoto, A., Sen, J., Stein, D., McNeill, H., Deng, W., Tworoger, M. & Ruohola-Baker, H. (2000) Nat. Genet 24, 429-433. [DOI] [PubMed] [Google Scholar]
- 15.Jin, Z., Zhang, J., Klar, A., Chedotal, A., Rao, Y., Cepko, C. & Bao, Z. (2003) Development (Cambridge, U.K.) 130, 1037-1048. [DOI] [PubMed] [Google Scholar]
- 16.Burglin, T. (1997) Nucleic Acids Res. 25, 4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao, D., Woolner, S. & Bownes, M. (2000) Dev. Genes. Evol. 210, 449-457. [DOI] [PubMed] [Google Scholar]
- 18.Irvine, K. (1999) Curr. Opin. Genet. Dev. 9, 434-441. [DOI] [PubMed] [Google Scholar]
- 19.Jennings, B, Tyler, D. & Bray, S. (1999) Mol. Cell. Biol. 19, 4600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollock, R. & Treisman, R. (1990) Nucleic Acids Res. 18, 6197-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey, T. & Elkan, C. (1994) Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28-36. [PubMed] [Google Scholar]
- 22.Watabe, T., Kim, S., Candia, A., Rothbacher, U., Hashimoto, C., Inoue, K. & Cho, K. (1995) Genes Dev. 9, 3038-3050. [DOI] [PubMed] [Google Scholar]
- 23.Wilson, D., Sheng, G., Lecuit, T., Dostatni, N. & Desplan, C. (1993) Genes Dev. 7, 2120-2134. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto, K., Nishihara, S., Kamimura, M., Shiraishi, T., Otoguro, T., Uehara, M., Maeda, Y., Ogura, K., Lumsden, A. & Ogura, T. (2004) Nat. Neurosci. 7, 605. [DOI] [PubMed] [Google Scholar]
- 25.Sosinsky, A., Bonin, C. P., Mann, R. S. & Honig, B. (2003) Nucleic Acids Res. 31, 3589-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.