Abstract
The prsA-like gene from Lactococcus lactis encoding its single homologue to PrsA, an essential protein triggering the folding of secreted proteins in Bacillus subtilis, was characterized. This gene, annotated pmpA, encodes a lipoprotein of 309 residues whose expression is increased 7- to 10-fold when the source of nitrogen is limited. A slight increase in the expression of the PrsA-like protein (PLP) in L. lactis removed the degradation products previously observed with the Staphylococcus hyicus lipase used as a model secreted protein. This shows that PmpA either triggers the folding of the secreted lipase or activates its degradation by the cell surface protease HtrA. Unlike the case for B. subtilis, the inactivation of the gene encoding PmpA reduced only slightly the growth rate of L. lactis in standard conditions. However, it almost stopped its growth when the lipase was overexpressed in the presence of salt in the medium. Like PrsA of B. subtilis and PrtM of L. lactis, the L. lactis PmpA protein could thus have a foldase activity that facilitates protein secretion. These proteins belong to the third family of peptidyl-prolyl cis/trans-isomerases (PPIases) for which parvulin is the prototype. Almost all PLP from gram-positive bacteria contain a domain with the PPIase signature. An exception to this situation was found only in Streptococcaceae, the family to which L. lactis belongs. PLP from Streptococcus pneumoniae and Enterococcus faecalis possess this signature, but those of L. lactis, Streptococcus pyogenes, and Streptococcus mutans do not. However, secondary structure predictions suggest that the folding of PLP is conserved over the entire length of the proteins, including the unconserved signature region. The activity associated with the expression of PmpA in L. lactis and these genomic data show that either the PPIase motif is not necessary for PPIase activity or, more likely, PmpA foldase activity does not necessarily require PPIase activity.
Lactococcus lactis is a gram-positive bacterium which is classified as “generally regarded as safe” following its long history of use in the production of fermented milk products. The potential for using L. lactis for new applications, such as in secretion of heterologous proteins in fermented food products or in the digestive tract of humans, is currently under active study (3, 27, 49, 50, 52). In a previous work, we expressed the Staphylococcus hyicus lipase in L. lactis in order to use lactococci as a lipase delivery vehicle (13). This goal could alleviate lipase deficiency in the digestive tract during digestion (steatorrhea) or improve flavor development in some cheese-making processes. In this study, we have identified several factors that affect the secretion of the S. hyicus lipase in L. lactis (13). In this heterologous host, about 50% of the lipase is not translocated through the cytoplasmic membrane and remains bound to the inner side of the membrane as unprocessed preprolipase. Most of the translocated lipase is associated to the cell wall and partially degraded at its N-terminal end (13). Proteolysis is a natural process required not only for protein turnover but also for the degradation of misfolded proteins (15). In the case of the S. hyicus lipase secretion in L. lactis, it was suggested that most of the translocated lipase is not correctly folded and is thus degraded (13).
L. lactis has recently been sequenced, allowing the identification of at least eight genes implicated in protein secretion (6). Furthermore, the membrane protease HtrA, involved in the degradation of hybrid exported proteins, has also been characterized (41). In addition, L. lactis possesses a lipoprotein homologous to PrsA of Bacillus subtilis, encoded by nlp4 (40) or pmpA (6). In B. subtilis, the model organism for protein secretion in gram-positive bacteria, the overproduction of PrsA increases the yield of several heterologous proteins, such as α-amylase (32) or antidigoxin single-chain antibody (55). This indicates that PrsA is a limiting factor in the secretion of heterologous proteins. Recently PrsA of B. subtilis has been shown to belong to a family of a class of proteins: the peptidyl cis/trans-isomerases (PPIases) (42, 46). PPIases are enzymes which catalyze the cis/trans isomerization of the peptidyl-proline bonds in oligopeptides and proteins. Such an isomerization could be important for the proper and efficient folding of proteins. The fact that several extracytoplasmic enzymes involved in the folding of external proteins, such as PrsA (31, 32), PrtM (carried on a plasmid) (24), SurA (4, 45), and PpiD (9), are members of this family suggests that PPIase activity is required for their activity. The PPIase activity was demonstrated for SurA and PpiD (9, 45), and the loss of isomerase-foldase activity was caused by directed mutations affecting the residues essential for PPIase activity (9), although the parvulin domain was shown to be dispensable for SurA chaperone activity (4, 45). Furthermore, inactivation of the surA gene decreased the level of major membrane proteins in the outer membrane (45). Finally, inactivation of either of the two genes induced a periplasmic stress response controlled by sigmaE (9, 37, 45), known to be specifically induced by misfolded proteins in the periplasm.
Here, we report the characterization of the gene that chromosomally encodes the unique PrsA-like protein (PLP), annotated pmpA for protein maturation protein in the genome of L. lactis IL-1403 (6). We show that this gene encodes a lipoprotein involved in L. lactis secretion machinery. Surprisingly, PmpA does not contain motifs required for PPIase activity (6), but it has a foldase activity because it is involved in improving the secretion of the heterologous S. hyicus lipase used as a model of incorrectly folded proteins. The corresponding gene is thus likely the L. lactis orthologue of prsA. Genome comparisons show that PLP with diversely conserved PPIase motifs are present in several species of the genus Streptococcus.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids are listed in Table 1. L. lactis strains were grown at 30°C in M17 glucose medium (51) or in chemically defined medium (CDM) (48) with 0.5% glucose, supplemented or not with Casitone (Sigma-Aldrich) or NaCl (Merck). Escherichia coli was grown in Luria-Bertani medium at 37°C (36). Chloramphenicol was used at a concentration of 10 μg/ml, and erythromycin was used at concentrations of 10 μg/ml for L. lactis and 1 mg/ml for E. coli. Nisin was used at a concentration of 0.5 μg/ml (13).
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| TG1 | supE thiD(lac-proAB) hsdD5 F+ traD36 proAB lacIZΔM15 | 19 |
| L. lactis | ||
| IL1403 | L. lactis subsp. lactis, plasmid-free | 8 |
| MG1363 | L. lactis subsp. cremoris, plasmid-free | 18 |
| NZ9000 | MG1363, pepN::nisRnisK | 12 |
| MG-Pnis::pmpA | Cmr, NZ9000 carrying pJIM2080 | This work |
| MG-Pnis::pmpA-Lip+ | Cmr, Eryr, MG-Pnis::pmpA carrying pJIM2430 | This work |
| MG-Pnis-Lip+ | Cmr, Eryr NZ9000 carrying pNZ8037 and pJIM2430 | This work |
| MG-pmpA | Eryr, MG1363 carrying pJIM2084 inactivating the pmpA gene | This work |
| MG-pmpA-Lip+ | Cmr, Eryr, MG-pmpA carrying pJIM2088 | This work |
| MG-PpmpA::lux | Cmr, MG1363 carrying pJIM2087 duplicating the pmpA promoter after integration | This work |
| MG-Lip+ | MG1363 carrying pJIM2088 | This work |
| Plasmids | ||
| pBluescript | Ampr | Biolabs |
| pGemT | Ampr | Promega |
| pBV502 | Eryr, Cmr, contains the cassette P5-cat-86 | 5 |
| pNZ8037 | Cmr, pNZ273 derivative carrying PnisA | 12 |
| pJIM2430 | Eryr, high-copy-number vector carrying the lip gene under the control of P44 | 13 |
| pJIM2374 | Eryr, integrative vector | 11 |
| pJIM3035 | Cmr, integrative vector carrying the luxA-luxB gene downstream from a multiple cloning site | E. Guédon |
| pJIM2078 | Ampr, pGemT carrying the structural gene encoding PmpA | This work |
| pJIM2080 | Cmr, derivative of pNZ8037 and pJIM2078 carrying the pmpA gene under the control of PnisA (Pnis::pmpA) | This work |
| pJIM2082 | Ampr, pBluescript containing an internal fragment of pmpA | This work |
| pJIM2084 | Eryr, pJIM2374 carrying the internal fragment of the pmpA gene from pJIM2082 | This work |
| pJIM2085 | Ampr, pBluescript carrying the promoter region of pmpA | This work |
| pJIM2087 | Cmr, derivative of pJIM3035 carrying the pmpA promoter region upstream the luxA-luxB genes (PpmpA::lux) | This work |
| pJIM2088 | Cmr, Emr, pJIM2430 (expressing the lip gene under the control of P44) carrying the P5-cat-86 cassette of pBV502 | This work |
DNA and RNA manipulations.
Plasmid DNA was isolated by standard methods for E. coli (25) and L. lactis (1). E. coli was transformed using the heat shock procedure (47). L. lactis was transformed by electroporation of cells grown in the presence of glycine (26). Restriction and modification enzymes were used according to the instructions of the supplier. RNA was prepared from cells grown exponentially in CDM. Total RNA was prepared as previously described for B. subtilis (20). Northern hybridizations were performed according to the method of Sambrook et al. (47).
Plasmid and strain constructions. (i) Overproduction of PmpA in L. lactis.
The structural gene corresponding to pmpA with its ribosome binding site was amplified from L. lactis subsp. lactis MG1363 with the oligonucleotides 5′-CATCGAATCATGCGAAAGGAGA-3′ and 5′-CTCACCATTTAGAGCTCTCTT-3′ designed from the complete genome sequence of L. lactis IL-1403 (6). The corresponding 1-kb PCR fragment was cloned on the pGemT (Promega) to yield pJIM2078. This plasmid was fused by NcoI site to pNZ8037 (12) that contains the inducible promoter PnisA to yield pJIM2079. The initial vector, pGemT, was then deleted by a PstI digestion followed by ligation. The resulting plasmid, pJIM2080, was then transformed in L. lactis subsp. cremoris NZ9000 (12), which contains nisRK, the products of which are necessary to activate PnisA.
pJIM2430 carrying the lip gene under the control of P44 was used as the model of incorrectly folded protein (13). The chloramphenicol resistance gene from pBV502 (5) was inserted in its XhoI site to yield pJIM2088.
(ii) Inactivation of pmpA by gene insertion.
A PCR fragment of 0.74 kb internal to the prsA-like gene was amplified using the oligonucleotides 5′-AGCAAGGATATCATCATCACAATG-3′ and 5′-GTATTTAGGAAGAATTCCACTTAC-3′ from the MG1363 chromosome. This fragment obtained was cloned by EcoRI and EcoRV present on the primers (boldface letters) in pBluescript (Biolabs) to yield pJIM2082. This plasmid was fused by SmaI to the integrative plasmid pJIM2374 (11). The initial vector, pBluescript, was then deleted by SalI to give pJIM2084 and transformed in L. lactis MG1363. Erythromycin-resistant clones containing pJIM2084 integrated into the chromosome were checked by PCR. The inactivation of pmpA was checked by radiolabeling with [3H]palmitic acid. One mutant, MG-pmpA, was chosen for further study.
(iii) Study of the L. lactis pmpA promoter.
The pmpA promoter region of 0.51 kb was amplified using the oligonucleotides 5′-AAATATATCGAATTCCTACCAA-3′ and 5′-CATTGTGATGATATCCTTGCT-3′ from the MG1363 chromosome. This fragment was inserted in EcoRI-EcoRV (boldface letters) of pBluescript to generate pJIM2085. This plasmid was fused by EcoRV in the integrative plasmid pJIM3035 (E. Guédon, personal communication), which contains the luciferase genes. The initial vector, pBluescript, was then deleted between SalI and NotI. The resulting plasmid, pJIM2087, yields chloramphenicol-resistant clones after transformation of L. lactis subsp. cremoris MG1363, among which MG-PpmpA::lux was chosen after PCR checking of its proper integration into the chromosome.
Preparation of cellular and supernatant fractions.
The cells were recovered from the medium by centrifugation for 10 min at 6,000 × g and treated as previously described (34). The supernatants were passed through filters (0.25 μ) to remove eventual cellular debris. The extracellular proteins, including the heterologous lipase, were precipitated by the addition of solid ammonium sulfate to the supernatant to 70% (wt/vol) (53). After agitation for 2 h at 4°C, the precipitate was collected by centrifugation for 30 min at 10,000 × g and 4°C. The pellets were resuspended in a Tris-HCl buffer (20 mM; pH 8) and dialyzed for 14 h at 4°C. Proteins were electrophoresed in sodium dodecyl sulfate (SDS)-polyacrylamide gels (33) and then either stained with Coomassie blue or immunoblotted with polyclonal rabbit antibodies antilipase (13) followed by ECL detection (Amersham). Quantification of the relative amount of protein in the bands was performed by densitometry.
Radiolabeling of PrsA.
L. lactis growing exponentially in CDM (1 ml) at 30°C was labeled with 50 μCi of [3H]palmitic acid (NEN) for one generation (32, 35). The cellular fractions were prepared as previously described, and samples were run in SDS-polyacrylamide gel electrophoresis. The gel was fixed in isopropanol-water-acetic acid (25:65:10) for 30 min and soaked in Amplify (Amersham) for 15 min. It was dried for 1 h at 80°C and mounted on X-ray film for fluorography.
Luciferase activity measurement.
The luciferase activity was measured immediately after the addition of 5 μl of nonaldehyde (Sigma) to either culture broth or a sample diluted in water (14, 44). Light emission was measured in a photometer (LB9501; Bertold).
RESULTS
Analysis of the prsA-like gene of L. lactis.
The sequence of genes homologous to prsA was searched in the complete genome sequence of L. lactis IL-1403 (6). Only one potential open reading frame (ORF) showed a low but significant level of homology to the prsA gene of B. subtilis, and its product was named PmpA. The previously characterized prtM gene encoding the PrtP maturase is harbored by a plasmid in lactococci (6) and encodes a PrsA homologue found in particular L. lactis strains. The most probable translational start of pmpA is a TTG codon, which is preceded by cgAAGGAGa, a sequence close to the ribosome binding site consensus (lowercase letters indicate sequence that does not fit the consensus). The L. lactis pmpA gene potentially encodes a protein of 309 amino acids that is 27% identical to PrsA from B. subtilis and 95% identical to 94 residues known from Nlp4 of L. lactis MG1363 (40). Its first 24 residues have the characteristic features of signal peptides for exported proteins. Moreover, it contains a consensus cleavage site of lipoprotein signal peptides producing an N-terminal Cys residue in the processed protein (30, 54). We demonstrated that this Cys residue is acylated by radiolabeling with palmitic acid (see below).
About 80 nucleotides upstream from the translational start is a potential promoter with a perfect −10 extended box but no −35 box, a feature already established in an L. lactis promoter (21). Some 70 nucleotides upstream from the potential promoter is the last codon of a previously identified ORF, which encodes a potential methyl transferase (CAM) found in the operon with the pepF2 gene (38). There is no typical rho-independent terminator structure at the end of this ORF in the IL-1403 sequence. Downstream from the prsA-like gene is an imperfect stem and loop structure that is not typical for rho-independent terminators. However, the structure is preceded by a sequence that perfectly matches a regulatory element known as the T-box (22). In gram-positive bacteria (including L. lactis), the T-box is involved in the transcriptional regulation of the termination-antitermination mechanism of tRNA synthetases and genes involved in amino acid biosynthesis. About 100 nucleotides downstream from this structure is an ORF sharing about 60% identity with alanyl tRNA synthetases from gram-positive bacteria, and that T-box could be involved in the regulation of this gene. A distinct 1.2-kb messenger band was revealed by Northern blot with a probe internal to the prsA-like gene (not shown), suggesting that pmpA is transcribed from the potential promoter described above down to the putative terminator attenuating the transcription of the alanyl tRNA synthetase.
Distribution and organization of prsA-like gene loci in the bacterial world.
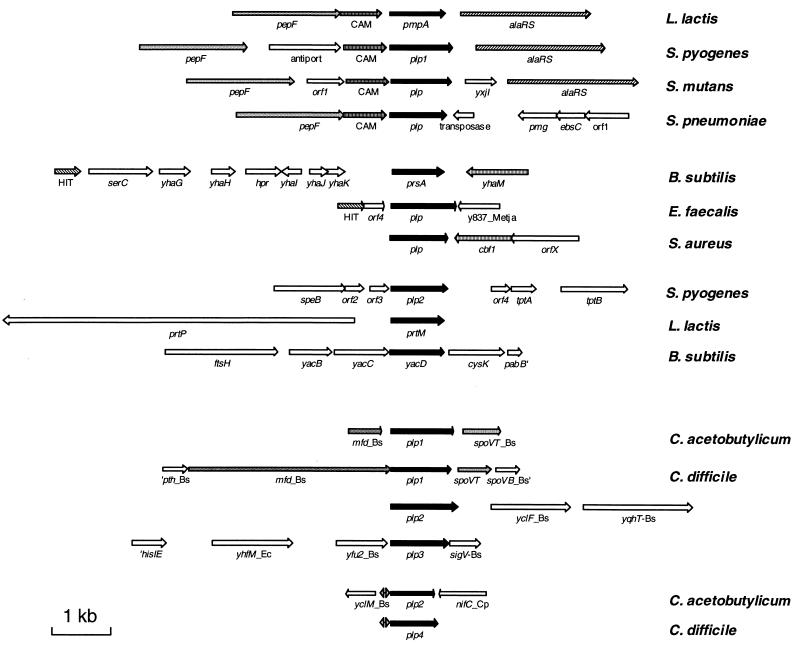
We searched for the homologues of PrsA in databanks (GenBank at NCBI and the unfinished genomes at The Institute for Genomic Research), paying special attention to gram-positive bacteria. Potential proteins sharing homology with PrsA were found in the unfinished genomes of Staphylococcus aureus, Enterococcus faecalis, Streptococcus pneumoniae, Streptococcus pyogenes (two copies), Streptococcus mutans, Clostridium difficile (four copies), and Clostridium acetobutylicum (two copies). Additional homologues were found in gram-negative bacteria and the archeon Cenarchaeum symbiosum. Some of these proteins may be secreted (such as Cbf2 from Campylobacter jejuni [7]), others may be periplasmic (such as SurA from E. coli [45]), intracellular (such as ParvA from E. coli [42]), or associated with the cytoplasmic membrane (such as PpiD, also from E. coli [9]). No homologues were detected in the complete genomes of Synechocystis, Chlamydia, and the six completely sequenced archeobacteria (Aeropyrum pernix strain K1, Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum, Methanococcus jannaschii, Pyrococcus abyssi, and Pyrococcus horikoshii). Concerning the gram-positive bacteria, PrsA homologues were not found in Mycoplasma and Mycobacterium genomes. The comparative organization of the prsA-like genes in gram-positive bacteria is presented in Fig. 1. The plp genes from three streptococci are found in a genetic environment similar to that of L. lactis, with the exception of the second copy of the gene in S. pyogenes. They are all downstream from the PepF-CAM genes and upstream of the alanyl tRNA synthetase gene, except in S. pneumoniae where an insertion sequence is present downstream of it. In contrast, the E. faecalis plp gene is not close to these genes. However, genes encoding histidine triad proteins are present 1 and 5 kb upstream of prsA in B. subtilis and its E. faecalis homologue. The S. aureus prsA-like gene is followed by a cbf1 gene, the product of which shares 52% identity with YhaM encoded by the gene following prsA in B. subtilis. The organization in the vicinity of the prsA-like genes from the two clostridia differs from the preceding ones. plp1 from both is preceded by gene homologous to mfd and followed by spoVT and spoVB from B. subtilis, while plp2 and plp4 from C. acetobutylicum and C. difficile, respectively, may be preceded by the same sequences, but the limited size of the plp4 contig does not allow confirmation of this possibility.
FIG. 1.
Comparative organization of different prsA-like genes. Arrows represent the open reading frames. Arrows with a common color or motif encode homologous products in the different genomes. Those discussed in the text are the following: plp (prsA-like protein), pepF (peptidase F), CAM (similar to a caffeoyl-CoA O-methyltransferase), alaRS (alanyl-tRNA synthetase), HIT (histidine triad proteins), speB (exotoxin SpeB), tpt (fibrial tuft organization from S. cristatus), plasmid-borne prtP (cell wall protease) and prtM (PtrP maturase), cbf1 and yhaM (similar to CMP-binding factor), spoVT (stage V sporulation protein T from B. subtilis), and mfd (transcription repair coupling factor mfd from B. subtilis). The double arrows indicate the conserved promoter region of plp2 and plp4 from C. acetobutylicum and C. difficile, respectively.
The signature motif for peptidyl-prolyl PPIases is absent in PLP of several streptococci.
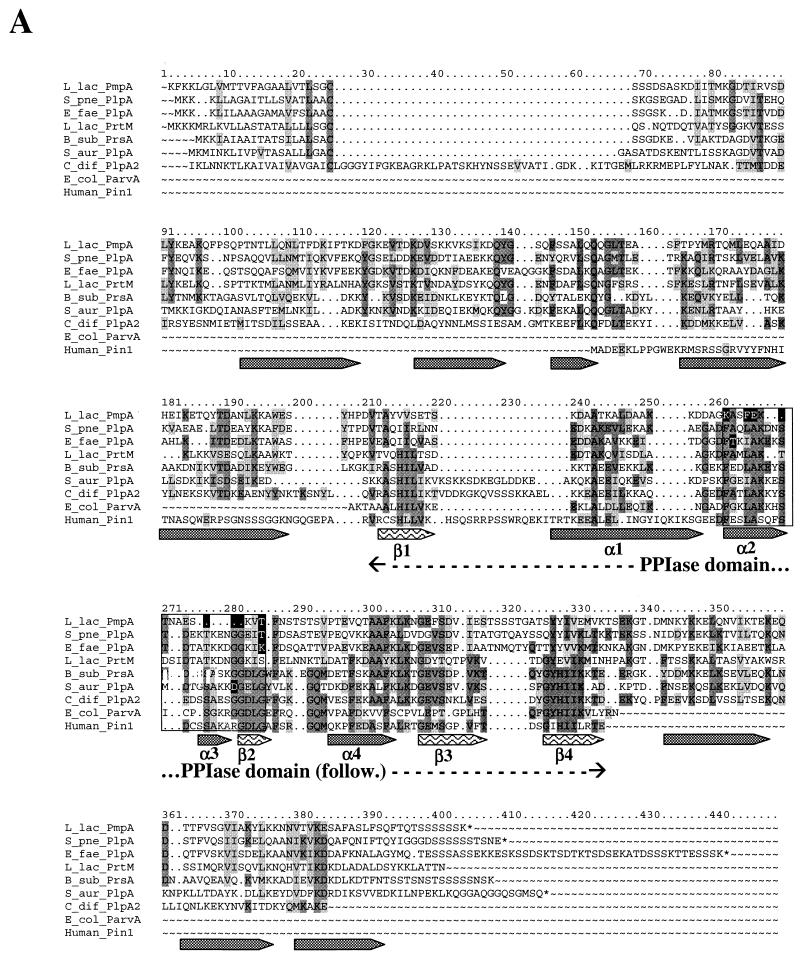
The alignments of the previously reported PLP with representative proteins from the PPIase family are presented in Fig. 2A and B. The identities between these proteins are restricted to the PPIase domain between PLP and the other PPIase (Fig. 2B) and remain low between the different PLP (Fig. 2C). For example, only 40 to 50% identical residues are found in the Streptococcus homologues. The identity falls to 20% or less between PLP from clostridia and those of the other gram-positive bacteria (Fig. 2C). Although identity is low between these proteins, they share common structural features. Most PLP from the gram-positive bacteria contain a peptide leader followed by a cysteine, suggesting that they are lipoproteins. Plp2 from C. acetobutylicum and Plp4 from C. difficile only appear to be intracellular, since they are not preceded by a potential leader peptide. The extracellular PLP show a similar composition and organization of three protein regions (Fig. 2A): (i) an N-terminal region of 140 to 150 residues, (ii) a central part corresponding to the E. coli PPIase Parvulin, and (iii) a C-terminal region of 50 to 100 residues. The predicted secondary structure of these proteins seems to be well conserved even in the first and the last regions that are less conserved on the amino acid sequence level. The signature of the PPIase motif F-[GSADEI]-x-[LVAQ]-A-x(3)-[ST]-x(3,4)-[STQ]-x(3,5)-[GER]-G-x-[LIVM]-[GS] (45) is preserved in the PLP from clostridia and differs by at most two residues in E. faecalis, S. pneumoniae, and S. aureus and by as much as five to seven residues in S. pyogenes (the two proteins), S. mutans, and L. lactis (shown as white letter on a black background in Fig. 2B). This suggests that the PPIase activity may be absent in the last four proteins.
FIG. 2.
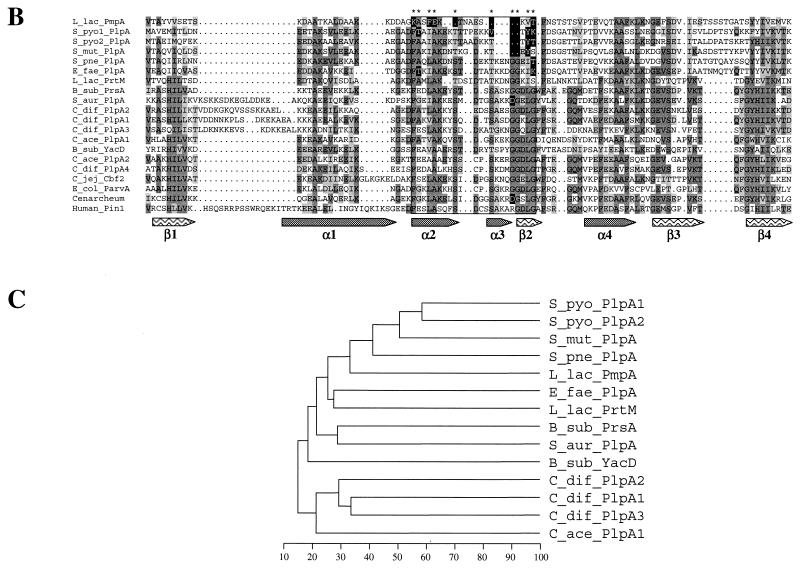
Multiple alignment of several PrsA-like proteins and PinA (A), more extensive alignment of PPIase domain of PLP with representative proteins belonging to the PPIase family (B), and phylogenetic dendrogram indicating the percentage of identity between the PLP of gram-positive bacteria (C). Protein sequences were aligned by using the PILEUP program (GCG). The alignments were edited manually and colored by Boxshade. Identical residues and conservative changes in more that half of the aligned sequences are on dark and light gray backgrounds, respectively. The boxed region contains the PPIase signature motif. Residues diverging from the PPIase signature are in white on a black background. The secondary structure of these proteins was predicted by the GORIV program (17). Alpha helix (gray arrows) and beta sheet (arrows with wave pattern) structures were indicated when predicted in more than 16 sequences at each position. The predicted structures fit with the known structure of the PPIase part of the human Pin1 protein is shown, for which the names of the domains are indicated under the arrows (α1 to α4 and β1 to β4) (6). The dendrogram was obtained by the successive use of the pileup, distances, and growtree programs of the GCG package. B_sub, B. subtilis; C_jej, C. jejuni; C_ace, C. acetobutylicum; C_dif, C. difficile; E_fae, E. faecalis; E_col, E. coli; L_lac, L. lactis; S_aur, S. aureus; S_mut, S. mutans; S_pne, S. pneumoniae; S_pyo, S. pyogenes.
Effect of L. lactis PmpA overproduction on lipase production.
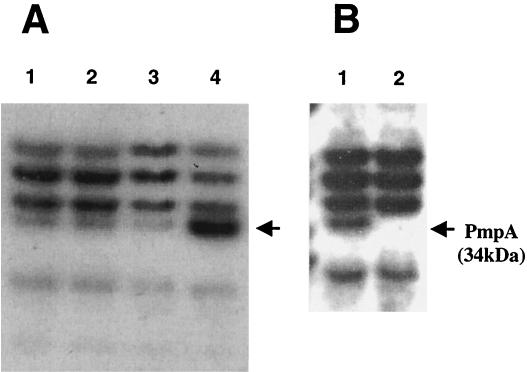
Since PmpA from L. lactis presents the most important divergence from the PPIase signature, we decided to check whether or not it still has a foldase activity similar to that of PrsA from B. subtilis. For this purpose, the L. lactis MG1363 pmpA gene was placed in pJIM2080 under the control of PnisA, a promoter inducible in L. lactis. The overproduction of PmpA was confirmed by the analysis of lipoproteins after radiolabeling the cells with tritiated palmitic acid (Fig. 3A). A protein with a mass of 34 kDa, whose size corresponds to that of PmpA, was clearly overproduced in MG-Pnis::pmpA induced by nisin (lane 4), whereas this band was less intense without nisin induction (lane 3) or in the control without the cloned gene (lanes 1 and 2). The relative amount of PmpA could be quantified by comparing the intensity of the band corresponding to PmpA with the bands corresponding to the other labeled lipoproteins. PmpA is overproduced about 2-fold and more than 10-fold in the presence of pJIM2080 without and with nisin, respectively.
FIG. 3.
SDS-polyacrylamide gel electrophoresis and fluorography of total cell extracts radiolabeled with [3H]palmitic acid. The loaded samples correspond to 0.1 U (0.1 U: quantity of cells in 100 μl of L. lactis culture at an optical density at 600 nm of 1). (A) Overproduction of PmpA in L. lactis. Lanes 1 and 2, NZ9000 (control strain without pJIM2080) noninduced and induced by nisin, respectively; lanes 3 and 4, MG-Pnis::pmpA (NZ9000 with pJIM2080) noninduced and induced by nisin, respectively. (B) Visualization of pmpA mutation. Lane 1, MG1363; lane 2, MG-pmpA.
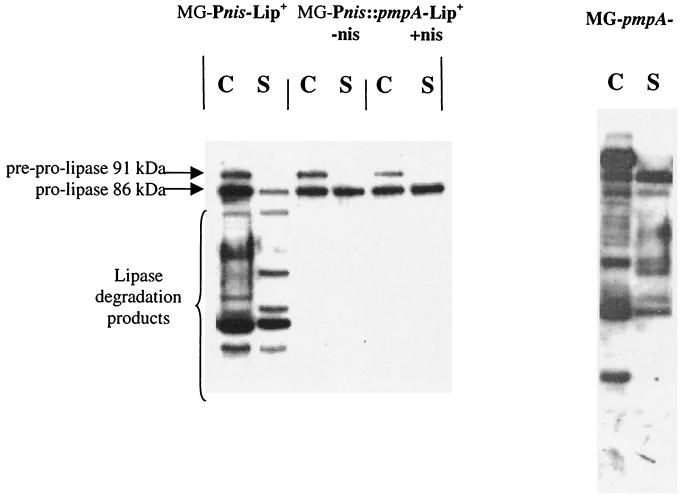
In a preceding work, we have shown that S. hyicus lipase was not correctly exported in L. lactis (13). In particular, degraded forms of the lipase were found to accumulate in the cell wall fraction. The lipase gene under the control of a strong constitutive L. lactis promoter carried on pJIM2430 was introduced in the strain MG-Pnis::pmpA carrying pmpA under the control of the nisin-inducible promoter. The production of the different forms of the lipase was followed by Western blotting with anti-CtermLIP serum (Fig. 4). Degradation products of the lipase were not found in the L. lactis MG1363 derivative carrying the pmpA gene under the control of PnisA. This effect did not require the addition of nisin, showing that a slight level of PmpA overexpression (less than twofold) was sufficient to produce this effect. The total amount of lipase seems lower in the presence of pJIM2080. However, the amount of the correctly translocated lipase present in the extracellular fraction is about fourfold higher in the PmpA-overproducing strain, suggesting that it is better secreted.
FIG. 4.
Western blot using anti-CtermLIP serum of total cell extract (C) (0.1 U) and supernatant (S) (0.5 U) of the control strain, MG-Pnis-Lip+, the strain overproducing PmpA with the lipase, MG-Pnis::pmpA-Lip+, and the pmpA mutant, MG-pmpA. The addition of the nisin inducer in the growing medium is indicated by -nis or +nis. See the legend to Fig. 3 for an explanation of U.
PmpA inactivation affects cells growth only when heterologous lipase is overproduced.
Since many previously known membrane-bound components of the bacterial secretion machinery including PrsA of B. subtilis are indispensable, it was of interest to test whether this would be the case in L. lactis. A fragment of 0.74 kb internal to the MG1363 pmpA gene was cloned into the integrative plasmid pJIM2374. Integration of this plasmid by simple crossing over is expected to interrupt the pmpA gene and yield erythromycin-resistant recombinants. Since pmpA is transcribed independently from upstream genes and not clustered to the downstream essential alaRS gene, no polar effect is expected upon pJIM2374 integration. Erythromycin-resistant clones were tested by PCR (data not shown) and radiolabeling with [3H]palmitic acid (Fig. 3B). In contrast with the case with B. subtilis (31, 32), the inactivation of the pmpA gene was easily obtained with L. lactis MG1363. Under normal growth conditions (in CDM at 30°C), a slight decrease of the growth rate of the MG-pmpA mutant was observed (Fig. 5A).
FIG. 5.
Growth of MG1363 (filled squares), MG-pmpA (open squares), MG-Lip+ (filled circles), and MG-pmpA-Lip+(open circles) at 30°C in CDM (A) and in CDM with NaCl 0.25 M (B).
To better define the phenotype associated with PmpA inactivation in L. lactis, we tested the ability of the resulting mutant to grow under different environmental conditions. There was no significant difference during its growth at 37°C, which is a high temperature for L. lactis (not shown). However, in osmotic stress induced by the addition of 0.25 M NaCl (Fig. 5B) or 0.6 M sorbitol (not shown), the generation time of MG-pmpA was threefold higher than that of MG1363 (210 min instead of 75 min).
To test whether PmpA is indispensable for the cells when heterologous proteins are secreted, pJIM2088 was transformed in MG1363 and in its pmpA mutant. When lipase was produced by the cell, the growth of the mutant was more strongly affected than that of the wild-type strain. This effect was dramatic when the cells were subject to an additional stress with 0.25 M NaCl, since the mutant strain no longer grew (Fig. 5A and B). Western blots were done to compare the different forms of the lipase produced with the wild-type and pmpA mutant strains (Fig. 4). The same bands were observed for the two strains, but the amount of the precursor form (preprolipase) relative to the processed prolipase increased about fourfold in the mutant.
Transcriptional regulation of pmpA.
The transcription of pmpA was studied by luciferase gene fusion with the pmpA promoter in pJIM2087. This plasmid was integrated into the chromosome of the MG1363 strain to produce MG-PpmpA::lux. The integration of the plasmid duplicates the pmpA promoter so that the strain has a wild-type phenotype. In the M17 medium, the luciferase activity per cell was almost constant for some 3 h and then decreased slightly (Fig. 6). In CDM, this activity increased about sevenfold during the mid-exponential growth phase and then decreased in the stationary phase. The addition of NaCl or sorbitol to CDM did not change the expression level or pattern (not shown). Moreover, during overproduction of the lipase the transcription of pmpA was not affected (not shown).
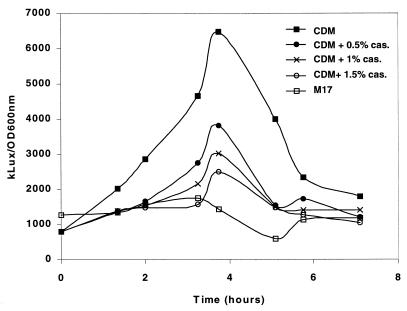
FIG. 6.
Measure of the lux activity from the pmpA promoter in the strain MG-PpmpA::lux grown in CDM with Casitone.
The increase of PmpA expression in CDM compared to M17 suggests that some components of M17 may repress its expression, or that components of CDM may induce it. M17 is rich in peptides, since it contains 5 g of each beef extract, yeast extract, and soja extract/liter. We thus tested the addition of Casitone, a peptide- and amino acid -containing nitrogen source, to CDM. Increasing amounts of Casitone progressively reduced the transcription of pmpA. In a 1% Casitone solution, the luciferase activity of the pmpA::lux fusion was at a level comparable to that produced in the M17 medium (Fig. 6). This indicates that a nitrogen source may repress pmpA expression. Since peptides are more rapidly metabolized by L. lactis than are amino acids, it is possible that they regulate pmpA expression.
DISCUSSION
PLP are not well conserved in gram-positive bacteria.
In this work we have characterized the unique chromosomal gene of L. lactis, pmpA, whose product is slightly homologous to PrsA of B. subtilis. The N-terminal sequence corresponding to its first 96 amino acids was previously detected by a systematic screen for exported proteins (40). A search of sequences in the available genomes revealed that homologous genes are present in all gram-positive bacteria, except mycoplasma and mycobacteria that do not have a typical cell wall. With gram-positive bacteria, almost all the PLP are potential lipoproteins, since they contain a cysteine residue next to the potential cleavage site of their leader peptide. The only two exceptions to this rule are two probable intracellular proteins encoded by plp2 and plp4 in C. acetobutylicum and C. difficile, respectively. Genes encoding PrsA-like lipoproteins are present in several copies in B. subtilis (prsA and yacD), S. pyogenes (2 copies), C. difficile (3 copies) and several strains of L. lactis (pmpA and prtM). Alignment of these proteins shows that the different copies present in C. difficile and S. pyogenes are more closely related to each other than to genes from closely related genomes, suggesting that they arose by gene duplication. This is not the case with PrsA and YacD in B. subtilis and PmpA and PrtM in L. lactis. For the latter, it could be expected that the presence of prtM is the result of a lateral transfer, since the gene is carried on a plasmid and is flanked by IS elements (24).
A striking feature of PLP is their low percentage of homology. For example, PLP from streptococci share only 40% to 50% identical residues, against 70 to 80% for most orthologous proteins (for example, about 75% for the CAM and PepF localized just upstream of pmpA). This difference is even more pronounced for more distantly related bacteria. The PLP of S. pyogenes is 36, 12, and 10% identical to those of E. faecalis, S. aureus, and clostridia, versus 57, 43, and 39% for PepF. This suggests (i) that the foldase activity of these proteins does not require a high level of conservation of the primary sequences and/or (ii) that the corresponding genes evolved rapidly under a strong selective pressure and/or, lastly, (iii) that these genes are not true orthologues. The fact that the genes in the vicinity of the prsA-like genes are conserved in bacterial families such as the Streptococcaceae (Streptococcus, Lactococcus) may support the first hypothesis.
PLP are thought to be involved in the folding of many exported proteins. The product of the second copy of the gene, when present, may have a more focused role. PrtM from L. lactis is required for the self-activation of the cell wall protease PrtP (24). The expression of Plp2 from S. pyogenes may be required for the proper export of the virulence-associated factors encoded upstream (the exotoxin SpeB) or downstream (homologous to tpt genes from Streptococcus cristatus involved in fimbrial tuft organization) in a region that appears to be a virulence island (Fig. 1).
The absence of the PPIase motif in L. lactis PmpA does not impair secretion-improving activity.
PLP previously described were shown to contain a PPIase signature domain (42, 46). A remarkable feature of some PLP presented here is their unconserved PPIase signature domain. This is particularly striking for PLP from Streptococcaceae, including L. lactis and the pathogenic streptococci with the exception of S. pneumoniae. Moreover, PLP from theses bacteria do not share additional conserved residues in the PPIase domain of the protein. This is obvious for the motifs forming β1, β4, and the turn between β2 and α4, and to a lesser extent in α4 and β3. This suggests that (i) these potential foldases do not have PPIase activity, and/or (ii) these particular residues do not need to be conserved for PPIase activity in Streptococcaceae. The secondary structure in these regions seems to be perfectly conserved in all of the aligned proteins except those which lack the PPIase motifs where a helix α3 seems to be absent (Fig. 2B). However, three-dimensional structure simulation of this region suggests that a coiled region could compensate this deletion, supporting the idea that the overall structure of the domain is conserved (not shown).
Our results strongly suggest that PmpA from L. lactis has a foldase activity, since its overproduction drastically decreased the accumulation of degradation products of an overproduced heterologous lipase and increase significantly the amount of correctly secreted prolipase. The simplest interpretation is that PmpA allowed the proper folding of the lipase, thus preventing its degradation by HtrA, the unique surface protease in L. lactis (41). Indeed, the extracellular form of the lipase is not degraded in an htrA mutant of IL-1403 (41). An alternative explanation would be that the overproduction of PmpA increased the activity of HtrA toward the lipase degradation products, either by changing their folding to forms more susceptible to proteolysis or by affecting the folding of HtrA. Since HtrA does not require the presence of PmpA to degrade the lipase in a pmpA mutant, we consider this alternative less likely. However, in both cases, PmpA would exert a foldase activity in L. lactis. This is particularly interesting since L. lactis PmpA lacks the motifs found in PPIases of the parvulin family. Since mutation of the PPIase domain of PpiD from E. coli (9) abolished their activity, we propose that either the L. lactis foldase does not require PPIase activity or its PPIase activity is carried out by a motif greatly different from those characterized previously. Further work is required to distinguish between these possibilities.
The sequence analysis data presented here may be further exploited to guide future research on this topic. PLP are formed by 3 regions: the central one may confer a PPIase activity in certain PLP, but the function of the N-terminal and carboxy-terminal domains remains unknown. The primary sequences of these regions are not always well conserved in all PLP proteins. However, in addition to common motifs, the secondary structures of these proteins seem to be similar, supporting the hypothesis that they play a similar function. It was suggested that these domains may increase the specific activity of the PPIase by providing specificity for a selected substrate (45). Alternatively, they may perform additional folding functions. The fact that L. lactis PmpA has a foldase activity without a conserved PPIase domain supports the second hypothesis. Other chaperones were found to exert a second activity, such as DnaJ, which is also a protein disulfide isomerase (10). Cyclophilin may have a chaperone role independent of its PPIase activity (16), although this was not later confirmed (28). Recently, it was also shown that the chaperone-like activity of FkpA in E. coli was independent of its PPIase activity (2, 43).
Potential role of PmpA in L. lactis.
The pmpA gene was deleted from L. lactis without a strong effect on the growth rate in a rich medium under normal environmental conditions. This suggests that the PmpA protein plays a limited role in L. lactis. In general, extracellular enzymes play a rather limited role in the life of these bacteria, living in a nutrient-rich environment (e.g., milk, mucosa, or vegetals), compared to the case with soil bacteria, such as B. subtilis, where PrsA is required for growth (31, 32). Interestingly, PLP from S. pneumoniae retained the PPIase motif, suggesting that this pathogenic bacteria exports proteins requiring a PPIase activity associated with PLP. The possibility of obtaining a null mutant of this gene in L. lactis may provide a useful tool for the molecular study of enzymes of this family.
The phenotypical effect of the pmpA mutation was more pronounced under salt stress, which induces the synthesis of at least 12 new proteins in L. lactis (29, 39). The production of the lipase during salt stress amplified the effect of the pmpA mutation, suggesting that PmpA became a limiting factor for the production of salt resistance systems in L. lactis. Since the transcription of pmpA is not stimulated by salt stress, it is possible that the role of PmpA in salt stress resistance is indirect. It cannot be ruled out that accumulation of unprocessed lipase in the membrane causes defects that result in an increased sensitivity of the cell. Interestingly, pmpA transcription is increased during exponential growth when the nitrogen source is limited, suggesting that its expression may be required for the folding of factors involved in nitrogen supply, such as the cell wall protease or the three peptide transport systems Opp, DtpT, and DtpP, that are induced under similar conditions (23).
Interestingly, the unprocessed preprolipase is accumulated in larger amounts in the pmpA mutant. In a previous work, we have shown that this unprocessed form of the lipase was present at the inner surface of the cytoplasmic membrane (13). The increased defect of lipase translocation in a pmpA mutant suggests that PmpA might have a role in the folding of some trigger factors involved in the translocation machinery. The amount of PmpA in the wild-type strain does not limit the activity of these factors, since its overproduction did not decrease the amount of the unprocessed lipase. However, a defect in the activity of such a factor in a pmpA mutant would explain the phenotype conferred by the pmpA mutation, in particular the increased salt sensitivity during lipase secretion caused by limiting the production of resistance systems as proposed above.
Acknowledgments
This study was financed by an MRE grant from the French government.
We thank Isabelle Poquet for useful discussions.
REFERENCES
- 1.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arie, J. P., N. Sassoon, and J. M. Betton. 2001. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol. Microbiol. 39:199-210. [DOI] [PubMed] [Google Scholar]
- 3.Arnau, J., E. Hjerl-Hansen, and H. Israelsen. 1997. Heterologous gene expression of bovine plasmin in Lactococcus lactis. Appl. Microbiol. Biotechnol. 48:331-338. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S., R. Maier, H. de Cock, F. X. Schmid, and C. A. Gross. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bojovic, B., G. Djordjevic, A. Banina, and L. Topisirovic. 1994. Mutational analysis of cat-86 gene expression controlled by lactococcal promoters in Lactococcus lactis subsp. lactis and Escherichia coli. J. Bacteriol. 176:6754-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burucoa, C., C. Fremaux, Z. Pei, M. Tummuru, M. J. Blaser, Y. Cenatiempo, and J. L. Fauchere. 1995. Nucleotide sequence and characterization of peb4A encoding an antigenic protein in Campylobacter jejuni. Res. Microbiol. 146:467-476. [DOI] [PubMed] [Google Scholar]
- 8.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 9.Dartigalongue, C., and S. Raina. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17:3968-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Crouy-Chanel, A., M. Kohiyama, and G. Richarme. 1995. A novel function of Escherichia coli chaperone DnaJ. Protein-disulfide isomerase. J. Biol. Chem. 270:22669-22672. [DOI] [PubMed] [Google Scholar]
- 11.Delorme, C., S. D. Ehrlich, and P. Renault. 1999. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 181:2026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 2000. Expression of the Staphylococcus hyicus lipase in Lactococcus lactis. Appl. Environ. Microbiol. 66:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 1999. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 65:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enfors, S. O. 1992. Control of in vivo proteolysis in the production of recombinant proteins. Trends Biotechnol. 10:310-315. [DOI] [PubMed] [Google Scholar]
- 16.Freskgard, P. O., N. Bergenhem, B. H. Jonsson, M. Svensson, and U. Carlsson. 1992. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science 258:466-468. [DOI] [PubMed] [Google Scholar]
- 17.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:540-553. [DOI] [PubMed] [Google Scholar]
- 18.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilson, T. J. 1984. Ph.D. thesis. University of Cambridge, Cambridge, England.
- 20.Glatron, M. F., and G. Rapoport. 1972. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie 54:1291-1301. [DOI] [PubMed] [Google Scholar]
- 21.Goupil-Feuillerat, N., G. Corthier, J. J. Godon, S. D. Ehrlich, and P. Renault. 2000. Transcriptional and translational regulation of alpha-acetolactate decarboxylase of Lactococcus lactis subsp. lactis. J. Bacteriol. 182:5399-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundy, F. J., and T. M. Henkin. 1994. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria. J. Mol. Biol. 235:798-804. [DOI] [PubMed] [Google Scholar]
- 23.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haandrikman, A. J., J. Kok, H. Laan, S. Soemitro, A. M. Ledeboer, W. N. Konings, and G. Venema. 1989. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J. Bacteriol. 171:2789-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 26.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 27.Horn, N., M. I. Martinez, J. M. Martinez, P. E. Hernandez, M. J. Gasson, J. M. Rodriguez, and H. M. Dodd. 1999. Enhanced production of pediocin PA-1 and coproduction of nisin and pediocin PA-1 by Lactococcus lactis. Appl. Environ. Microbiol. 65:4443-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kern, G., D. Kern, F. X. Schmid, and G. Fischer. 1994. Reassessment of the putative chaperone function of prolyl-cis/trans-isomerases. FEBS Lett. 348:145-148. [DOI] [PubMed] [Google Scholar]
- 29.Kilstrup, M., S. Jacobsen, K. Hammer, and F. K. Vogensen. 1997. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63:1826-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein, P., R. L. Somorjai, and P. C. Lau. 1988. Distinctive properties of signal sequences from bacterial lipoproteins. Protein Eng. 2:15-20. [DOI] [PubMed] [Google Scholar]
- 31.Kontinen, V. P., P. Saris, and M. Sarvas. 1991. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol. Microbiol. 5:1273-1283. [DOI] [PubMed] [Google Scholar]
- 32.Kontinen, V. P., and M. Sarvas. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol 8:727-737. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1998. A nine-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J. Bacteriol. 180:1895-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leskela, S., E. Wahlstrom, V. P. Kontinen, and M. Sarvas. 1999. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the Lgt gene. Mol. Microbiol. 31:1075-1085. [DOI] [PubMed] [Google Scholar]
- 36.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Missiakas, D., J. M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871-884. [DOI] [PubMed] [Google Scholar]
- 38.Nardi, M., P. Renault, and V. Monnet. 1997. Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis. J. Bacteriol. 179:4164-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obis, D., A. Guillot, J. C. Gripon, P. Renault, A. Bolotin, and M. Y. Mistou. 1999. Genetic and biochemical characterization of a high-affinity betaine uptake system (BusA) in Lactococcus lactis reveals a new functional organization within bacterial ABC transporters. J. Bacteriol. 181:6238-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poquet, I., V. Saint, E. Seznec, N. Simoes, A. Bolotin, and A. Gruss. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 42.Rahfeld, J. U., K. P. Rucknagel, B. Schelbert, B. Ludwig, J. Hacker, K. Mann, and G. Fischer. 1994. Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases. Amino acid sequence and recombinant production of parvulin. FEBS Lett. 352:180-184. [DOI] [PubMed] [Google Scholar]
- 43.Ramm, K., and A. Pluckthun. 2000. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. II. Isomerase-independent chaperone activity in vitro. J. Biol. Chem. 275:17106-17113. [DOI] [PubMed] [Google Scholar]
- 44.Renault, P., G. Corthier, N. Goupil, C. Delorme, and S. D. Ehrlich. 1996. Plasmid vectors for gram-positive bacteria switching from high to low copy number. Gene 183:175-182. [DOI] [PubMed] [Google Scholar]
- 45.Rouviere, P. E., and C. A. Gross. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10:3170-3182. [DOI] [PubMed] [Google Scholar]
- 46.Rudd, K. E., H. J. Sofia, E. V. Koonin, G. Plunkett III, S. Lazar, and P. E. Rouviere. 1995. A new family of peptidyl-prolyl isomerases. Trends Biochem. Sci. 20:12-14. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Sissler, M., C. Delorme, J. Bond, S. D. Ehrlich, P. Renault, and C. Francklyn. 1999. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. USA 96:8985-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steidler, L., K. Robinson, L. Chamberlain, K. M. Schofield, E. Remaut, R. W. Le Page, and J. M. Wells. 1998. Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine. Infect. Immun. 66:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steidler, L., J. M. Wells, A. Raeymaekers, J. Vandekerckhove, W. Fiers, and E. Remaut. 1995. Secretion of biologically active murine interleukin-2 by Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 61:1627-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terzaghi, B., and W. E. Sandine. 1975. Improved medium for lactic Streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van de Guchte, M., J. Kodde, J. M. van der Vossen, J. Kok, and G. Venema. 1990. Heterologous gene expression in Lactococcus lactis subsp. lactis: synthesis, secretion, and processing of the Bacillus subtilis neutral protease. Appl. Environ. Microbiol. 56:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Oort, M. G., A. M. Deveer, R. Dijkman, M. L. Tjeenk, H. M. Verheij, G. H. de Haas, E. Wenzig, and F. Gotz. 1989. Purification and substrate specificity of Staphylococcus hyicus lipase. Biochemistry 28:9278-9285. [DOI] [PubMed] [Google Scholar]
- 54.von Heijne, G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2:531-534. [DOI] [PubMed] [Google Scholar]
- 55.Wu, S. C., R. Ye, X. C. Wu, S. C. Ng, and S. L. Wong. 1998. Enhanced secretory production of a single-chain antibody fragment from Bacillus subtilis by coproduction of molecular chaperones. J. Bacteriol. 180:2830-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]