Abstract
The gram-positive bacterium Renibacterium salmoninarum produces relatively large amounts of a 57-kDa protein (p57) implicated in the pathogenesis of salmonid bacterial kidney disease. Antigenic variation in p57 was identified by using monoclonal antibody 4C11, which exhibited severely decreased binding to R. salmoninarum strain 684 p57 and bound robustly to the p57 proteins of seven other R. salmoninarum strains. This difference in binding was not due to alterations in p57 synthesis, secretion, or bacterial cell association. The molecular basis of the 4C11 epitope loss was determined by amplifying and sequencing the two identical genes encoding p57, msa1 and msa2. The 5′ and coding sequences of the 684 msa1 and msa2 genes were identical to those of the ATCC 33209 msa1 and msa2 genes except for a single C-to-A nucleotide mutation. This mutation was identified in both the msa1 and msa2 genes of strain 684 and resulted in an Ala139-to-Glu substitution in the amino-terminal region of p57. We examined whether this mutation in p57 altered salmonid leukocyte and rabbit erythrocyte binding activities. R. salmoninarum strain 684 extracellular protein exhibited a twofold increase in agglutinating activity for chinook salmon leukocytes and rabbit erythrocytes compared to the activity of the ATCC 33209 extracellular protein. A specific and quantitative p57 binding assay confirmed the increased binding activity of 684 p57. Monoclonal antibody 4C11 blocked the agglutinating activity of the ATCC 33209 extracellular protein but not the agglutinating activity of the 684 extracellular protein. These results indicate that the Ala139-to-Glu substitution altered immune recognition and was associated with enhanced biological activity of R. salmoninarum 684 p57.
Renibacterium salmoninarum is a gram-positive bacterial pathogen that causes significant losses of salmonid fish (reviewed in references 22 and 50). This bacterium is prevalent throughout most regions of the world where salmon live wild or are cultured. R. salmoninarum causes a chronic bacteremia termed bacterial kidney disease that is characterized by focal lesions in the viscera, particularly the kidney (22, 50). Bacterial kidney disease is difficult to control as R. salmoninarum is a slowly growing, facultatively intracellular pathogen (27, 54) that is transmitted from parent to progeny via intracellular infection of eggs (8, 10, 18), as well as from fish to fish during cohabitation (32). Successful control of bacterial kidney disease has been achieved in part through extensive chemotherapy combined with culling of infected broodstock (reviewed in reference 16).
Diagnosis of R. salmoninarum infection in salmon is often based on detection of the 57-kDa (p57) protein, mRNA, or DNA (6, 11, 13, 34, 37, 41). p57 is a good marker of active infection as this protein is the predominant cell surface and secreted protein produced by R. salmoninarum (23, 26, 51). However, little is known about whether antigenic variation exists in p57 or how variation may affect the biological functions of p57.
The gene encoding p57 has been cloned and designated msa (major soluble antigen) (12), and two copies of the msa gene have been identified in the R. salmoninarum genome (36). The msa1 and msa2 genes have identical coding regions and are both present in all strains of R. salmoninarum that have been examined (36). The msa1 and msa2 gene sequences diverge 40 bp 5′ to the open reading frame, while sequences 3′ to the open reading frame are essentially identical for at least 225 bp, except for a single base substitution 37 nucleotides (nt) downstream of the stop codon (36).
While the precise role of p57 in pathogenesis is unclear, we and other workers have demonstrated that secreted p57 has both agglutinating and immunomodulatory activities (7, 21, 42, 47, 52). p57 concentrated from bacterial culture supernatant binds and agglutinates salmonid leukocytes, as well as red blood cells from a number of mammalian species (14, 52). p57 does not agglutinate salmonid erythrocytes or mouse leukocytes, suggesting that binding occurs via a specific receptor(s). Two murine monoclonal antibodies (MAbs), MAbs 4C11 and 4H8, block the leukocyte-agglutinating activity, while the same antibodies, as well as MAb 4D3, also block p57-mediated agglutination of rabbit red blood cells (52). Furthermore, MAbs 4C11, 4H8, and 4D3 recognize epitopes on the amino-terminal portion of p57 since these antibodies bind a recombinant amino-terminal fragment but do not bind a recombinant carboxy-terminal fragment of p57 (48). These data suggest that the leukocyte-binding domain(s) may be associated with the amino-terminal portion of p57. At present it is unclear if these antibodies inhibit one leukocyte-binding site or multiple binding sites.
In this study we screened natural isolates of R. salmoninarum using MAbs to determine whether antigenic variation occurs in p57 and to facilitate characterization of the precise epitopes recognized by the agglutination-blocking MAbs. We describe a Norwegian R. salmoninarum isolate, strain 684, that produces p57 but lacks the 4C11 epitope. Full-length p57 is produced, and only a single nucleotide substitution was identified in the coding regions of both the msa1 and msa2 genes. p57 isolated from 684 has an increased capacity to bind leukocytes, indicating that the mutation, in addition to altering an antigenic epitope, is also associated with enhanced functional activity. To our knowledge, this is the first molecular characterization of antigenic variation produced by mutations in the msa genes of R. salmoninarum.
MATERIALS AND METHODS
Animals.
Chinook salmon (Oncorhynchus tshawytscha) were maintained in sand-filtered, UV-treated lake water at the Western Fisheries Research Center, Seattle, Wash.
R. salmoninarum strains and culture.
R. salmoninarum isolates K50, K70, K28, Little Goose, and D6 were provided by C. Banner (Department of Fish and Wildlife, Corvallis, Oreg.) and are recognized by anti-p57 MAbs 4D3 and 2G5 (51). R. salmoninarum strains ATCC 33209 and ATCC 33739 were obtained from the American Type Culture Collection (Manassas, Va.). R. salmoninarum isolate 684 was provided by O. B. Dale (National Veterinary Institute, Oslo, Norway). Isolate 684 was isolated from a clinically diseased brown trout (Salmo trutta) that was obtained from a hatchery in Aurland Sognefjord, Norway. The hatchery had experienced previous clinical outbreaks of bacterial kidney disease, and the Gram stain results, growth characteristics, and API-ZYM test results for isolate 684 were consistent with classification of this organism as R. salmoninarum (O. B. Dale, personal communication). Frozen (−70°C) aliquots of R. salmoninarum strains were initially cultured in KDM-2 medium containing 10% fetal bovine serum (17). For large-scale culture, starter cultures of ATCC 33209 and 684 were first prepared from freezer stocks and grown in KDM-2 broth medium modified so that it contained 0.05% (wt/vol) cysteine and no fetal bovine serum. The cultures were stirred continuously while they were incubated at 15°C for 10 days. Bacteria were enumerated by using the membrane filtration-fluorescent antibody technique (15), and equivalent numbers of R. salmoninarum ATCC 33209 and 684 were inoculated into separate 1.25-liter volumes of the modified KDM-2 broth medium. In the first experiment (preparation 1), the initial inoculum densities were 1 × 105 cells ml−1, and in the second experiment (preparation 2), the initial inoculum densities were 8 × 103 cells ml−1. Cultures were grown for an additional 12 days (preparation 1) or 14 days (preparation 2), and the numbers of viable bacteria were determined by plate counting. To harvest cultures, cells were pelleted by centrifugation at 5,000 × g, and both cell pellets obtained at 4°C and supernatants were frozen at −70°C.
Chromosomal DNA isolation and Southern blotting.
Genomic DNA was prepared by using a modified procedure described in the Molecular Biology Protocols (U.S. Department of Commerce/NOAA/NMFS/NWFSC website at http://www.nwfsc.noaa.gov/protocols/GramPosDNA.html) that was originally adapted from the work of Flamm et al. (20). Briefly, 100 μl of a 500-mg (wet weight) ml−1 suspension of bacterial cells was added to 0.9 ml of 0.01 M sodium phosphate-buffered saline (PBS) containing 20% sucrose, 2.5 mg of lysozyme (Sigma), and 1 mM EDTA and incubated overnight at 37°C. Nine milliliters of lysis buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 10 mg of proteinase K [Gibco BRL, Grand Island, N.Y.] ml−1, 1% sodium dodecyl sulfate [SDS]) was added, and the preparation was incubated for 3 h at 37°C. DNA was extracted with phenol-chloroform, ethanol precipitated, isolated by using a glass rod, and resusupended in 10 mM Tris-HCl-1 mM EDTA. For Southern blotting, genomic DNA was cut with BamHI, HindIII, EcoRI, and XhoI restriction endonucleases for 6 h at 37°C. Digested DNA (7 μg) was resolved on a 0.75% agarose gel for 21 h. DNA was transferred to a nitrocellulose membrane and heated for 45 min at 80°C as described previously (43). A probe comprising a 629-nt PCR fragment obtained by using primers 7 and 8 (Fig. 1) was labeled with [α-32P]dCTP by using a random hexamer labeling kit (Roche) according to the manufacturer's directions (19). Southern hybridization and washes of the blot were performed by standard methods (43).
FIG. 1.
(A) Linear depiction of the msa1 and msa2 genes of R. salmoninarum ATCC 33209. The 5′ region and the 3′ single nucleotide that are different in the ATCC 33209 msa1 and msa2 genes are indicated by boxes. (B) Primers used for PCR and sequencing of p57 msa1 and msa2. Primers 1, 3, 5, and 6 are specific for either msa1 or msa2, while all other primers do not distinguish between the known msa gene sequences. Translational start (ATG) and stop (TAA) sites are indicated.
ECP preparation.
Extracellular protein (ECP) was concentrated from medium by ultrafiltration followed by ammonium sulfate precipitation as previously described (52). The predominant components of ECP are intact p57 and associated proteolytic degradation fragments, as determined by SDS-polyacrylamide gel electrophoresis (PAGE) (42, 52). Total protein was determined by using the Bradford protein assay (Pierce, Rockford, Ill.) with bovine serum albumin as the standard. p57 was filter sterilized (pore size, 0.22 μm) and stored at −70°C or kept at 4°C for immediate use.
Amplification and sequencing of msa1 and msa2.
The entire coding sequences of the msa1 and msa2 genes were amplified from R. salmoninarum genomic DNA by using 5′ region forward primer 1, 3, 5, or 6 in combination with 3′ reverse primer 2 (Fig. 1). PCR amplification was carried out by using high-fidelity PfuTurbo polymerase as recommended by the manufacturer (Stratagene, La Jolla, Calif.). PCR amplification cycles (95°C for 5 min, followed by 30 cycles of 95°C for 40 s, 58°C for 40 s, and 72°C for 4 min) and final extension for 10 min at 72°C were performed with a Perkin-Elmer 9700 thermocycler. Reactions were carried out in a total volume of 50 μl. PCR products were resolved on a 1% agarose gel (Metaphor agarose; BioWhittaker Molecular Applications, Rockland, Maine) and were purified by QIAquick gel extraction (Qiagen Inc., Valencia, Calif.). Four 5′ primers (primers 1, 3, 5, and 6) and an internal 3′ region reverse primer (primer 4) were used to sequence the msa1 and msa2 PCR amplicons directly from the 5′ region through 726 nt of the coding region. Subsequently, the entire coding sequence of 684 msa1 and msa2 was determined by sequencing both strands of cloned msa1 and msa2 genes with primers 1 to 4 and 7 to 15 (Fig. 1). Nucleotide sequencing was performed by using an ABI Prism terminator cycle sequencing kit and AmpliTaq DNA polymerase according to the manufacturer's directions (Applied Biotechnology, Inc., Foster City, Calif.). Reactions were performed with an Applied Biosystems 377 PRISM automated DNA sequencer (PE Applied Biosystems, Foster City, Calif.) at the Nucleic Acid Core Facility of the Department of Molecular Microbiology and Immunology at Oregon Health and Science University, Portland.
Cloning of msa1 and msa2 from strain 684.
msa1 and msa2 PCR products were ligated into the pT7Blue-1 vector (Novagen, Madison, Wis.) and transformed into NovaBlue Singles Competent Escherichia coli according to the manufacturer's directions (Novagen).
MAbs.
MAbs 3H1, 4D3, 4C11, and 4H8 bind p57 and have been described previously (52). Cells were grown in BALB/c mice as ascites, and antibody was purified by using protein A-Sepharose.
Gel electrophoresis.
SDS-PAGE was performed as previously described (53).
Dot and Western blotting.
Frozen R. salmoninarum cells were thawed, pelleted, washed in 10 mM PBS (pH 7.4), and finally resuspended in an equal volume of PBS. For dot blotting, bacterial cells were diluted 1:500 in PBS, and 100 μl of the suspension was applied to a dot blot apparatus and incubated overnight at 17°C. Wells were blocked with 3% bovine serum albumin-Tris-buffered saline (pH 8.0) for 1 h at room temperature. Wells were washed three times with 150 μl of Tris-buffered saline, 100 μl of a 10-μg ml−1 solution of primary antibody was added, and the preparation was incubated for 1 h at room temperature. Wells of the dot blot were washed three times, and the nitrocellulose was removed. Dot blots were incubated in a 1:500 dilution of peroxidase-conjugated goat anti-mouse immunoglobulin (HyClone, Logan, Utah) for 1 h at room temperature. After three washes, blots were incubated with a 4-chloro-1-naphthol substrate (Bio-Rad Laboratories, Hercules, Calif.).
Leukocyte agglutination assays.
Salmonid anterior kidney leukocytes were isolated and separated by using 51% Percoll as previously described (1). About 1 × 106 cells were applied per well in 10% fetal bovine serum in a total volume of 100 μl. Leukocytes were incubated with twofold dilutions of p57 (200 to 1.6 μg ml−1). While agglutination occurred rapidly, the minimum concentration of p57 that exhibited agglutinating activity was determined after 24 h by microscopic examination. For inhibition experiments, protein A-purified MAb 4C11 or control antibody PCG1-1 [immunoglobulin G2b(κ) (49)] was preincubated with ECP prior to addition of cells.
p57 leukocyte binding assay.
Peripheral blood or anterior kidney leukocytes (5 × 106 cells) in tissue culture medium containing 20% fetal bovine serum were incubated with 25 μg of p57 for 1 h at 4°C in a total volume of 150 μl. The cells were washed three times with tissue culture medium (1 ml) and then lysed with a buffer containing 1% bovine serum albumin, 0.05% Tween 20, and 0.25% NP-40 in PBS for 30 min. The lysates were heated for 5 min at 85°C to release bound p57, and the insoluble material was pelleted by centrifugation (14,000 × g for 2 min). A quantitative enzyme-linked immunosorbent assay (ELISA) was used to determine the concentration of soluble p57 in the supernatant.
Quantitative ELISA for p57.
A quantitative ELISA for p57 was performed as described previously (41), with the following modifications. Briefly, half-area microplate wells were coated with 50 μl of a 5-μg ml−1 MAb 3H1 solution, and following the antigen incubation step wells received 50 μl of a 1-μg ml−1 biotinylated MAb 4D3 solution. The biotinylated MAb was detected by using a 1:500 dilution of streptavidin-alkaline phosphatase (Southern Biotechnology, Birmingham, Ala.) and incubation with a p-nitrophenylphosphate substrate (Sigma).
Nucleotide sequence accession numbers.
Sequences of the cloned strain 684 msa1 and msa2 genes have been deposited in the GenBank database under accession numbers AF458101 and AF458102.
RESULTS
Epitope variation in R. salmoninarum.
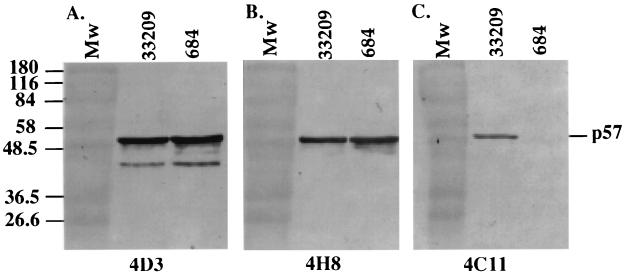
We examined whether antigenic variation exists in bacterial cell-associated p57 by probing bacteria with four MAbs that recognize spatially separate epitopes. Eight strains of R. salmoninarum, obtained from geographically diverse locations, were reacted with MAbs 3H1, 4D3, 4H8, and 4C11 by using the dot blot assay (Fig. 2). MAb 3H1 recognizes an epitope localized in the central region of p57 (52), while MAbs 4D3, 4H8, and 4C11 recognize distinct epitopes located in the amino-terminal region of p57 (48). Notably, Norwegian R. salmoninarum strain 684 lacked the 4C11 epitope, while all other strains contained epitopes recognized by the 3H1, 4D3, 4H8, and 4C11 MAbs. To determine if the loss of the 4C11 epitope was due to a large deletion in p57 or due to occlusion with other R. salmoninarum cellular constituents, we conducted Western blot experiments in which bacterial lysates were first resolved by using reducing SDS-PAGE and then probed with MAbs 4D3, 4H8, and 4C11 (Fig. 3). p57 produced by strain 684 exhibited the same relative migration as p57 produced by ATCC 33209, indicating that full-length protein was produced by strain 684 (Fig. 3A and B). Furthermore, the quantities of 684 p57 produced appeared to be similar to the quantities of ATCC 33209 p57 produced, as judged by the intensities of MAb 4D3 and 4H8 immunoreactivities (Fig. 3A and B) and Coomassie blue protein staining (data not shown). In agreement with the dot immunoblotting results, MAb 4C11 failed to bind 684 p57 as determined by Western blotting, while MAb 4C11 bound ATCC 33209 p57 (Fig. 3C). The failure of MAb 4C11 to bind 684 p57 under reducing and denaturing conditions suggested that the 4C11 epitope was directly altered rather than occluded by noncovalently associated R. salmoninarum cellular constituents.
FIG. 2.
Screening R. salmoninarum isolates with MAbs 3H1, 4D3, 4H8, and 4C11. Washed bacterial cells were dot blotted onto nitrocellulose and individually probed with 10 μg of each MAb ml−1. Binding was detected with peroxidase-conjugated goat anti-mouse immunoglobulin. The designation and origin of each isolate are indicated on the right.
FIG. 3.
R. salmoninarum strains 684 and ATCC 33209 express similar amounts of p57 with indistinguishable electrophoretic migration positions as determined by Western blotting with MAbs 4D3 (A) and 4H8 (B). p57 from strain 684 lacks the 4C11 epitope (C). Equal amounts of R. salmoninarum cells were boiled in reducing SDS-PAGE sample buffer, separated on an SDS-12% PAGE gel, transferred to nitrocellulose overnight, and subjected to immunoblotting with 5 μg of MAb 4D3, 4H8, or 4C11 ml−1. Peroxidase-conjugated goat anti-mouse immunoglobulin was used to detect MAb binding. The molecular masses (in kilodaltons) of prestained markers (lanes Mw) are indicated on the left. The results are representative of one of three experiments.
Both the msa1 and msa2 genes are present in strain 684.
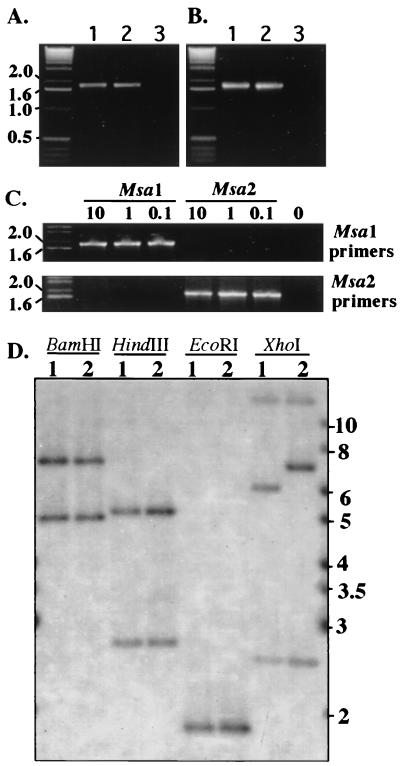
All strains of R. salmoninarum that have been examined contain two identical copies of the msa gene (36). At present, it is not known whether one or both of the msa genes are transcribed. Antigenic variation could be due to exclusive expression of one mutated gene. Alternatively, the same mutation may be present in both genes. To distinguish between these possibilities, we first examined whether the msa1 and msa2 genes were present in the genome of strain 684 by performing PCR amplification with gene-specific primers. The 5′ regions of msa1 and msa2 diverge 40 nt upstream of the transcription start site (36); thus, primers were designed that specifically hybridize to the 5′ region of either msa1 (primers 1 and 6) (Fig. 1) or msa2 (primers 3 and 5) (Fig. 1). Because only a single nucleotide difference in the noncoding region is known to exist between the msa1 and msa2 genes (36), a 3′ reverse primer (primer 2) (Fig. 1) that recognizes both msa1 and msa2 was used in all amplifications. Genomic DNA from either ATCC 33209 or 684 was subjected to 30 cycles of PCR amplification with the high-fidelity TurboPfu polymerase. A PCR product corresponding to the predicted 1.87-kb fragment was generated from both R. salmoninarum ATCC 33209 and 684 DNA with either msa1-specific primers (Fig. 4A) or msa2-specific primers (Fig. 4B), indicating that both genes are present in both strains. The specificity of the primers for either msa1 or msa2 was demonstrated by the fact that msa1 primers amplified cloned msa1 but not cloned msa2, while msa2 primers amplified cloned msa2 but not the msa1 gene when a range of input plasmid concentrations was used (Fig. 4C). To confirm the presence and examine the genomic organization of the msa genes, Southern blotting was performed with a 5′ region msa gene probe. Genomic DNA from ATCC 33209 and 684 were cut with restriction enzymes BamHI, HindIII, EcoRI, and XhoI and probed with a 629-nt probe amplified from ATCC 33209 by using primers 7 and 8 (Fig. 1). In agreement with the data of O'Farrell and Strom (36), BamHI digestion of ATCC 33209 genomic DNA produced 7.2-kb (msa2) and 5.3-kb (msa1) hybridizing bands. BamHI digestion of 684 genomic DNA produced bands with migration positions identical to those of ATCC 33209 bands. Similarly, HindIII digestion produced 5.4-kb (msa2) and 2.8-kb (msa1) hybridizing bands that were present in both ATCC 33209 and 684 digests. EcoRI digestion of ATCC 33209 and 684 DNA produced a single 1.8-kb band known to contain the entire coding sequence of both msa1 and msa2 except for 92 nt of the 5′ coding sequence. These data confirm that both full-length msa1 and full-length msa2 are present in the genome of R. salmoninarum 684, in agreement with PCR data. While the restriction enzyme digestion patterns for the genomic DNA are similar for the two strains, they are not identical, as digestion of ATCC 33209 DNA with XhoI produced a 6.2-kb band, while digestion of 684 DNA produced a 6.9-kb hybridizing band. A third band, at ∼14 kb, was likely a product of cross-hybridization of the 5′ probe to the second A repeat present in both the msa1 and msa2 genes, as a 3′ probe amplified from ATCC 33209 by using primers 11 and 2 hybridized only with the ∼14-kb band (Wiens, unpublished data). Taken together, these data demonstrate that the msa1 and msa2 genes are present in isolate 684 and that the flanking DNA sequence of the msa1 and msa2 genes in isolate 684 is similar, but not identical, to the flanking DNA sequence in ATCC 33209, as judged by restriction enzyme analysis.
FIG. 4.
Strain 684 contains both msa1 and msa2 as determined by PCR (A to C) and Southern blotting (D). (A and B) PCR was performed as described in Materials and Methods by using primers 1 and 2 to amplify msa1 (A) and primers 3 and 2 to amplify msa2 (B). The target DNA in PCR amplifications were as follows: lanes 1, 10 ng of ATCC 33209 genomic DNA; lanes 2, 10 ng of 684 genomic DNA; lanes 3, control (no DNA). (C) PCR amplification performed with either msa1- or msa2-specific primers and 0.1 to 10 ng of target plasmid DNA containing either a cloned msa1 gene or a cloned msa2 gene. (D) Southern blotting of genomic DNA from strain ATCC 33209 (lane 1) or strain 684 (lane 2) that was probed with a 629-nt PCR fragment obtained with primers 7 and 8 and labeled with [α-32P]dCTP. The migration positions of the molecular size markers (in kilobases) are indicated on the left in panels A through C and on the right in panel D.
Genetic basis of the loss of the 4C11 epitope.
The molecular basis of the loss of the 4C11 epitope was determined by directly sequencing PCR-amplified msa1 and msa2 genes. Since the location of the 4C11 epitope was previously demonstrated to be in the first 243 amino acids (48), the sequences of the 5′ noncoding regions through amino acid 243 were determined by direct sequencing of 684 msa1 and msa2 PCR amplicons (Fig. 5 and data not shown) obtained by using four independent primers unique to the divergent 5′ region of each gene (primers 1, 3, 5, and 6) in combination with the 3′ region primer (primer 2) that binds to a sequence common to the two genes. (Fig. 1). A single C-to-A nucleotide difference in the coding region was identified that resulted in an Ala139-to-Glu amino acid substitution in both the msa1 and msa2 gene products (Fig. 5). The unique sequence in the 5′ region of each PCR product confirmed the identity as either the msa1 or msa2 gene. Amplification and direct sequencing of the ATCC 33209 msa1 and msa2 genes that were amplified in parallel with the 684 msa genes did not reveal any differences from the reported sequences, indicating the fidelity of the amplification and sequencing process. The presence of the single mutation in both the msa1 and msa2 genes from strain 684 was confirmed by cloning the full-length 684 msa1 and msa2 genes and sequencing the entire coding regions, as well as the immediate 5′ and 3′ flanking regions. The single C-to-A mutation was present in both cloned genes, while the 5′ region and the rest of the coding sequence were identical to those of the corresponding ATCC 33209 msa1 and msa2 genes. Interestingly, the single nucleotide difference between the ATCC 33209 msa1 and msa2 genes, located 37 nt downstream of the stop codon (Fig. 1), was not present in 684. The isolate 684 msa1 and msa2 genes both contained a C at this position, like the ATCC 33209 msa2 gene. A cytosine at this position in both msa1 and msa2 of isolate 684 was also confirmed by direct sequencing of PCR products (data not shown).
FIG. 5.
DNA sequences and predicted amino acids of p57 encoded by msa1 and msa2 from R. salmoninarum strains ATCC 33209 and 684. The 5′ primers used to amplify msa1 or msa2 are underlined. Dashes indicate identity with ATCC 33209 msa1. Only one sequence for ATCC 33209 msa1 and msa2 is shown as the sequences are identical starting 38 bp 5′ of the ATG start site through the entire coding region.
Agglutinating activity of p57 from 684.
To investigate whether p57 from strain 684 has altered biological activity, we prepared two independent ECP preparations from strains 684 and ATCC 33209 grown under identical conditions. Both protein preparations were tested for binding and agglutinating activities by using salmonid leukocytes and rabbit red blood cells. Surprisingly, ECP from strain 684 exhibited an approximately twofold increase in agglutinating activity for both chinook salmon anterior kidney leukocytes (Table 1) and rabbit erythrocytes (data not shown). The minimum leukocyte-agglutinating concentration of 684 ECP was 12.5 μg ml−1, while the minimum leukocyte-agglutinating concentration of ATCC 33209 ECP was 25 μg ml−1. To confirm these results, a quantitative binding assay was performed. In this assay, ECP was incubated with leukocytes, unbound proteins were removed by washing, and the amount of bound p57 was determined by a quantitative ELISA that specifically detects p57. Significantly more strain 684 p57 (89 ± 11 ng; n = 3) than strain ATCC 33209 p57 (60 ± 6 ng; n = 3) was bound per 1 × 106 leukocytes in 1 h (Fig. 6A). A twofold increase in strain 684 binding activity to rabbit red blood cells was also observed with this assay (Fig. 6B). The difference in p57 binding activity was not due to differences in the amounts of p57 in the extracellular preparations as measured by a p57-specific ELISA (Fig. 6C) and by SDS-PAGE (data not shown). In addition, ELISA analysis confirmed the lack of the 4C11 epitope on p57 isolated from strain 684 and the presence of the 4C11 epitope on p57 from ATCC 33209 (Fig. 6D). Binding and agglutination were specific to p57 as MAb 4C11 inhibited leukocyte agglutination mediated by ATCC 33209 ECP, while MAb 4C11 did not inhibit agglutination mediated by 684 ECP (Table 2). This indicates that the loss of the 4C11 epitope on p57 resulted in a functional inability of MAb 4C11 to block agglutinating activity. A control MAb, MAb PCG1-1, did not inhibit leukocyte agglutination mediated by either ATCC 33209 or 684 ECP, indicating that the inhibition by MAb 4C11 of ATCC 33209 ECP was specific. These data indicate that p57 produced by strain 684 has greater binding activity per microgram of protein than an equivalent amount of p57 produced by ATCC 33209 and suggest that the enhanced activity is due to the single Ala139-to-Glu substitution.
TABLE 1.
ECP from isolate 684 agglutinates salmonid leukocytes with twofold higher activity than ECP from ATCC 33209
| ECP (μg ml−1) | Agglutination
|
|||||
|---|---|---|---|---|---|---|
| Fish 1
|
Fish 2
|
Fish 3
|
||||
| ATCC 33209 | 684 | ATCC 33209 | 684 | ATCC 33209 | 684 | |
| 500 | +a | + | + | + | + | + |
| 100 | + | + | + | + | + | + |
| 50 | + | + | + | + | + | + |
| 25 | + | + | + | + | + | + |
| 12.5 | − | + | − | + | − | + |
| 6.25 | − | − | − | − | − | − |
| 0 (PBS) | − | − | − | − | − | − |
+, agglutinating activity; −, no agglutinating activity. The data are data from one of two experiments that yielded similar results.
FIG. 6.
(A) p57 produced by R. salmoninarum strain 684 has 50% increased binding activity for chinook salmon anterior kidney leukocytes compared to the binding activity of an equivalent amount of p57 isolated from R. salmoninarum strain ATCC 33209. The data are averages ± standard errors of the means for binding of p57 to leukocytes from three salmon. The asterisk indicates that the results were significantly different at a P value of <0.05 (Student's t test). ECP was purified from culture supernatants of bacteria grown at the same time under identical conditions. The purification procedure and binding assay are described in Materials and Methods. (B) p57 from strain 684 exhibits a twofold increase in binding to rabbit red blood cells. The three asterisks indicate that the results were significantly different at a P value of <0.001 (Student's t test). (C) Identical amounts of immunoreactive p57 are present, as determined by a capture ELISA. MAb 3H1 was coated onto a plate, followed by dilutions of the ECP containing p57 from either strain ATCC 33209 or strain 684. Bacterial protein concentrations were determined by the Bradford protein assay. Biotinylated MAb 4D3 was used as a capture antibody, and this was followed by detection with a strepavadin-alkaline phosphatase conjugate. O.D., optical density. (D) Secreted p57 produced by strain 684 lacks the 4C11 epitope. The ELISA was identical to the ELISA whose results are shown in panel C, except that biotinylated MAb 4C11 was used instead of biotinylated MAb 4D3. The data are representative of the data from two experiments performed with the two preparations of ATCC 33209 and 684 ECP.
TABLE 2.
MAb 4C11 inhibits ATCC 33209 p57-mediated agglutination of leukocytes but not 684 p57-mediated agglutination of leukocytesa
| Inhibitor | Concn (μg ml−1) | Agglutination
|
|||
|---|---|---|---|---|---|
| Preparation 1
|
Preparation 2
|
||||
| ATCC 33209 | 684 | ATCC 33209 | 684 | ||
| Control | +b | + | + | + | |
| 4C11 | 80 | − | + | − | + |
| 40 | − | + | − | + | |
| 20 | ± | + | ± | + | |
| 10 | + | + | ± | + | |
| 5 | + | + | + | + | |
| PCG1-1 | 80 | + | + | + | + |
ECP (50 μg ml−1) was preincubated with dilutions of protein A-purified MAb 4C11, PCG1-1 (antibody of the same isotype that does not bind p57), or tissue culture medium alone (control).
+, agglutination; −, no agglutination; ±, weak agglutination.
DISCUSSION
R. salmoninarum infects salmonid fish over a wide geographic area, including North America, South America, Europe, and Japan. However, isolates from these diverse locations have been quite similar, as determined by serological, biochemical, and genetic analyses (9, 23, 26, 45, 51). Here we report the occurrence of antigenic variation in p57 produced by Norwegian R. salmoninarum strain 684. Strain 684 p57 lacks the 4C11 epitope and contains an Ala139-to-Glu mutation in the products of both the msa1 and msa2 genes. Cook and Lynch (13) have reported a single nucleotide change in the msa gene of R. salmoninarum strain K2A2. Interestingly, the mutation in strain K2A2 was identical to that in strain 684 (B. Lynch, personal communication); however, whether the mutation occurred in both msa genes of strain K2A2 was not determined. The presence the same mutation in both strain K2A2 and strain 684 suggests that these isolates may have a common origin or that the second nucleotide of the Ala139 codon is a hot spot for mutation. A common origin would be surprising as strain K2A2 was isolated from an Atlantic salmon on the Margaree River in Nova Scotia, while strain 684 was isolated from a brown trout in Norway. Recently, genetic analysis of an exact tandem repeat locus (ETR-A) and the sequence of the 16S-23S rRNA intergenic spacer have provided a method to determine the relatedness of R. salmoninarum isolates (24-26). We carried out an analysis of these loci in strain 684 and determined that strain 684 possesses only one copy of the ETR-A locus (TR1) and that the sequence of the 16S-23S rRNA intergenic spacer belongs to sequevar 4 (Wiens, unpublished data). The TR1 and sequevar 4 genotypes resemble those of other R. salmoninarum strains isolated from Norway, Iceland, and Scotland and differ from those of most United States, England, Wales, mainland Europe, and Canadian strains, which belong to sequevar 1 and contain the TR2 allele (25, 26). Taken together, these data argue for a Scandinavian origin of strain 684 and may implicate transfer of R. salmoninarum to eastern Canada via infected eggs or fish. It will be interesting to examine additional R. salmoninarum isolates from the eastern United States and Canada and Scandinavia for nucleotide substitutions in the Ala139 codon to determine the prevalence of the Glu substitution and to investigate whether these strains share other genetic loci indicative of a common origin.
A surprising finding was that the two 684 msa genes had a common mutation, while the remaining coding sequences of 684 msa1 and msa2 were indistinguishable. With the exception of a single nucleotide change, the 684 msa1 and msa2 coding sequences were also identical to the reported coding sequences of ATCC 33209 msa1 and msa2 (36). O'Farrell and Strom (36) have proposed that the msa genes arose via duplication of an ancestral msa gene and that the presence of the two genes provides a selective advantage to R. salmoninarum. Here, we confirmed and extended the findings of these authors by demonstrating the presence of msa1 and msa2 in both ATCC 33209 and a Norwegian R. salmoninarum isolate. Examination of the genomic organization of the msa genes by BamHI, HindIII, and EcoRI digestion followed by Southern blotting identified hybridizing DNA fragments that were the same size in both 684 and ATCC 33209. These data suggest that the genomic organization of msa1 and msa2 is similar in the two strains and that two msa genes are present in R. salmoninarum isolates from diverse geographic areas. However, there are also differences, as XhoI digestion produced a 6.2-kb band in ATCC 33209 and a 6.9-kb hybridizing band in 684. Perhaps this difference and the presence of the 4C11 epitope may be useful for further molecular classification of R. salmoninarum isolates. It should be noted that only five isolates, including isolate 684, have been examined to date for the presence of msa1 and msa2 and that a more extensive survey is required to determine how prevalent these genes are in other R. salmoninarum isolates. The primers that we have identified which selectively amplify either msa1 and msa2 may facilitate further analysis of these genes.
The mechanism by which the identical changes were introduced into msa1 and msa2 of strain 684 is unclear. One possibility is that a spontaneous mutation occurred in one of the msa genes, which was followed by nonreciprocal recombination with the other msa gene. In support of this mechanism, the single nucleotide difference 3′ of the stop codon in ATCC 33209 (Fig. 1) is absent in isolate 684. In ATCC 33209, msa1 contains a G while msa2 contains a C at this position. Like the ATCC 33209 msa2 gene, the 684 msa1 and msa2 genes both contain a C located 37 bp 3′ of the stop codon. If the nonreciprocal recombination mechanism hypothesis is correct, then in the simplest scenario the mutation may have been first introduced into msa2 and then introduced by recombination into the msa1 locus, thus resulting in identical coding sequences as well as identical 3′ sequences. It should be noted that the 5′ sequences, at least as far as the −69 (msa1) and −73 (msa2) nucleotides 5′ of the translational initiation codon, resemble those determined for ATCC 33209 msa1 and msa2 and thus may not be involved in recombination. It is notable that IS3-like insertion sequences have been reported to flank the msa copies at a distance of 2 to 3 kb and thus may have been involved in msa gene duplication, potentially though recombination (36, 40). Recombination and unidirectional transfer of DNA between a pseudogene(s) and a functional gene(s) or between complete gene copies appear to be a common mechanism of antigenic variation in a number of pathogenic microorganisms (5, 30, 31, 35, 38, 46). A well-studied example is Neisseria gonorrhoeae, in which pilin antigenic variation can occur by unidirectional transfer of DNA sequences from a silent pilin locus to the expressed pilin gene through high-frequency recombination events (28, 29, 33, 44). At present we do not know if both msa genes are expressed or the frequency of the putative recombination process in the R. salmoninarum msa genes. The divergent 5′ sequences of msa1 and msa2 suggest that the msa genes may be differentially regulated. In addition to recombination as a mechanism of antigenic variation, we do not exclude the possibility that the identical mutations may be due to a bias in the mutational machinery or repair process targeting this region of the msa genes.
A surprising finding was that p57 isolated from isolate 684 culture supernatant displayed increased agglutinating and binding activity for salmonid leukocytes and rabbit erythrocytes. We envisage several possible mechanisms by which the mutation enhances binding activity. One possibility is that the mutation may increase the stability of p57. We have previously demonstrated that p57 is highly susceptible to digestion by an R. salmoninarum-produced serine protease (39, 42, 48). In addition, proteolytic activity may reside within the p57 protein itself (3). Alternatively, the mutation may increase the binding affinity for the putative receptor(s) on fish leukocytes and rabbit red blood cells. Enhanced binding could be due to an altered conformation of p57 or a change in the actual binding site. It is of interest that the mutation occurs close to the A1 repeat in p57 (48). The A1 and A2 direct repeats are 81 amino acids long, and each repeat contains a transcription factor-immunoglobulin (TIG)-like domain (2), also called an IPT domain (immunoglobulin-like fold shared by plexins and transcription factors [4]). The TIG/IPT domain is predicted to form an unusual type E immunoglobulin fold and is found in the extracellular regions of members of the plexin family of adhesion-repulsion molecules. If the A1 and A2 repeats are involved in binding to fish leukocytes, it is intriguing to speculate that the mutation at position 139 may alter these domains or their function. A final possibility is that the mutation may alter the binding of a cofactor involved in the binding of p57 to leukocytes. Investigations are under way to distinguish among these possibilities and fully map the binding site(s) on p57.
Although the role of p57 in pathogenesis is unclear, this protein is a well-accepted marker of infection by R. salmoninarum (6, 11, 13, 34, 37, 41). The correlation of cell-associated p57 with isolate virulence, the high levels of synthesis by R. salmoninarum, the in vitro binding activity with fish leukocytes, and the maintenance of duplicated msa genes support the hypothesis that p57 plays an important role in the pathogenesis of bacterial kidney disease. In this study we identified a p57 antigenic variant that has lost the 4C11 epitope and has increased in vitro binding and agglutinating activities. If this binding activity is relevant in pathogenesis, then loss of a neutralizing epitope may help R. salmoninarum escape a salmonid immune response. Alternatively, the mutation may be involved in tropism among tissues or among different salmonid species. A novel finding of this work was the demonstration of identical mutations in the coding regions of both the msa1 and msa2 genes. The development of isogenic strains and in vivo challenge experiments are required to determine the contribution of the p57 Ala139-to-Glu mutation to R. salmoninarum virulence.
Acknowledgments
Support for this research was provided by USDA National Research Initiative Competitive Grants Program award 2001-02229 to G.D.W.
We thank B. Wiens, M. Rittenberg, T. Welch, and J. Crosa for critical reviews of the manuscript. We thank T. Welch for help with Southern blotting and S. Alcorn for isolation of salmon leukocytes.
REFERENCES
- 1.Alcorn, S. W., A. L. Murray, and R. J. Pascho. 2002. Effects of rearing temperature on immune functions in sockeye salmon (Oncorhynchus nerka). Fish Shellfish Immunol. 12:303-334. [DOI] [PubMed]
- 2.Aravind, L., and E. V. Koonin. 1999. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287:1023-1040. [DOI] [PubMed] [Google Scholar]
- 3.Barton, T. A., L. A. Bannister, S. G. Griffiths, and W. H. Lynch. 1997. Further characterization of Renibacterium salmoninarum extracellular products. Appl. Environ. Microbiol. 63:3770-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork, P., T. Doerks, T. A. Springer, and B. Snel. 1999. Domains in plexins: links to integrins and transcription factors. Trends Biochem. Sci. 24:261-263. [DOI] [PubMed] [Google Scholar]
- 5.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, L. L., T. P. T. Evelyn, G. K. Iwama, and W. S. Nelson. 1995. Bacterial species other than Renibacterium salmoninarum cross-react with antisera against R. salmoninarum but are negative for the p57 gene of R. salmoninarum as detected by the polymerase chain reaction (PCR). Dis. Aquat. Organisms 21:227-231. [Google Scholar]
- 7.Brown, L. L., G. K. Iwama, and T. P. T. Evelyn. 1996. The effect of early exposure of Coho salmon (Oncorhynchus kisutch) eggs to the p57 protein of Renibacterium salmoninarum on the development of immunity to the pathogen. Fish Shellfish Immunol. 6:149-165. [Google Scholar]
- 8.Bruno, D. W. 1986. Histopathology of bacterial kidney disease in laboratory infected rainbow trout, Salmo gairdneri (Richardson), and Atlantic salmon, Salmo salar L., with reference to naturally infected fish. J. Fish Dis. 9:523-537. [Google Scholar]
- 9.Bruno, D. W., and A. L. S. Munro. 1986. Uniformity in the biochemical properties of Renibacterium salmoninarum isolates obtained from several sources. FEMS Microbiol. Lett. 33:247-250. [Google Scholar]
- 10.Bullock, G. L., H. M. Stuckey, and D. Mulcahy. 1978. Corynebacterial kidney disease: egg transmission following iodophore disinfection. Fish Health News 7:51-52. [Google Scholar]
- 11.Chase, D. M., and R. J. Pascho. 1998. Development of a nested polymerase chain reaction for amplification of a sequence of the p57 gene of Renibacterium salmoninarum that provides a highly sensitive method for detection of the bacterium in salmonid kidney. Dis. Aquat. Organisms 34:223-229. [DOI] [PubMed] [Google Scholar]
- 12.Chien, M. S., T. L. Gilbert, C. Huang, M. L. Landolt, P. J. O'Hara, and J. R. Winton. 1992. Molecular cloning and sequence analysis of the gene coding for the 57-kDa major soluble antigen of the salmonid fish pathogen Renibacterium salmoninarum. FEMS Microbiol. Lett. 75:259-265. [DOI] [PubMed] [Google Scholar]
- 13.Cook, M., and W. H. Lynch. 1999. A sensitive nested reverse transcriptase PCR assay to detect viable cells of the fish pathogen Renibacterium salmoninarum in Atlantic salmon (Salmo salar L.). Appl. Environ. Microbiol. 65:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly, J. G., and R. M. Stevenson. 1990. Characterization of the Renibacterium salmoninarum haemagglutinin. J. Gen. Microbiol. 136:949-953. [DOI] [PubMed] [Google Scholar]
- 15.Elliott, D. G., and T. Y. Barila. 1987. Membrane filtration-fluorescent antibody staining procedure for detecting and quantifying Renibacterium salmoninarum in coelomic fluid of chinook salmon. Can. J. Fish. Aquat. Sci. 44:206-210. [Google Scholar]
- 16.Elliott, D. G., R. J. Pascho, and G. L. Bullock. 1989. Developments in the control of bacterial kidney disease of salmonid fishes. Dis. Aquat. Organisms 6:201-215. [Google Scholar]
- 17.Evelyn, T. P. T. 1977. An improved growth medium for the kidney disease bacterium and some notes on using the medium. Bull. Int. Epizool. 87:511-513. [Google Scholar]
- 18.Evelyn, T. P. T., L. Prosperi-Porta, and J. E. Ketcheson. 1986. Experimental intra-ovum infection of salmonid eggs with Renibacterium salmoninarum and vertical transmission of the pathogen with such eggs despite their treatment with erythromycin. Dis. Aquat. Organisms 1:197-202. [Google Scholar]
- 19.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 20.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredriksen, A., C. Endresen, and H. I. Wergeland. 1997. Immunosuppressive effect of a low molecular weight surface protein from Renibacterium salmoninarum on lymphocytes from Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 7:273-282. [Google Scholar]
- 22.Fryer, J. L., and J. E. Sanders. 1981. Bacterial kidney disease of salmonid fish. Annu. Rev. Microbiol. 35:273-298. [DOI] [PubMed] [Google Scholar]
- 23.Getchell, R. G., J. S. Rohovec, and J. L. Fryer. 1985. Comparison of Renibacterium salmoninarum isolates by antigenic analysis. Fish Pathol. 20:149-159. [Google Scholar]
- 24.Grayson, T. H., S. M. Alexander, L. F. Cooper, and M. L. Gilpin. 2000. Renibacterium salmoninarum isolates from different sources possess two highly conserved copies of the rRNA operon. Antonie Leeuwenhoek 78:51-61. [DOI] [PubMed] [Google Scholar]
- 25.Grayson, T. H., F. A. Atienzar, S. M. Alexander, L. F. Cooper, and M. L. Gilpin. 2000. Molecular diversity of Renibacterium salmoninarum isolates determined by randomly amplified polymorphic DNA analysis. Appl. Environ. Microbiol. 66:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grayson, T. H., L. F. Cooper, F. A. Atienzar, M. R. Knowles, and M. L. Gilpin. 1999. Molecular differentiation of Renibacterium salmoninarum isolates from worldwide locations. Appl. Environ. Microbiol. 65:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutenberger, S. K., R. J. Duimstra, J. S. Rohovec, and J. L. Fryer. 1997. Intracellular survival of Renibacterium salmoninarum in trout mononuclear phagocytes. Dis. Aquat. Organisms 28:93-106. [Google Scholar]
- 28.Haas, R., and T. F. Meyer. 1986. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 44:107-115. [DOI] [PubMed] [Google Scholar]
- 29.Hagblom, P., E. Segal, E. Billyard, and M. So. 1985. Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 315:156-158. [DOI] [PubMed] [Google Scholar]
- 30.Howell-Adams, B., and H. S. Seifert. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37:1146-1158. [DOI] [PubMed] [Google Scholar]
- 31.Kenri, T., R. Taniguchi, Y. Sasaki, N. Okazaki, M. Narita, K. Izumikawa, M. Umetsu, and T. Sasaki. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 67:4557-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKibben, C. L., and R. J. Pascho. 1999. Shedding of Renibacterium salmoninarum by infected chinook salmon Oncorhynchus tschawytscha. Dis. Aquat. Organisms 38:75-79. [DOI] [PubMed] [Google Scholar]
- 33.Mehr, I. J., C. D. Long, C. D. Serkin, and H. S. Seifert. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miriam, A., S. G. Griffiths, J. E. Lovely, and W. H. Lynch. 1997. PCR and probe-PCR assays to monitor broodstock Atlantic salmon (Salmo salar L.) ovarian fluid and kidney tissue for presence of DNA of the fish pathogen Renibacterium salmoninarum. J. Clin. Microbiol. 35:1322-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noormohammadi, A. H., P. F. Markham, A. Kanci, K. G. Whithear, and G. F. Browning. 2000. A novel mechanism for control of antigenic variation in the haemagglutinin gene family of Mycoplasma synoviae. Mol. Microbiol. 35:911-923. [DOI] [PubMed] [Google Scholar]
- 36.O'Farrell, C. L., and M. S. Strom. 1999. Differential expression of the virulence-associated protein p57 and characterization of its duplicated gene msa in virulent and attenuated strains of Renibacterium salmoninarum. Dis. Aquat. Organisms 38:115-123. [DOI] [PubMed] [Google Scholar]
- 37.Pascho, R. J., and D. Mulcahy. 1987. Enzyme-linked immunosorbent assay for a soluble antigen of Renibacterium salmoninarum, the causative agent of salmonid bacterial kidney disease. Can. J. Fish. Aquat. Sci. 44:183-191. [Google Scholar]
- 38.Peterson, S. N., C. C. Bailey, J. S. Jensen, M. B. Borre, E. S. King, K. F. Bott, and C. A. Hutchison 3rd. 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. USA 92:11829-11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piganelli, J. D., G. D. Wiens, and S. L. Kaattari. 1999. Elevated temperature treatment as a novel method for decreasing p57 on the cell surface of Renibacterium salmoninarum. Dis. Aquat. Organisms 36:29-35. [DOI] [PubMed] [Google Scholar]
- 40.Rhodes, L. D., T. H. Grayson, S. M. Alexander, and M. S. Strom. 2000. Description and characterization of IS994, a putative IS3 family insertion sequence from the salmon pathogen Renibacterium salmoninarum. Gene 244:97-107. [DOI] [PubMed] [Google Scholar]
- 41.Rockey, D. D., L. L. Gilkey, G. D. Wiens, and S. L. Kaattari. 1991. Monoclonal antibody based analysis of the Renibacterium salmoninarum P57 protein in spawning chinook and coho salmon. J. Aquat. Anim. Health 3:23-30. [Google Scholar]
- 42.Rockey, D. D., P. S. Turaga, G. D. Wiens, B. A. Cook, and S. L. Kaattari. 1991. Serine proteinase of Renibacterium salmoninarum digests a major autologous extracellular and cell-surface protein. Can. J. Microbiol. 37:758-763. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Southern hybridization, p. 6.33-6.64. In Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Segal, E., P. Hagblom, H. S. Seifert, and M. So. 1986. Antigenic variation of gonococcal pilus involves assembly of separated silent gene segments. Proc. Natl. Acad. Sci. USA 83:2177-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starliper, C. E. 1996. Genetic diversity of North American isolates of Renibacterium salmoninarum. Dis. Aquat. Organisms 27:207-213. [Google Scholar]
- 46.Sung, S. Y., J. V. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect. Immun. 68:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turaga, P. S. D., G. D. Wiens, and S. L. Kaattari. 1987. Bacterial kidney disease: the potential role of soluble protein antigen(s). J. Fish Biol. 31:191-194. [Google Scholar]
- 48.Wiens, G. D., M. S. Chien, J. R. Winton, and S. L. Kaattari. 1999. Antigenic and functional characterization of p57 produced by Renibacterium salmoninarum. Dis. Aquat. Organisms 37:43-52. [DOI] [PubMed] [Google Scholar]
- 49.Wiens, G. D., K. A. Heldwein, M. P. Stenzel-Poore, and M. B. Rittenberg. 1997. Somatic mutation in VH complementarity-determining region 2 and framework region 2: differential effects on antigen binding and Ig secretion. J. Immunol. 159:1293-1302. [PubMed] [Google Scholar]
- 50.Wiens, G. D., and S. L. Kaattari. 1999. Bacterial kidney disease (Renibacterium salmoninarum), p. 269-301. In P. T. K. Woo and D. W. Bruno (ed.), Fish diseases and disorders: viral, bacterial and fungal infections, vol. 3. CAB International, Wallingford, Oxon, United Kingdom. [Google Scholar]
- 51.Wiens, G. D., and S. L. Kaattari. 1989. Monoclonal antibody analysis of common surface protein(s) of Renibacterium salmoninarum. Fish Pathol. 24:1-7. [Google Scholar]
- 52.Wiens, G. D., and S. L. Kaattari. 1991. Monoclonal antibody characterization of a leukoagglutinin produced by Renibacterium salmoninarum. Infect. Immun. 59:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiens, G. D., P. S. D. Turaga, and S. L. Kaattari. 1990. Western blot analysis of a fish pathogen, p. 87-94. In T. C. F. J. S. Stolen, D. P. Anderson, B. S. Roberson, and W. B. van Muiswinkel (ed.), Techniques in fish immunology. SOS Publications, Fair Haven, N.J.
- 54.Young, C. L., and G. B. Chapman. 1978. Ultrastructural aspects of the causative agent and renal histopathology of bacterial kidney disease in brook trout (Salvelinus fontinalis). J. Fish. Res. Board Can. 35:1234-1248. [Google Scholar]