Abstract
To examine the dissemination of Shiga-toxigenic Escherichia coli (STEC) within cattle groups, dairy calves on two farms utilizing different calf-rearing practices were exposed to a traceable STEC strain. Test strain dissemination differed significantly between farms, with a higher prevalence being associated with group penning. Pen floors and calf hides may be the main environmental mechanisms of transmission. Dairy calf husbandry represents a control point for reducing on-farm STEC prevalence.
Shiga toxin-producing Escherichia coli (STEC) organisms are an important public health threat, causing hemorrhagic colitis and hemolytic uremic syndrome (13). STEC isolated from such cases is generally referred to as enterohemorrhagic E. coli (EHEC), and E. coli O157:H7 is considered to be the definitive EHEC strain due to its high association with morbidity (13, 15). Ruminant livestock, particularly cattle, are considered the primary reservoir for STEC and E. coli O157:H7, with transmission to humans being ostensibly foodborne and also caused by direct human contact with cattle or exposure to farm environments (2, 13).
Differences in fecal excretion of STEC between cattle have been observed, and many factors are proposed to explain this phenomenon, including the ages, diets, climate conditions, and management of the animals or herd factors such as stocking density, waste management, and housing systems (7, 9, 11, 21). The degree or likelihood of initial host inoculation or reinoculation with STEC is proposed as an important factor of STEC presence on farms. The aim of this study was to investigate how group dynamics and variable exposure to STEC affect fecal excretion by calves and contamination of their environment. To avoid typical shortfalls of epidemiological surveys (11) yet examine STEC transmission under natural circumstances, index calves on two dairy farms with different calf-rearing practices were inoculated with a marked STEC strain and each calf unit was tested for inoculation and environmental contamination. Calf-rearing units were studied because of high STEC and E. coli O157 prevalence in calves and suggestions that calf management practices significantly modulate shedding (7, 10).
This research was approved by the University of Queensland Animal Ethics Committee (certificate no. MICRO/PARA/076/00/UQPGRS/CSIRO/PHD).
Experimental calves.
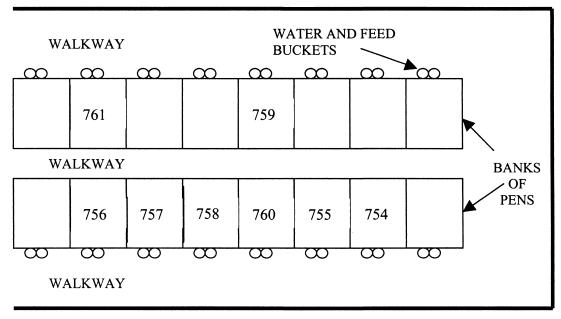
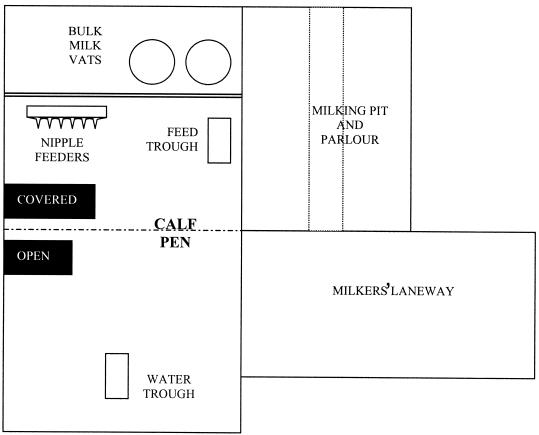
Calf cohorts consisted of eight weaning calves (2 to 8 weeks of age), including an index calf inoculated with the STEC test strain. Farms A and B, commercial dairy operations in South East Queensland, Australia, were of equal size and utilized standard husbandry for pasture-fed dairy cattle with no significant management differences apart from calf-rearing practices. On Farm A, the calves were housed in individual covered pens isolated from the rest of the herd from approximately 7 to 110 days of age (Fig. 1). Pens had wire mesh walls that allowed limited contact between immediately adjacent calves and raised wire mesh floors. Younger calves were fed milk from buckets until around 80 days, with access to solid feed (commercial calf starter ration) and water ad libitum. Calves fed and drank from individual buckets. Each day, feed and water were changed, and pens and concrete floors were hosed out. On Farm B, calves were housed in a group of 20 to 25 from 7 to 90 days of age in a single pen immediately adjacent to the milking parlor that allowed some contact with adult cattle through pen rails (Fig. 2). Milk was fed once or twice daily from silicone nipple feeders. Water and calf starter ration were available ad libitum from shared troughs in the pen. Concrete floors were hosed twice daily.
FIG. 1.
Design of the calf-rearing unit at Farm A. Calf 754 was the index calf, inoculated with the test strain.
FIG. 2.
Design of the calf-rearing unit at Farm B.
Bacterial inoculation.
The inoculum, designated EC596, was a nalidixic acid-resistant (Nalr) E. coli O136:H16 with Shiga toxin 1 (stx1) and enterohemolysin (ehx) genes derived from EC144, a cattle fecal isolate (Food Science Australia culture collection). Nalr was selected by successive plating on nutrient agar (Oxoid, Basingstoke, Hampshire, United Kingdom) incorporating 20 μg of nalidixic acid (Sigma, Castle Hill, NSW, Australia) per ml. A 200-μl volume of Luria-Bertani broth subculture (static at 37°C for 18 h) of EC596 was suspended in 20 ml of phosphate buffered saline and used to orally dose index calves via the retropharynx. Inocula were enumerated on modified hemorrhagic colitis agar (22) incorporating 20 μg of nalidixic acid per ml (mHC+Nal) prior to inoculation.
Sample collection.
Feces were collected immediately prior to (controls) and for 10 days following inoculation, day 1 being 24 h after inoculation. Feces were collected from each calf and from 10 randomly selected cows from the milking herd daily. Samples were collected daily from the pen floor, calf feed and water, hides and saliva (oral swabs), and water from the milking-cow water troughs. Feces collected via anal swabs using sterilized cotton-tipped applicators (Medical Wire and Equipment, Corsham, United Kingdom) were placed in 10 ml of modified E. coli broth incorporating 0.02 mg of novobiocin (mEC+n [16]) per ml. Pen floors were sampled by swabbing an area approximating 200 cm2 beneath the pen of the index calf or its immediate neighbors on Farm A and around the nipple feeders on Farm B, corresponding to areas of likely maximal contamination. Feed (starter ration and milk) and water samples were collected in 70-ml sterile sample jars (Laboratory Supply, Brisbane, Queensland, Australia) from individual calf (index calf and in-contact calves) buckets on Farm A and from nipple and creep feeders or communal water troughs on Farm B. Hides were sampled by swabbing an area approximating 100 cm2 around the paralumbar fossa area with no selection for or avoidance of obvious coat soiling. All samples were transported immediately to the laboratory for further processing.
Detection of the inoculation strain.
Calf fecal swabs were enumerated by using spread plates of thoroughly vortexed, noncultured enrichment broth serially diluted on mHC+Nal and incubated 18 h at 37°C. Colonies were confirmed by slide agglutination with O136 antiserum (Denka Seiken, Tokyo, Japan). CFU per gram of feces were calculated assuming an average fecal sample of 0.145 g per swab (determined prior to experimentation). The limit of enumeration was 69 CFU/g of feces. Calf fecal samples with no growth following enumeration, cow fecal swabs, and saliva, hide, and pen floor swabs were vortexed and enriched in mEC+n statically at 37°C for 18 h. Twenty milliliters of water was added to an equal volume of 2× mEC+n, 2 ml of milk was added to 98 ml of modified tryptone soy broth incorporating 0.02 mg of novobiocin per ml (mTSB+n) (16), 2 g of solid feed was added to 98 ml of mEC+n, and all mixtures were incubated statically for 18 h at 37°C. Two hundred microliters of enrichment broth was streaked onto mHC+Nal, and Nalr colonies were tested for O136 agglutination. Prevalence data were compared with Minitab 12.1 software using the chi-square test for independence with statistical significance set at the 95% confidence level (P < 0.05) unless otherwise stated. Bacterial counts were analyzed by using analysis of variance with a general linear model (Minitab Inc., State College, Pa.).
Fecal dissemination of the inoculation strain.
No Nalr E. coli O136 organisms were isolated from control feces preinoculation. Neither of the two inoculated calves displayed clinical evidence of enteric disease. Calves 754 on Farm A and 703 on Farm B (index calves) were inoculated with 1.5 × 108 CFU and 1.4 × 108 CFU of EC596, respectively, on day 0 of the trial. Fecal excretion of EC596 by these and the cohorted calves for Farms A and B is described in Tables 1 and 2, respectively. Index calves on Farms A and B shed the STEC test strain continually during the 10-day study period. Levels of shedding were comparable to those of other inoculation studies, considering the lower inoculation dose used in the present study (3, 5). The decline in fecal EC596 counts was not entirely linear. Fluctuations in fecal STEC counts over time have been noted in other inoculation studies (3, 4) and may be due to ecological equilibration of the introduced organism within the gastrointestinal tract, reinoculation by the test strain from the calves' environment, or sampling error. Although analysis of anal swabs is generally not as sensitive as analysis of larger amounts of feces (14), it was more practical under the circumstances and provided a limit of enumeration (69 CFU/g) that compared well with those of similar studies (1, 20).
TABLE 1.
Detection and/or enumeration of STEC EC596 in calf fecal samples on Farm A for 10 days following inoculation of an index calf
| Calf no. | Log10 EC596/g of feces on day:
|
No. of days positive | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| 754 | 5.6 | 5.3 | 4.4 | 4.0 | 5.5 | 2.7 | 4.4 | 4.3 | 4.9 | 2.7 | 10 |
| 755 | −a | +b | − | − | − | − | − | − | − | − | 1 |
| 756 | − | − | + | − | − | − | − | − | − | − | 1 |
| 757 | − | − | − | − | 3.2 | − | − | − | − | − | 1 |
| 758 | − | − | − | − | 2.7 | − | − | − | − | − | 1 |
| 759 | − | − | − | − | − | − | − | − | − | − | 0 |
| 760 | − | − | − | − | 2.9 | − | − | − | − | − | 1 |
| 761 | − | − | − | − | − | − | − | − | − | − | 0 |
| No. of calves positive (% of total) | 1 (13) | 2 (25) | 2 (25) | 1 (13) | 4 (50) | 1 (13) | 1 (13) | 1 (13) | 1 (13) | 1 (13) | 15 (188) |
−, EC596 not isolated from sample.
+, inoculation strain was detected after enrichment only.
TABLE 2.
Detection and/or enumeration of STEC EC596 in calf fecal samples on Farm B for 10 days following inoculation of an index calf
| Calf no. | Log10 EC596/g of feces on day:
|
No. of days positive | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| 703 | 5.5 | 3.3 | 2.3 | +b | 3.4 | 2.9 | 3.2 | 4.1 | 3.6 | 3.6 | 10 |
| 704 | −a | − | − | + | 1.8 | − | 3.0 | + | 2.8 | 1.8 | 6 |
| 705 | − | 3.1 | 2.5 | 1.8 | − | − | − | − | + | − | 4 |
| 706 | − | 3.4 | − | − | 3.0 | 4.0 | + | − | 2.7 | 2.7 | 6 |
| 707 | 2.5 | 2.5 | − | − | − | − | − | + | 2.7 | − | 4 |
| 708 | 2.2 | + | 2.5 | − | − | − | 2.5 | 3.0 | 2.7 | − | 6 |
| 709 | + | 2.2 | − | − | − | − | − | + | 2.7 | + | 5 |
| 710 | − | 2.7 | 2.3 | − | − | + | 1.9 | − | + | + | 6 |
| No. of calves positive (% of total) | 4 (50) | 7 (88) | 4 (50) | 3 (38) | 3 (38) | 3 (38) | 5 (63) | 5 (63) | 8 (100) | 5 (63) | 47 (59) |
−, EC596 not isolated from sample.
+, inoculation strain was detected after enrichment only.
Patterns of transmission of EC596 from index to nonindex calves on Farms A and B differed considerably, suggesting different mechanisms or dynamics of STEC spread among calves under each management system. On Farm B, all calves were positive for the test strain at some time, daily EC596 prevalence was high, and an apparently random incidence of infection was evident. EC596 was passed to seven of eight in-contact calves within 2 days, contrasting with the relatively limited rate of transmission to nonindex calves on Farm A. Overall prevalence rates were significantly different on each farm (P < 0.001), with more nonindex calves excreting EC596 on Farm B (46%) than Farm A (6.4%) (P < 0.001) and daily prevalence being generally higher on Farm B. If transmission patterns can be extrapolated to E. coli O157:H7 and other EHEC serotypes, this confirms that farms that group calves at or before the time of weaning have an increased chance of a herd being STEC positive compared to farms that group calves after weaning only (10, 21). More farms need to be studied to further confirm this. The mean fecal count for EC596 was significantly less on Farm B than Farm A (P < 0.01), which may be due to the smaller number of in-contact calves (with generally lower excretion levels than the index calf) shedding on Farm A.
Environmental dissemination of the inoculation strain.
Although only a limited number of nonfecal samples were collected and the number of each sample differed slightly overall and day to day due to sampling constraints, trends in the dissemination of EC596 from the index calves were noted. Combining farm data, EC596 was most frequently detected on pen floors (15 of 24 samples) and hides (14 of 29). Feed (1 of 25) and water (1 of 27) had significantly lower contamination rates (P < 0.001). Isolation rates differed between farms, although their patterns for the two units matched. Water and feed have been considered an important means of STEC and E. coli O157:H7 transmission (7, 18, 21). In this study, the primary environmental reservoirs for EC596 appeared to be pen floors and calf hides, particularly those of index calves, and this likely reflects their relatively heavy fecal contamination. The persistence of E. coli O157:H7 on dairy farms has similarly been found more readily in samples with a high fecal load (17, 18). Hide prevalence varied on each farm, being higher on Farm A (4 of 14 samples) than Farm B (10 of 15). Hide contamination is an important means of animal-to-carcass contamination (6), and from this study it appears that this is an important mechanism for horizontal transmission between animals. The inoculated strain was present in 5 of 29 saliva samples and more frequent in the saliva of Farm A calves (4 of 15) than Farm B calves (1 of 14). Saliva may play a significant role in STEC transmission, particularly through feed and water contamination, or from one calf to another via intersucking and hides (3, 21). Saliva may become contaminated by STEC following ingestion of polluted feed and water, though the low prevalence of the test strain in water and feed samples in this study suggests otherwise. It is possible that rumination and regurgitation inoculate saliva, as E. coli O157:H7 has been identified in ruminal contents (5). The role of saliva in STEC transmission requires further examination.
Farms A and B differed in the prevalence of EC596 in nonfecal samples; Farm B samples (22 of 68) were more often contaminated than those from Farm A (14 of 66). This is likely related to the increased range of movement of the index calf and also to faster and more widespread transmission to nonindex calves, resulting in the rapid contamination of the pen environment. This is exemplified by differences in floor contamination: 4 of 11 samples for Farm A and 11 of 13 for Farm B were found positive. Contaminated samples on Farm A were associated with the index calf or its immediate surroundings, not from nonindex calves. On Farm B, however, hide samples from calves other than the index calf were contaminated with the inoculation strain on two occasions.
General considerations.
Differences in dissemination of the inoculation strain on Farms A and B are likely to relate to specific mechanisms of transmission. The dissemination of EC596 on Farm A appeared to be less widespread and progress more slowly than on Farm B. Calves 759 and 761 did not excrete the inoculation strain at any time, which may be due to their spatial separation from the index calf (Fig. 1). The small burst of shedding by calves 757, 758, and 760 on Farm A may be correlated with their proximity to the index calf and other calves that were EC596 positive immediately before them. A different farmhand was made responsible for feeding calves and cleaning pens on days 4 and 5. He confirmed that he had tended the index calf before neighboring calves in the same pen bank, suggesting a role for animal handlers as a means of STEC transmission between livestock. Infection with the inoculation strain may relate to exposure to feces during the hosing of pens. STEC has been demonstrated to survive for prolonged periods of time in bovine feces (8), and E. coli O157:H7 can also survive on inorganic surfaces for extended periods (19). Garber et al. (9) commented that flushing dairy alleys with water may help disseminate E. coli O157:H7. The use of shared nipple feeders rather than individual milk feeding has also been demonstrated to be a risk factor for STEC infection and is likely to be a factor in transmission between calves on Farm B (10). These aspects of calf management may represent specific control points for reducing STEC spread within dairy units or other animal-rearing areas and are worthy of ongoing scrutiny. Considering the high prevalence of EC596 on pen floors and hides, these are likely to be the most significant means of indirect fecal-oral STEC dissemination among housed calves. Under conditions where calves have a higher STEC prevalence and are given greater contact with the milking herd, the herd STEC prevalence might be expected to reflect that of the calves. In this study, no transmission of EC596 from weaning heifers to the milking herd was evident on either farm. While this may indicate limited spread of calf STEC strains to the herd in general, further studies employing improved sampling strategies aimed at this hypothesis are necessary.
Longitudinal surveys have suggested that maintenance of E. coli O157:H7 in cattle herds relies on continual reinoculation of individual cattle (18). Shere et al. (21) concluded that a common source was probably responsible for E. coli O157:H7 dissemination on dairy farms, while other authors have considered that multiple sources are more likely and that horizontal transmission was an important feature of on-farm STEC ecology (7, 12). Relatively few researchers have used inoculation trials as a means of examining STEC transmission or dissemination in animal populations. Such trials have typically employed sheep or small animal cohorts as the experimental model (4, 14), which may not adequately represent actual on-farm cattle-to-cattle transmission. Others have inoculated cattle, though they were primarily interested in analyzing the strains' clonal turnover and shedding duration or in the effect of reinoculation or diet change (1, 20). The present study used inoculation of calves within a natural setting to more realistically demonstrate that how cattle interact at a group level can influence the dissemination of a novel STEC strain. Control of STEC is required at the farm level and may be achieved through a reduction of horizontal transmission within cattle groups, thus decreasing STEC prevalence. If this test strain is indicative of STEC in general or E. coli O157:H7 in particular, then the use of segregated penning systems rather than group housing of weaning calves may reduce the prevalence of these potential pathogens within the calf unit. If this results in a reduction in the general herd or farm STEC prevalence, then such changes in calf-rearing practices may offer a control point for preharvest STEC risk on the dairy farm.
Acknowledgments
We are greatly indebted to the farm owners and herdspeople for allowing inoculation of their cattle and assistance in sampling. Special thanks to Jocelyn Midgley for strain EC596.
REFERENCES
- 1.Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 184:79-83. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, G. L., J. Hollingsworth, and J. G. J. Morris. 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29-51. [DOI] [PubMed] [Google Scholar]
- 3.Buchko, S. J., R. A. Holley, W. O. Olson, V. P. Gannon, and D. M. Veira. 2000. The effect of different grain diets on fecal shedding of Escherichia coli O157:H7 by steers. J. Food Prot. 63:1467-1474. [DOI] [PubMed] [Google Scholar]
- 4.Cornick, N. A., S. L. Booher, T. A. Casey, and H. W. Moon. 2000. Persistent colonization of sheep by Escherichia coli O157:H7 and other E. coli pathotypes. Appl. Environ. Microbiol. 66:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukushima, H., K. Hoshina, and M. Gomyoda. 1999. Long-term survival of Shiga toxin-producing Escherichia coli O26, O111, and O157 in bovine feces. Appl. Environ. Microbiol. 65:5177-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber, L., S. Wells, L. Schroeder-Tucker, and K. Ferris. 1999. Factors associated with fecal shedding of verotoxin-producing Escherichia coli O157 on dairy farms. J. Food Prot. 62:307-312. [DOI] [PubMed] [Google Scholar]
- 10.Garber, L. P., S. J. Wells, D. D. Hancock, M. P. Doyle, J. Tuttle, J. A. Shere, and T. Zhao. 1995. Risk factors for fecal shedding of Escherichia coli O157:H7 in dairy calves. J. Am. Vet. Med. Assoc. 207:46-49. [PubMed] [Google Scholar]
- 11.Herriott, D. E., D. D. Hancock, E. D. Ebel, L. V. Carpenter, D. H. Rice, and T. E. Besser. 1998. Association of herd management factors with colonization of dairy cattle by Shiga toxin-positive Escherichia coli O157. J. Food Prot. 61:802-807. [DOI] [PubMed] [Google Scholar]
- 12.Heuvelink, A. E., F. L. A. M. Van Den Biggelaar, J. T. M. Zwartkruis-Nahuis, R. G. Herbes, R. Huyben, N. Nagelkerke, W. J. G. Melchers, L. A. H. Monnens, and E. De Boer. 1998. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36:3480-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1995. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl. Environ. Microbiol. 61:1363-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neaves, P., J. Deacon, and C. Bell. 1994. A survey of the incidence of E. coli O157 in the UK dairy industry. Int. Dairy J. 4:679-696. [Google Scholar]
- 17.Porter, J., K. Mobbs, C. A. Hart, J. R. Saunders, J. R. Pickup, and C. Edwards. 1997. Detection, distribution and probable fate of Escherichia coli O157 from asymptomatic cattle on a dairy farm. J. Appl. Microbiol. 83:297-306. [DOI] [PubMed] [Google Scholar]
- 18.Rahn, K., S. A. Renwick, R. P. Johnson, J. B. Wilson, R. C. Clarke, D. Alves, S. McEwen, H. Lior, and J. S. Spika. 1997. Persistence of Escherichia coli O157:H7 in dairy cattle and the dairy farm environment. Epidemiol. Infect. 119:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randall, L. P., C. Wray, and R. H. Davies. 1999. Survival of verocytotoxin-producing Escherichia coli O157 under simulated farm conditions. Vet. Rec. 145:500-501. [DOI] [PubMed] [Google Scholar]
- 20.Sanderson, M. W., T. E. Besser, J. M. Gay, C. C. Gay, and D. D. Hancock. 1999. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet. Microbiol. 69:199-205. [DOI] [PubMed] [Google Scholar]
- 21.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szabo, R. A., E. C. D. Todd, and A. Jean. 1986. Method to isolate Escherichia coli O157:H7 from food. J. Food Prot. 49:768-772. [DOI] [PubMed] [Google Scholar]