Abstract
In Schizosaccharomyces pombe, the Sty1 mitogen-activated protein kinase and the Atf1 transcription factor control transcriptional induction in response to elevated salt concentrations. Herein, we demonstrate that two repressors, Tup11 and Tup12, and the Prr1 transcription factor also function in the response to salt shock. We find that deletion of both tup genes together results in hypersensitivity to elevated cation concentrations (K+ and Ca2+) and we identify cta3+, which encodes an intracellular cation transporter, as a novel stress gene whose expression is positively controlled by the Sty1 pathway and negatively regulated by Tup repressors. The expression of cta3+ is maintained at low levels by the Tup repressors, and relief from repression requires the Sty1, Atf1, and Prr1. Prr1 is also required for KCl-mediated induction of several other Sty1-dependent genes such as gpx1+ and ctt1+. Surprisingly, the KCl-mediated induction of cta3+ expression occurs independently of Sty1 in a tup11Δ tup12Δ mutant and so the Tup repressors link induction to the Sty1 pathway. We also report that in contrast to a number of other Sty1- and Atf1-dependent genes, the expression of cta3+ is induced only by high salt concentrations. However, in the absence of the Tup repressors this specificity is lost and a range of stresses induces cta3+ expression.
INTRODUCTION
Exposure of cells to environmental stress triggers a rapid increase in the transcription of genes whose products have protective functions (Toone and Jones, 1998). Key to this response are stress-activated protein kinase (SAPK) pathways that transmit the signal from stress sensors to the transcription factors that regulate gene expression. These pathways are evolutionarily conserved, and homologs of the mammalian SAP kinases, p38/RK/CSBP (Marshall, 1994), are present in both Saccharomyces cerevisiae (Hog1) (Brewster et al., 1993) and Schizosaccharomyces pombe (Sty1/Spc1) (Miller et al., 1995; Shiozaki and Russell, 1995). The Hog1 pathway in S. cerevisiae is activated essentially by hyperosmolarity (Brewster et al., 1993), whereas the S. pombe Sty1 pathway, like mammalian p38, is activated by a range of adverse conditions (Millar et al., 1995; Shiozaki and Russell, 1996; Degols and Russell, 1997; Buck et al., 2001).
Models of SAPK-dependent regulation of transcription have been almost exclusively based upon the positive control of activators. However, recent analysis of S. cerevisiae has demonstrated that the Sko1 repressor regulates the expression of Hog1-dependent osmostress genes, such as ENA1 and GRE2, via recruitment of the Ssn6(Cyc8)-Tup1 global corepressor complex (Marquez et al., 1998; Proft and Serrano, 1999; Garcia-Gimeno and Struhl, 2000; Proft et al., 2001). Ssn6-Tup1 mediates its function via the organization of repressive chromatin structures (Cooper et al., 1994; Edmondson et al., 1996; Watson et al., 2000; Bone and Roth, 2001; Wu et al., 2001) and by inhibition of the basal transcription machinery (Redd et al., 1997; Papamichos-Chronakis et al., 2000; Zaman et al., 2001). This global repressor controls the expression of numerous genes through interaction with a variety of site-specific DNA binding proteins (Smith and Johnson, 2000). Relief from this repression is achieved by control of the proteins that serve to tether the complex to DNA; for example, Sko1 is phosphorylated by Hog1 at three sites in its N-terminal region, disrupting the interaction with Ssn6-Tup1 (Proft et al., 2001). Therefore, a component of the osmotic induction of some genes occurs via derepression rather than by activation.
In fission yeast, Sty1 operates via the transcriptional activators Atf1/Gad7 (Takeda et al., 1995; Kanoh et al., 1996) and Pap1 (Toda et al., 1991). Atf1 is phosphorylated in a Sty1-dependent manner and loss of Atf1 results in hypersensitivity to osmotic stress, high levels of calcium, and an inability to respond to deteriorating nutritional conditions (Takeda et al., 1995; Kanoh et al., 1996; Ohmiya et al., 1999b). In addition, Atf1 forms a heterodimeric complex with Pcr1, a related ATF/CREB factor, which is also required for transcriptional induction of some stress genes (Watanabe and Yamamoto, 1996). Pap1 activates transcription in response to oxidative stress, and its subcellular localization is regulated in a Sty1-dependent manner (Toone et al., 1998). Recently, Prr1, a homolog of Skn7 in S. cerevisiae (Brown et al., 1993), has also been implicated in the transcriptional response to oxidative stress (Ohmiya et al., 1999a). Skn7 and Prr1 have heat-shock factor-like DNA binding domains and also share homology with bacterial “two-component” response regulators that are controlled by histidine-to-aspartate phosphorelay systems (Appleby et al., 1996).
Herein, we have addressed the roles of the Tup-like repressors Tup11 and Tup12 (Mukai et al., 1999; Janoo et al., 2001) in the response to stress in S. pombe. We find that deletion of both tup genes in combination results in hypersensitivity to KCl and CaCl2, and we also identify cta3+ as a novel stress gene that is negatively regulated by Tup11-Tup12. The expression of cta3+ is rapidly and specifically induced in response to salt shock in a Sty1- and Atf1-dependent manner, but the dependence on the Sty1 pathway for induction is lost in a tup11Δ tup12Δ mutant. Furthermore, Tup11 and Tup12 proteins function as specificity factors by preventing induction of cta3+ in response to inappropriate stresses such as heat and oxidative stress. We also reveal a new role for the “response regulator” protein Prr1 and demonstrate that it is required for proper KCl-mediated transcriptional induction of Sty1-dependent genes such as cta3+, ctt1+, and gpx1+.
MATERIALS AND METHODS
Strains
Routine culture of S. pombe and general genetic methods were performed as described in Moreno et al. (1991). The strains used in this study are described in Table 1. The cta3+ gene was disrupted using a polymerase chain reaction (PCR)-based approach as described by Bahler et al. (1998). Oligonucleotides 5′ KO (5′-TTTGATTTTACTTATATTTCTCCCCTTCTACTCATCCCGATATATTCTTACTTCCTTGATTCAATCTCAAATATTGTTCAGCTTAGCTACAAATCCCACT-3′) and KO 3′ (5′-ATAAATCCTTTACGATTTGTCGGTTCTGTGAAAACGATACACTCACGCATATTCATATACATATTCATGGCAAGAAAACATCTGACATAAAACGCCTAGG-3′) were used to amplify a 1.6-kb ura4+-containing fragment from pRep42. The amplified fragment was used to transform strain NT5 strain to Ura+, creating strain SW95. Integration at the correct locus was confirmed by PCR analysis.
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| NT4 | h+ ade6-M210 leu1-32 ura4-D18 | Zhu et al. (1994) |
| NT5 | h− ade6-M216 leu1-32 ura4-D18 | Zhu et al. (1994) |
| SW54 | h+ ade6-M210 leu1-32 ura4-D18 tup11∷ura4+ | Janoo et al. (2001) |
| BSP03 | h+ ade6− leu1-32 ura4-D18 tup12∷ura4+ | Janoo et al. (2001) |
| RJP12 | h− ura4∷fbp1-lacZ tup11∷ura4+ tup12∷ura4+ | Janoo et al. (2001) |
| SW76 | h+ ade6− tup11∷ura4+ tup12∷ura4+ ura4-D18 leu1-32 | This study |
| HAI003 | h− ade6−M216 leu1-32 ura4-D18 cta3-lacZ∷ura4+ | Nishikawa et al. (1999) |
| SW107 | h− ade6− leu1-32 ura4-D18 tup11∷ura4+ tup12∷ura4+ cta3-lacZ∷ura4+ | This study |
| KS14709 | h− atf1HA6H∷ura4+ leu1-32 ura4-D18 | Shiozaki & Russell (1996) |
| JM1144 | h− sty1-1 leu1-32 ura4-D18 | Millar et al. (1995) |
| JM1160 | h− ade6-M216 leu1-32 ura4-D18 sty1∷ura4+ | Millar et al. (1995) |
| NT147 | h90 leu1-32 ura4-D18 atf1∷ura4+ | Takeda et al. (1995) |
| JX125 | h90 ade6− leu1-32 ura4-D18 pcr1∷ura4+ | Kanoh et al. (1996) |
| RJP59 | h+ his7-366 ura4∷fbp1-lacZ pcr1∷ura4+ tup11∷ura4+ tup12∷ura4+ | Janoo et al. (2001) |
| SW89 | h+ ade6− leu1-32 ura4-D18 tup11∷ura4+ sty1-1 | This study |
| SW88 | h+ leu1-32 ura4-D18 tup12∷ura4+ sty1-1 | This study |
| SW90 | h+ ade6− leu1-32 ura4-D18 tup11∷ura4+ tup12∷ura4+ sty1-1 | This study |
| SW91 | h− leu1-32 ura4-D18 tup11∷ura4+ tup12∷ura4− sty1∷ura4+ | This study |
| SW95 | h− ade6-M216 leu1-32 ura4-D18 cta3∷ura4+ | This study |
| SW93 | h+ ade6− leu1-32 ura4-D18 cta3∷ura4+ tup11∷ura4+ tup12∷ura4+ | This study |
| SW92 | h+ leu1-32 ura4-D18 atf1∷ura4+ tup11∷ura4+ tup12∷ura4+ | This study |
| RJP59 | h+ ura4∷fbp1-lacZ pcr1∷ura4+ tup11∷ura4+ tup12∷ura4+ his7-336 | Janoo et al. (2001) |
| SW97 | h+ ade6-M216 ura4-D18 his7-336 leu1-32 prr1∷ura4+ | This study |
| SW96 | h+ ade6− ura4-D18 leu1-32 prr1∷ura4+ tup11∷ura4+ tup12∷ura4+ his7-336 | This study |
Plasmids
The tup11+ coding sequence was amplified by PCR from a cDNA library by using the following primers: 5′-GCACGGATCCCATGGCGTCAGTGGAGGATGC-3′ and 5′-CTAGGGATCCAATTCAA-GGAGATGCAGGGTC-3′. The tup12+ coding sequence was amplified using primers 5′-GCACGGATCCCATGATTACTGTCCGCCAATC-3′ and 5′-CTGCTAGGCATATGGCGCTCATGAAACAAACCG-3′. Fragments were cleaved with BamHI and cloned into the BamHI site of derivatives of pRep41 and pRep42 vectors that allow the expression of proteins as N-terminal HA or 6HisMyc fusions (Craven et al., 1998).
RNA Analysis
RNA samples were prepared from 0.25 to 0.5 × 109 cells. Pellets were washed in H2O and resuspended in 200 μl of RNA buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 50 mM EDTA pH 8.0, and 0.25% SDS) with 200 μl of phenol/chloroform. Cells were disrupted with 0.75 ml of glass beads (0.5 mm; Biospec Products, Bartlesville, OK) in a Ribolyser (Hybaid, Middlesex, United Kingdom). A further 0.75 ml of RNA buffer was added followed by spinning in a microfuge for 10 min. The aqueous layer was subjected to two further phenol/chloroform extractions before the RNA was precipitated with 0.1 volume of sodium acetate, pH 5.2, and 0.6 volume of isopropanol. RNA pellets were washed in 70% ethanol and resuspended in H2O. RNA analysis was as described by White et al. (1986). Briefly, a 10–15-μg sample of total RNA was denatured with glyoxal, separated on a 1.2% agarose gel prepared in 15 mM sodium phosphate, pH 6.5, and transferred to a GeneScreen hybridization membrane (PerkinElmer Life Sciences, Boston, MA). The his3+ probe has been described previously (Baum et al., 1997). Other gene-specific probes were produced by PCR amplification from genomic DNA by using the appropriate primers. All probes were labeled with [α-32P]dCTP by using a Prime-a-Gene labeling kit (Promega, Madison, WI). Transcript levels were quantified relative to the loading control using a PhosphorImager BAS-1500 (Fuji Photo Film, Tokyo, Japan).
β-Galactosidase Assays
Assays were performed as described previously (Takeda et al., 1995).
Coprecipitations
Whole cell extracts were prepared as described by Whitehall et al. (1999) with some modification. Cultures were grown to mid-log phase (OD595 = 0.25–0.5) in EMM medium. Cells were harvested washed once and snap frozen. Pellets were washed in 1 ml of lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% NP-40, 10 mM imidazole, 2 μg/ml pepstatin, 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 100 μg/ml phenylmethylsulfonyl fluoride). Cells were disrupted with 2 ml of glass beads by vortexing twice for 45 s with 1-min incubation on ice in between. Protein extracts were recovered and centrifuged at 13,000 rpm for 10 min at 4°C. Protein precipitations were performed by adding 25 μl of nickel-agarose (50% slurry in lysis buffer) to 1 mg of whole protein extract and incubating at 4°C for 1 h with gentle agitation. Precipitates were recovered by centrifugation and washed four times with lysis buffer containing 200 mM NaCl and 20 mM imidazole. Samples were analyzed by SDS-PAGE and proteins were transferred to nitrocellulose membrane and subjected to Western blotting by using monoclonal hemagglutinin (HA) (12CA5) antibody (Babco, Berkeley, CA).
Electrophoretic Mobility Shift Assays (EMSAs)
Whole cell extracts were prepared as described above except that cells were grown in YE5S medium and extracts were prepared in buffer containing 25 mM HEPES pH 7.6, 0.1 mM EDTA, 150 mM KCl, 0.1% Triton X-100, 25% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride 2 μg/ml pepstatin, 2 μg/ml leupeptin, and 2 μg/ml aprotinin. Radiolabeled DNA fragments were prepared using PCR amplification as described in Zhu et al. (1994). The oligonucleotides used for amplification of probe 1 were 5′-TAAAACACCGACATGTAGCC-3′ and 5′-TTGAGAGAAACTAACCAAGG-3′. The oligonucleotides for probe 2 were 5′-CTCTGTCATGGAAATCCACAC-3′ and 5′-ATAAGCAGCAAAGCTTGCCTG-3′. Binding reactions were performed by adding 15 μg of whole cell extract to 20-μl reactions containing 25 mM HEPES pH 7.6, 34 mM KCl, 5 mM MgCl2, and 2 μg of poly[d(I-C)]. Reactions were incubated for 10 min at room temperature before the addition of ∼0.5 ng of radiolabeled probe DNA followed by a further 20-min incubation. Samples were analyzed by electrophoresis through 4% polyacrylamide gels run in 0.5× Tris-borate-EDTA buffer. Antibody supershift was performed by adding 0.2 μg of monoclonal HA antibody (12CA5) (Babco) 10 min after the addition of probe DNA.
RESULTS
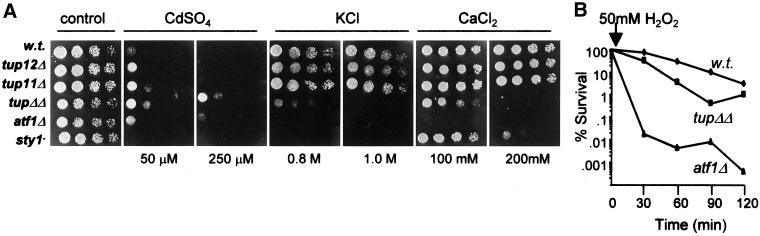
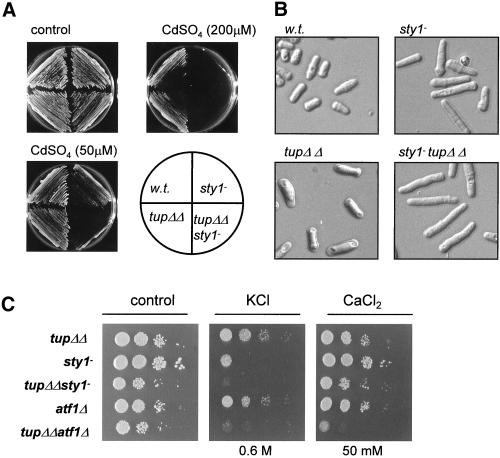
To address the role of repressors in the transcriptional response to stress in fission yeast we examined whether cells lacking the tup genes tup11+ and tup12+ exhibited any stress-related phenotypes. We found that single and double tup mutant strains exhibited an increased tolerance to cadmium but that the tup11Δ tup12Δ mutant strain had decreased tolerance to elevated levels of Ca2+ and K+ ions. The degree of sensitivity to these salt stresses was similar to that associated with loss of either the Atf1 transcription factor or the Sty1 MAP kinase that are known to control the induction of genes in response to elevated cation concentrations (Shiozaki and Russell, 1996; Wilkinson et al., 1996). The tup11Δ tup12Δ double mutant strain was only slightly less sensitive to KCl than the sty1-1 and atf1Δ strains and the tup11Δ tup12Δ strain was actually more sensitive to CaCl2 than strain lacking Sty1 (Figure 1A). In contrast, the double tup mutant strain had wild-type levels of tolerance to high sorbitol concentrations, indicating that although they are K+- and Ca2+-intolerant they are not osmosensitive (our unpublished data).
Figure 1.
Stress-related phenotypes of cells lacking Tup repressors. (A) Exponentially growing cultures (∼0.4 × 107 cells/ml) were diluted serially, spotted onto YE5S agar, and incubated for 2–3 d at 30°C or spotted onto YE5S agar supplemented with CaCl2, KCl, or CdSO4 at the indicated concentration and incubated for 3–4 d at 30°C. (B) Exponentially growing cultures of wild type (w.t.) (NT4), atf1Δ (NT147), and tup11Δ tup12Δ (SW76) strains were treated with H2O2 (to a final concentration of 50 mM), and viable cell numbers were determined by plating onto YE5S agar.
The similarity in the sensitivities of tup11Δ tup12Δ and atf1Δ strains to elevated K+/Ca2+ ions was unexpected because Tup11 and Tup12 have been previously demonstrated to be repressors (Mukai et al., 1999; Janoo et al., 2001), whereas Atf1 is primarily a transcriptional activator. We therefore investigated whether tup11Δ tup12Δ cells shared any other phenotypes with atf1Δ cells. It has recently been demonstrated that atf1Δ cells are sensitive to an acute oxidative stress (Nguyen et al., 2000; Quinn et al., 2002). When challenged with a high dose of H2O2 (50 mM) atf1Δ cells rapidly lose viability (Figure 1B). In contrast, tup11Δ tup12Δ cells were only slightly more sensitive than wild-type cells in this assay. Furthermore, although atf1Δ cells conjugate poorly (Takeda et al., 1995), a tup11Δ tup12Δ strain conjugates in nutrient-rich media (Janoo et al., 2001). Hence, tup11Δ tup12Δ cells and atf1Δ cells share only a subset of phenotypes. Taken together, these findings are consistent with Tup11 and Tup12 having overlapping functions and indicate that Tup11 and Tup12 play roles in the cellular response to stress.
Tup11 and Tup12 Negatively Regulate Expression of Salt-Stress gene cta3+
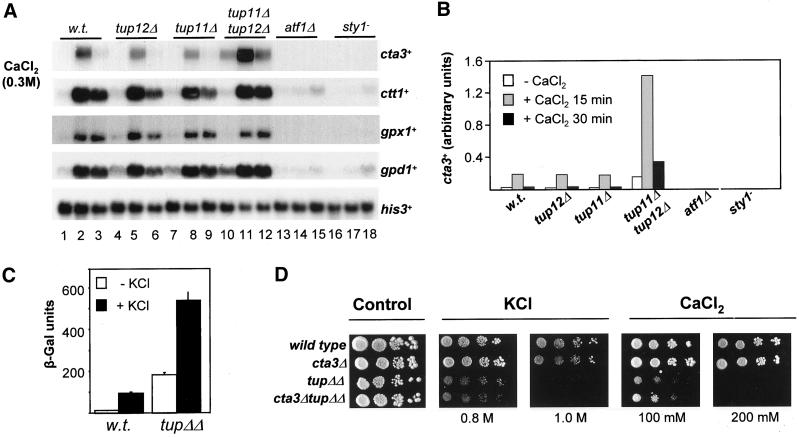
Cells lacking atf1+ and both tup genes have similar sensitivities to salt stress and so we examined whether the expression of genes known to be induced by exposure to salt stress, via Atf1, were also regulated by the Tup repressors (Figure 2, A and B). We found that expression of cta3+, which encodes a cation-transporting P-type ATPase (Ghislain et al., 1990; Halachmi et al., 1992; Benito et al., 2002), was markedly influenced by loss of the Tup proteins; deletion of both tup genes together resulted in a large increase in the basal level of expression (Figure 2, A and B). Loss of both Tup proteins also resulted in a large increase in expression after exposure to CaCl2 (Figure 2, A and B) and KCl (Figures 5, 7, and 8). Thus, Tup11 and Tup12 function in a partially redundant manner to repress cta3+ expression and limit the level of induction. As previously observed (Nishikawa et al., 1999), the induction of cta3+ expression is completely dependent upon both the Sty1 MAP kinase and the Atf1 transcription factor, and thus cta3+ displays a novel pattern of stress regulation that is positively controlled by Sty1 and Atf1 but negatively regulated by the Tup repressors. Indeed, deletion of the tup genes either singly or in combination had only minor effects on the expression levels of other Sty1-dependent genes such as gpd1+, ctt1+, and gpx1+ in unstressed cells and in cells subjected to a CaCl2 shock (Figure 2, A and B).
Figure 2.
Tup11 and Tup12 repress the transcription of the salt stress gene cta3+. (A) Strains used are indicated above the lane and were wild type (w.t.) (NT4), tup12Δ (BSP03), tup11Δ (SW53), tup11Δ tup12Δ (SW76), atf1Δ (78 Τ147), and sty1-1 (JM1144). Log phase cultures growing at 30°C in YE5S (lanes 1, 4, 7, 10, 13, and 16) were incubated with CaCl2 (to final concentration of 0.3 M) for 15 min (lanes 2, 5, 8, 11, 14, and 17) and 30 min (lanes 3, 6, 9, 12, 15, and 18). Total RNA was extracted, separated by electrophoresis, and Northern blots were analyzed with the indicated probes. The level of his3+ mRNA was used as a loading control. (B) Quantification of cta3+ mRNA levels in A. (C) Influence of Tup proteins on the activity of a cta3-lacZ reporter. β-Galactosidase assays were performed on extracts derived from exponentially growing cells (open bars) and cells treated with KCl to 0.6 M for 60 min (black bars). The strains used were wild type (w.t.) (HAI003) and tup11Δ tup12Δ (SW107). Data is the mean of three independent cultures. (D) Deletion of cta3+ does not rescue the salt sensitivity of tup11Δ tup12Δ cells. Exponentially growing cultures (∼0.4 × 107 cells/ml) were diluted serially, spotted onto YE5S agar and incubated for 2–3 d at 30°C or spotted onto YE5S agar supplemented with CaCl2 or KCl (at the indicated concentration) and incubated for 3–4 d at 30°C. Strains used were wild type (w.t.) (NT4), tup11Δ tup12Δ (SW76), cta3Δ (SW95), and cta3Δ tup11Δ tup12Δ (SW93).
Figure 5.
Deletion of tup11+ and tup12+ allows Sty1-independent transcriptional induction of cta3+. (A) Strains used are indicated above the lanes and were wild type (w.t.) (NT4), sty1-1 (JM1144), sty1-1 tup12Δ (SW88), sty1-1 tup11Δ (SW89), tup11Δ tup12Δ (SW76), and sty1-1 tup11Δ tup12Δ (SW90). Log phase cultures growing at 30°C in YE5S (lanes 1, 3, 5, 7, 9, and 11) were incubated with KCl (to a final concentration of 0.6 M) for 15 min (lanes 2, 4, 6, 8, 10, and 12). Total RNA was extracted, separated by electrophoresis, and analyzed by Northern blotting with the indicated probes. The level of his3+ mRNA was used as a loading control. (B) Quantification of the cta3+ mRNA levels in A. (C) Kinetics of the induction of cta3+ mRNA in response to KCl shock. Log phase cultures growing at 30°C in YE5S (lanes 1, 7, and 13) were incubated with KCl (to a final concentration of 0.6 M) for 10 min (lanes 2, 8, and 14), 20 min (lanes 3, 9, and 15), 30 min (lanes 4, 10, and 16), 40 min (lanes 5, 11, and 17), or 50 min (lanes 6, 12 and 18). The strains used were wild type (w.t.) (NT4), tup11Δ tup12Δ (SW76), and sty1-1 tup11Δ tup12Δ (SW90). Total RNA was extracted, separated by electrophoresis, and analyzed by Northern blotting with the indicated probes. (D) Quantification of the cta3+ mRNA levels in C.
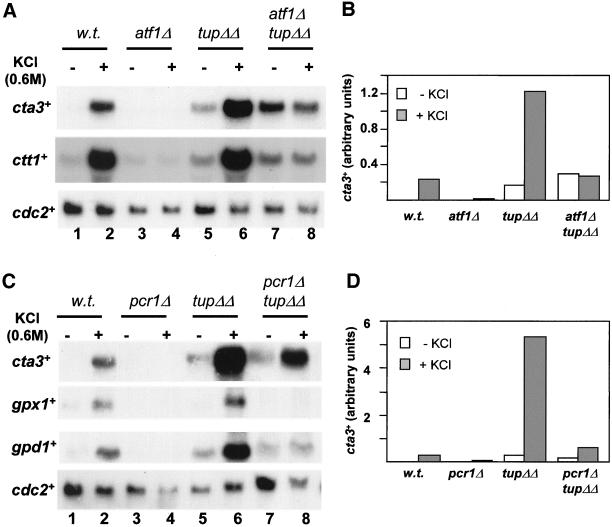
Figure 7.
Transcription induction of cta3+ in cells lacking Tup11 and Tup12 is Atf1 dependent. (A) Strains used are indicated above the lanes and were wild type (w.t.) (NT4), atf1Δ (NT147), tup11Δ tup12Δ (SW76), and atf1Δ tup11Δ tup12Δ (SW92). Log phase cultures growing at 30°C in YE5S (lanes 1, 3, 5, and 7) were incubated with KCl (to a final concentration of 0.6 M) for 15 min (lanes 2, 4, 6, and 8). Total RNA was extracted, separated by electrophoresis and subjected to Northern analysis with the indicated probes. The level of cdc2+ mRNA was used as a loading control. (B) Quantification of cta3+ mRNA levels in A. (C) As for A, except strains used were wild type (w.t.) (NT4), pcr1Δ (JX25), tup11Δ tup12Δ (SW76), and pcr1Δ tup11Δ tup12Δ (RJP59). (D) Quantification of cta3+ mRNA levels in C.
Figure 8.
Prr1 is involved in the regulation of gene expression in response to high salt. (A) Strains used are indicated above the lanes and were wild type (w.t.) (NT4) and prr1Δ (SW97). Log phase cultures growing at 30°C in YE5S (lanes 1 and 3) were incubated with KCl (to a final concentration of 0.6 M) for 15 min (lanes 2 and 4). Total RNA was prepared and subjected to Northern analysis with the indicated probes. The level of his3+ mRNA was used as a loading control. (B) Quantification of the mRNA levels in A. (C) Strains used are indicated above the lanes and were tup11Δ tup12Δ (SW76) and prr1Δ tup11Δ tup12Δ (SW96), and the treatment was as described in A. (D) Quantification of the mRNA levels in C.
To confirm that increased level of cta3+ transcripts associated with deletion of tup genes was due to an effect on transcription and not mRNA stability, we measured the expression of an integrated cta3+ promoter—lacZ reporter (Nishikawa et al., 1999). It is highly unlikely that Tup repressors would specifically influence the stability of lacZ transcripts. Consistent with the Northern analysis, deletion of both the tup genes resulted in 14-fold increase in expression of the lacZ reporter relative to the wild-type control (Figure 2C). Furthermore, exposure of cells to high KCl concentrations (0.6 M for 1 h) increased the level of expression sevenfold in wild-type cells and threefold in the cells lacking the Tup repressors (Figure 2C). These results suggest that S. pombe Tup proteins exert their effects at the level of transcription.
To determine whether the high level of cta3+ expression observed in the tup11Δ tup12Δ double mutant confers the increased sensitivity of this strain to elevated K+ and Ca2+ concentrations we examined the effect of deleting the cta3+ gene in the presence or absence of the tup genes. Loss of Cta3 function has previously been reported to result in increased sensitivity to elevated Ca2+ concentrations (Ghislain et al., 1990). In contrast, Nishikawa et al. (1999) found that cta3 null cells did not exhibit any detectable change in resistance to K+, Ca2+, or Na+ ions. In agreement with the latter study our cta3Δ mutant exhibited wild-type levels of resistance to both Ca2+ and K+ (Figure 2D). Moreover, deletion of the cta3+ gene in a tup11Δ tup12Δ strain did not rescue the salt-sensitive phenotype associated with the loss of the tup genes (Figure 2D), and plasmid-mediated overexpression of cta3+ in wild-type cells did not result in any increased sensitivity to KCl or CaCl2 (our unpublished data). These results indicate that the salt sensitivity of tup− cells is not simply due to the elevated expression of the cta3+ gene.
To date, the fbp1+ gene encoding fructose 1,6-bisphosphatase is the only gene that has been identified as a target gene for Tup11-Tup12–mediated repression (Mukai et al., 1999; Janoo et al., 2001). The expression of fbp1+ is also positively regulated by the Sty1 pathway (Takeda et al., 1995; Kanoh et al., 1996; Stettler et al., 1996), but its expression is induced by carbon limitation (Hoffman and Winston, 1991) and not by other acute stresses that activate Sty1, such as heat shock, oxidative stress, and osmotic shock. Furthermore, the cAMP pathway negatively regulates the expression of fbp1+, and mutations that disrupt this pathway result in increased expression under repressing (glucose-rich) conditions (Hoffman and Winston, 1991). In contrast, growing cells under carbon-limiting conditions did not induce the expression of cta3+ nor were mRNA levels influenced by a deletion of git2+ that encodes adenylate cyclase (our unpublished data).
Formation of Protein Complexes on the cta3+ Promoter
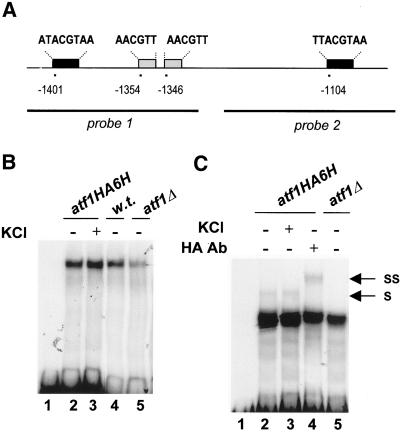
Studies have indicated that Atf1 binds constitutively to a CRE binding site in the gpd1+ promoter (Wilkinson et al., 1996). In contrast, EMSAs of the fbp1+ promoter have demonstrated that Atf1 associates with a CRE-like element in UAS1 only under activating (glucose-limiting) conditions (Neely and Hoffman, 2000). We therefore examined the ability of Aft1 to bind to the cta3+ promoter. Inspection of the DNA sequence revealed the presence of a number of potential CRE-like Atf1 binding sites located between −1111 and −1401 relative to the initiation codon (Figure 3A). We performed EMSAs by using whole cell extracts and a DNA fragment corresponding to the −1477 to −1297 region of the promoter. This region includes a near consensus CRE element and two CRE-like sequences containing the highly conserved ACGT core sequence. A major slow-migrating complex was formed on this probe (Figure 3B). This binding activity was not changed by subjecting cells to stress (KCl 0.6 M for 15 min) before extract preparation. The complex was also present in extracts derived from atf1Δ cells, indicating that it does not require Atf1. Furthermore, the mobility of the complex was unchanged when HA antibody was included in reactions containing HA epitope-tagged Atf1 (our unpublished data). Next, we examined the ability of complexes to form on a probe corresponding to the −1249 and −1058 region of the promoter that contains a single CRE element. In this case, we also observed a binding activity that was Atf1-independent (Figure 3C). However, we also detected a slow-migrating complex that was absent in reactions lacking Atf1. Also, the mobility of this complex was reduced by the addition of the HA antibody to reactions containing HA epitope-tagged Atf1. This Atf1-dependent complex was present in reactions using extracts derived from unstressed and stressed cells, indicating that at least under these experimental conditions Atf1 binds constitutively to this region of the cta3+ promoter. We were unable to properly assess the role of Tup proteins on DNA binding activity; when tup− extracts were used a marked reduction in the level of complex formation on both probes was observed. However, this seemed to be due to difficulties in preparing extracts from these cells rather than a specific effect because we found that extracts lacking Tup proteins also showed a reduced ability to form complexes on a DNA probe unrelated to the cta3+ promoter (our unpublished data).
Figure 3.
Complexes formed on the cta3+ promoter. (A) Schematic of the CRE-like elements in the cta3+ promoter and the probes used for EMSAs. Consensus or near consensus CRE elements are shaded black, and the CRE-like sequences containing an ACGT core are shaded gray. Their locations are given relative to the initiation codon. (B) EMSAs were performed with probe 1 by using whole cell extracts derived from exponentially growing atf1HA (KS14709), w.t. (NT4), or atf1Δ (NT147) cells as indicated above the lanes. The extract used in lane 3 was derived from atf1HA6H cells treated with KCl (0.6 M for 15 min) immediately before extract preparation. Lane 1 is a probe alone control. (C) EMSAs were performed with probe 2 by using extracts derived from exponentially growing cells (lanes 2, 4, and 5) or cells treated with KCl (0.6 M for 15 min) immediately before extract preparation (lane 3). The identity of the extract is indicated above the lane. The reaction in lane 4 contained 0.2 μg of HA antibody. Lane 1 is a probe alone control. Arrows mark the positions of the Atf1-dependent shifted (S) and supershifted (SS) complexes.
Tup11 and Tup12 Interact
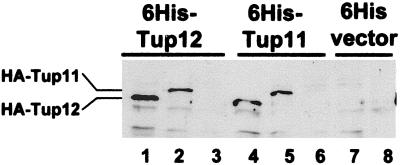
Our data indicate that Tup11 is capable of repressing cta3+ expression in the absence of Tup12 and vice versa. It is also known that S. cerevisiae Tup1 tetramerizes through its N-terminal domain (Varanasi et al., 1996; Jabet et al., 2000) and based on homology it is very likely that the S. pombe Tup proteins form homotetramers. However, it is possible that in addition to functioning in homomeric complexes Tup11 and Tup12 may also function in a heteromeric complex. Therefore, we investigated the ability of Tup11 and Tup12 to interact using a coprecipitation assay. Whole cell extracts were prepared from wild-type cells that expressed 6His-tagged Tup11 (or Tup12) and coexpressed HA-tagged Tup11 (or Tup12). Ni2+-agarose was then used to precipitate His-tagged Tup proteins, and the presence of HA-tagged Tup proteins was examined by Western blotting (Figure 4). In these experiments, Tup11 copurified with Tup12 and vice versa, indicating that Tup11 and Tup12 physically interact. The specificity of this interaction was demonstrated by the absence of HA-tagged Tup proteins in control precipitates derived from cells extracts expressing the empty 6His vector (Figure 4, lanes 7 and 8). Thus, Tup11 and Tup12 have the potential to regulate gene expression in the same protein complex.
Figure 4.
Tup11 and Tup12 coprecipitate. Whole cell extracts were prepared from wild-type cells containing plasmids expressing epitope-tagged Tup proteins: pRep42-HisMycTup12 (lanes 1–3), pRep42-HisMycTup11 (lanes 4–6), pRep42-HisMyc empty vector (lanes 7 and 8), pRep41-HATup12 (lanes 1, 4, and 7), and pRep41-HATup11 (lanes 2, 5, and 8). Extracts were precipitated with Ni2+-agarose analyzed on 8% SDS polyacrylamide gels and subjected to Western blotting by using HA monoclonal antibody.
Tup Repressors Link Transcriptional Induction to the Sty1 Pathway
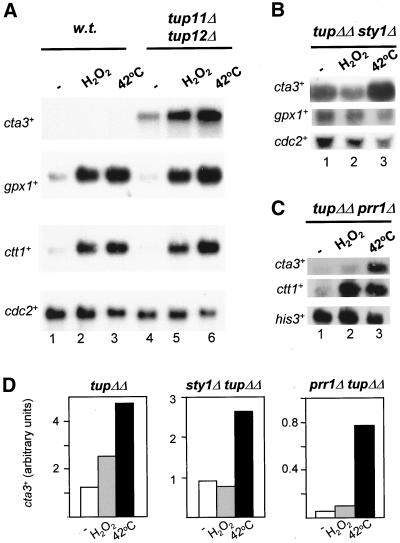
To test whether the high level of cta3+ expression observed in the absence of the Tup repressors was dependent upon the Sty1 MAP kinase cta3+ mRNA levels were examined in a strain that lacks both Sty1 and Tup function (sty1-1 tup11Δ tup12Δ). In this mutant the level of cta3+ transcripts was similar to that observed in tup11Δ tup12Δ cells, indicating that Sty1 is not required for basal levels of expression (Figure 5, A and B). Surprisingly, exposure of this strain to a KCl-mediated shock resulted in induction of cta3+ expression, indicating that in the absence of the Tup proteins the Sty1 MAP kinase is not required for the stress-mediated induction of cta3+. Consistent with these observations the expression of cta3+ was also induced by salt shock (0.6 M KCl) in a sty1Δ tup11Δ tup12Δ strain (our unpublished data). The expression of other genes such as pyp2+ and gpd1+ was not induced in the sty1-1 tup11Δ tup12Δ triple mutant, although deletion of the tup genes did restore the basal level of expression in sty1− cells (Figure 5A). The kinetics of KCl-mediated induction of cta3+ were similar in wild-type and tup11Δ tup12Δ cells, with mRNA levels peaking at 20 min but elevated mRNA levels persisted for a greater length of time in cells lacking the Tup proteins (Figure 5, C and D). In the sty1-1 tup11Δ tup12Δ triple mutant strain induction was delayed and peak mRNA levels were not observed until 30 min after the addition of KCl.
We next determined whether removal of Tup11 and Tup12 rescued any of the other phenotypes associated with loss of Sty1. We examined the ability of cells to grow on medium supplemented with cadmium. Deletion of the tup genes in a sty1+ background increases resistance to cadmium (Figure 1A) but unexpectedly deletion of tup11+ and tup12+ in a sty1-1 background reduced cadmium tolerance (Figure 6A). Thus, the resistance of tup− cells to cadmium depends on Sty1 function and in its absence they become hypersensitive. The elongated cell morphology of sty1-1 cells that is indicative of a G2 cell cycle delay was slightly exacerbated by deletion of the tup genes (Figure 6B). Furthermore, deletion of the tup genes in an aft1Δ or a sty1-1 background resulted in a small increase in sensitivity to KCl and the tup11Δ tup12Δ atf1Δ triple mutant strain was slightly less tolerant to CaCl2 than the parental strains (Figure 6C).
Figure 6.
Genetic interactions. (A) Deletion of the tup genes does not rescue phenotypes associated with the sty1-1 mutation. The indicated strains were subcultured onto YE5S agar (control) or subcultured onto YE5S agar supplemented with CdSO4 (to the indicated concentration) and incubated at 30°C for 3–4 d. (B) Comparison of the morphology of wild type (w.t.) (NT4), sty1-1 (JM1144), tup11Δ tup12Δ (SW76), and sty1-1 tup11Δ tup12Δ (SW90) cells. (C) Exponentially growing cultures were diluted serially, spotted onto YE5S agar or YE5S agar supplemented with CaCl2 or KCl (at the indicated concentration), and incubated for 2 d at 30°C. The strains used were wild type (w.t.) (NT4), tup11Δ tup12Δ (SW76), sty1-1 (JM1144), sty1-1 tup11Δ tup12Δ (SW90) atf1Δ (NT147), and atf1Δ tup11Δ tup12Δ (SW92).
We next addressed whether removal of Tup11-Tup12 repression rendered the induction of cta3+ independent of Atf1. Deletion of atf1+ in a tup11Δ tup12Δ mutant strain resulted in a further increase in cta3+ transcript levels in unstressed cells, suggesting that nonactivated Atf1 may have a repressive effect on transcription that is independent of Tup11 and Tup12 (Figure 7, A and B). A similar effect has been observed previously; the decrease in the basal level of ctt1+ mRNA associated with loss of Sty1 function is suppressed by deletion of atf1+ (Degols and Russell, 1997). In the atf1Δ tup11Δ tup12Δ background, cta3+ mRNA levels did not increase after exposure to KCl (0.6 M), indicating that Atf1 is absolutely required for induction of cta3+ in response to a salt shock.
The bZIP transcription factor Pcr1 that can heterodimerize with Atf1 (Kanoh et al., 1996) is also required for stress-mediated induction of cta3+ expression (Figure 7, C and D). Examination of cta3+ mRNA levels in a pcr1Δ tup11Δ tup12Δ strain revealed that Pcr1 is not required for the high level of basal expression, and furthermore in this strain expression of cta3+ was partially induced in response to a salt shock. Thus, the Tup repressors ensure that induction of cta3+ remains dependent upon the Sty1 MAP kinase and to a lesser extent, the activator Pcr1.
Prr1 Is Involved in Regulation of Gene Expression in Response to Elevated K+ Ions
Our analysis suggests that another factor may regulate transcription of cta3+ independently of Sty1. The Prr1 transcription factor is known to regulate oxidative stress responsive genes (Ohmiya et al., 1999a), but there is no evidence that Sty1 regulates its activity directly. Therefore, we analyzed mRNA levels in a prr1Δ strain and found that the level of cta3+ transcripts after exposure to KCl was significantly reduced in comparison with the wild-type strain (Figure 8, A and B). Furthermore, the influence of Prr1 was not confined to the cta3+ gene because KCl-mediated induction of both ctt1+ and gpx1+ expression was also significantly reduced in the prr1Δ mutant strain. This was surprising because Prr1 has previously been reported not to be involved in the transcriptional response to high salt (Ohmiya et al., 1999a) and indeed KCl-mediated induction of some genes such as gpd1+ occurs independently of Prr1 (Ohmiya et al., 1999a; Figure 8, A and B). To determine the role that Prr1 plays in control of cta3+ expression, we measured cta3+ mRNA levels in a tup11Δ tup12Δ prr1Δ triple mutant strain. In this background the expression of cta3+ was induced by exposure to high concentrations of KCl (0.6 M) (Figure 8, C and D). However, the deletion of prr1+ in a tup11Δ tup12Δ background resulted in a decrease in the basal level of cta3+ mRNA (Figure 8, C and D). Thus, Prr1 activity contributes to the high basal level of expression that is associated with loss of the Tup repressors.
In vitro experiments have demonstrated that recombinant Prr1 binds to a heat shock-like element in the ste11+ promoter (Ohmiya et al., 1999b). Analysis of the cta3+ promoter revealed the presence of such an element (GGAAAATTC) located at −2068 relative to the initiation codon. However, in assays using this region of the promoter and whole cell extracts we were unable to detect a Prr1-dependent binding activity (our unpublished data). Therefore, we cannot exclude the possibility that the role of Prr1 in regulation of cta3+ expression is indirect.
Tup11 and Tup12 Prevent Induction in Response to Inappropriate Stresses
Sty1, and thus in turn Atf1-Pcr1, is activated in response to a number of environmental insults. Accordingly, the expression of Atf1- and Pcr1-dependent genes such as ctt1+, pyp2+, and gpx1+ are induced in response to a variety of stresses such as UV irradiation, heat shock, and hyperosmolarity and an oxidative stress elicited by exposure to high concentrations of H2O2 (Shiozaki and Russell, 1996; Wilkinson et al., 1996; Degols and Russell, 1997; Nguyen et al., 2000; Quinn et al., 2002). However, some Atf1-Pcr1 target genes are induced only by a subset of these stresses. For example, we found that cta3+ expression was induced specifically in response to salt shock but not by oxidative stress (6 mM H2O2) or by heat shock (15 min at 42°C) (Figure 9, A and D). In contrast, ctt1+ and gpx1+ mRNA levels were both induced by these treatments, indicating that Atf1 (and Pcr1) were active under these conditions. This indicates that activation of the Sty1 pathway per se is not sufficient to induce the expression of cta3+ and mechanisms must exist to prevent induction of gene expression in response to such “inappropriate stresses.” We wanted to examine the possibility that Tup11 and Tup12 play a role in this process. Therefore, we measured the levels of cta3+ mRNA after exposing a tup11Δ tup12Δ strain to an oxidative stress (6 mM H2O2) and a heat shock (15 min at 42°C). In contrast to the wild-type strain, the expression of cta3+ was significantly induced by heat stress and by exposure to high levels of H2O2. In addition, the level of cta3+ transcripts was induced by hypotonic conditions in a tup11Δ tup12Δ strain but not in a wild-type strain (our unpublished data). We also examined cta3+ transcript levels in a sty1Δ tup11Δ tup12Δ triple mutant strain (Figure 9, B and D). This revealed that the induction in expression in response to heat shock was partly independent of the MAP kinase. In contrast, the induction of cta3+ expression in response to oxidative stress mediated by H2O2 was completely dependent upon Sty1, suggesting a difference in the mechanism of induction. Further analysis indicated that the response to heat shock was independent of Prr1 (Figure 9, C and D) but dependent upon Atf1 (our unpublished data). Taken together, these findings indicate that the Tup repressors function as part of the mechanism that ensures the specificity of stress-mediated transcriptional induction at the cta3+ promoter.
Figure 9.
Tup11 and Tup12 prevent induction in response to inappropriate stresses. (A) Strains used were wild type (w.t.) (NT4) and tup11Δ tup12Δ (SW76). Mid log cultures growing at 30°C in YE5S (lanes 1 and 4) were incubated with H2O2 (final concentration 6 mM) for 15 min (lanes 2 and 5) or shifted to 42°C for 15 min (lanes 3 and 6). Total RNA was prepared and subjected to Northern analysis with the indicated probes. The level of cdc2+ mRNA was used as a loading control. (B) Strain used was sty1Δ tup11Δ tup12Δ (SW91) and the treatments were as described in A. (C) Strain used was prr1Δ tup11Δ tup12Δ (SW91) and the treatments were as described in A. (D) Quantification of the cta3+ mRNA levels in A, B, and C. The strains are indicated above the graphs and the treatments are indicated below.
DISCUSSION
In this study, we reveal roles for S. pombe Tup11 and Tup12 in the cellular response to elevated K+ and Ca2+ levels. We identify cta3+ as a novel stress-induced gene whose transcription is coregulated by the Sty1 MAP kinase pathway and the Tup repressors. Our results indicate that the Tup repressors fulfill a number of functions in the control of cta3+ expression. First, they maintain low levels of basal expression and limit the level of induction. Second, they ensure that induction of expression is linked to the Sty1 pathway. And third, they maintain the specificity of induction. We also reveal a new role for the response regulator Prr1 and demonstrate that it functions to regulate gene expression in response to elevated salt concentrations. Prr1 is known to contribute to the regulation of several genes whose expression is induced by oxidative stress via the Pap1 transcription factor (Ohmiya et al., 1999a), and so Prr1 regulates gene expression in response to a number of stresses.
Tup11–Tup12 Interaction
Our data and that of others (Mukai et al., 1999; Janoo et al., 2001) suggest that Tup11 and Tup12 can function in homomeric complexes. In addition, we demonstrate that Tup11 and Tup12 have the potential to form a heteromeric complex. This is significant because full function requires both repressors; some derepression of a fbp1::lacZ reporter is observed upon deletion of a single tup gene (Janoo et al., 2001). Furthermore, single tup mutants have demonstrable phenotypes such as increased resistance to cadmium. Thus, regulation of some genes may depend upon both repressors and the formation of heteromeric Tup complexes.
Relief of Tup11-Tup12–mediated Repression
The Hog1 MAP kinase in S. cerevisiae plays a direct role in relieving Ssn6-Tup1–mediated repression at osmostress genes; Hog1 phosphorylates Sko1, reducing its affinity for the corepressor complex (Proft et al., 2001). It is possible that the Sty1 MAP kinase may similarly antagonize the action of Tup11-Tup12; however, our results demonstrate that the Atf1, Pcr1, and Prr1 transcription factors are required for relief from Tup-mediated repression at the cta3+ promoter. It is probable that the S. pombe Tup proteins function, at least in part, through the organization of repressive chromatin structures (Mukai et al., 1999), and therefore it is possible that Atf1-Pcr1 and Prr1 overcome this repression by recruiting positive-acting chromatin remodeling complexes such as Swi-Snf or histone acetylase complexes (HATs). In support of this, DNA binding by the Atf1-Pcr1 heterodimer is known to alter local nucleosome positioning at the ade6-M26 hotspot and thereby promote meiotic recombination (Mizuno et al., 1997; Kon et al., 1998). Moreover, genes such as SUC2 in S. cerevisiae are regulated by the interplay between Ssn6-Tup1 repression and Swi-Snf–mediated activation (Gavin and Simpson, 1997).
In S. cerevisiae, Hog1-dependent transcriptional induction of HAL1 requires the Gcn4 activator that relieves Tup1-Ssn6–mediated repression by competing with Sko1 for the occupancy of a single CRE binding site (Pascual-Ahuir et al., 2001). This CRE element functions as a dual control element and integrates both positive and negative regulatory signals. Furthermore, analysis of the S. pombe fbp1+ promoter, which is regulated by both Atf1-Pcr1 and Tup11-Tup12, has demonstrated the presence of a control element (called UAS2) that contains a CRE-like sequence and is bound by multiple activators and repressors (Neely and Hoffman, 2000). Interestingly, the Atf1-Pcr1 transcription factor does not bind to UAS2 directly, but it does influence the protein complexes that assemble on it (Neely and Hoffman 2000). The cta3+ promoter contains a number of CRE-like elements at least one of which mediates Atf1 binding (Figure 3C). It will be interesting to determine the contributions of these elements to activation and repression of cta3+ transcription.
Tup Repressors Maintain Specificity of Induction
The advent of stressful conditions results in the rapid and Sty1-dependent phosphorylation of Atf1 (Shiozaki and Russell, 1996; Wilkinson et al., 1996). Although the precise role of Atf1 phosphorylation remains obscure, it is evident that transcriptional activation by Atf1 is dependent upon Sty1. However, deletion of the tup genes allows transcriptional induction of cta3+ to occur in sty1− cells. Furthermore, in a tup11Δ tup12Δ mutant induction of cta3+ expression does not require Pcr1 or Prr1. Thus, the Tup repressors function to “wire” induction to the Sty1 pathway, insulating it from interfering signals. These results also suggest that Atf1 activity can be “uncoupled” from Sty1 in this specific case and that an additional mechanism for activating transcription that requires Atf1 exists. The finding that Prr1 also controls expression of cta3+ suggests that it may function as part of this mechanism.
The Sty1 pathway in S. pombe is fundamentally different to the Hog1 pathway in S. cerevisiae because it is triggered by exposure to a wide range of adverse environmental conditions. As a consequence, a large number of Sty1 target genes are up-regulated by multiple stresses. The products of such genes may comprise a set of “general stress response proteins” that are necessary because a single environmental insult may result in multiple classes of intracellular stress (Rep et al., 2001). Nonetheless, discrete stimuli also produce distinct transcriptional outputs, because there are subsets of Sty1-dependent genes, such as cta3+, that are induced only by specific stresses. A major question to be addressed is the mechanism by which Sty1 signaling is integrated into the regulation of such genes. Expression of cta3+ is not induced by oxidative stress, heat shock, carbon limitation, or sexual differentiation (Figure 6; our unpublished data), and furthermore cta3+ is only poorly induced by an osmotic shock mediated by high sorbitol concentrations (Nishikawa et al., 1999). Thus, the transcriptional response is triggered essentially by elevated intracellular cation concentrations rather than by an osmotic effect (i.e., decrease in turgor pressure across the plasma membrane). The cta3+ gene encodes a putative intracellular P-type ATPase transporter that is involved in cation extrusion or sequestration into intracellular compartments. Loss of function leads to an accumulation of cytoplasmic Ca2+ levels (Ghislain et al., 1990; Halachmi et al., 1992), although recent evidence suggests that Cta3 is primarily a K+ ion pump (Benito et al., 2002). It is thus consistent that it is salt stress that specifically that triggers its transcriptional induction. However, removal of the constraints imposed by Tup repressors allows cta3+ to be induced in response to other stresses such as elevated temperature and oxidative stress. Thus, the Tup repressors function as a part of a mechanism that adds specificity to Sty1-dependent transcriptional induction.
Our results also indicate that activation of the Sty1 pathway alone is insufficient to induce cta3+ expression and implies that an elevated cation concentration triggers an additional pathway that is required to circumvent repression (Figure 10). In this respect it may be significant that Prr1 is involved in the regulation of cta3+ expression because its structure suggests that it may be one part the target of a histidine-aspartate phosphorelay pathway. Recent work has identified several of these pathways in fission yeast (Nguyen et al., 2000; Buck et al., 2001), and current experiments are addressing its contribution to the regulation of Prr1 in the response to stress.
Figure 10.
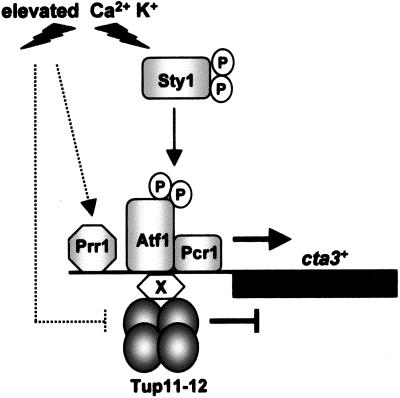
Model for the regulation of cta3+ expression. Under nonstress conditions, cta3+ expression is repressed by Tup11 and/or Tup12 that are tethered to the promoter through interaction with a site-specific DNA binding protein “X.” Activation of the Sty1 pathway alone is insufficient to induce expression, and the Tup repressors prevent activation by Atf1-Pcr1 and Prr1. Elevated Ca2+ or K+ concentrations trigger the activity of other pathways (indicated by dashed lines) that interfere with Tup repression and/or facilitate activation via the response regulator Prr1 and Atf1-Pcr1. In cells lacking the Tup repressors, specificity is lost and expression is induced in response to a range of stresses.
ACKNOWLEDGMENTS
We thank Janet Quinn, Mark Toone, and Burk Braun for advice on the manuscript. We also thank Burk Braun, Hirofumi Aiba, and Jonathan Millar for providing strains. This work was supported by a National Institutes of Health grant GM-46226 (to C.S.H.), by a Medical Research Council Ph.D. research studentship (to P.M.), a Medical Research Council Career Establishment Grant (to B.A.M.), and by a Biotechnology and Biological Sciences Research Council project grant (13/P11981) (to S.K.W.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–12–0568. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–12–0568.
REFERENCES
- Appleby JL, Parkinson JS, Bourret RB. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Baum B, Wuarin J, Nurse P. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 1997;16:4676–4688. doi: 10.1093/emboj/16.15.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito B, Garciadeblas B, Rodriguez-Navarro A. Potassium- or sodium-efflux ATPase, a key enzyme in the evolution of fungi. Microbiology. 2002;148:933–941. doi: 10.1099/00221287-148-4-933. [DOI] [PubMed] [Google Scholar]
- Bone JR, Roth SY. Recruitment of the yeast Tup1p-Ssn6p repressor is associated with localized decreases in histone acetylation. J Biol Chem. 2001;276:1808–1813. doi: 10.1074/jbc.M008668200. [DOI] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Brown JL, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck V, Quinn J, Soto Pino T, Martin H, Saldanha J, Makino K, Morgan BA, Millar JB. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol Biol Cell. 2001;12:407–419. doi: 10.1091/mbc.12.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Roth SY, Simpson RT. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- Craven RA, Griffiths DJF, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- Degols G, Russell P. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:3356–3363. doi: 10.1128/mcb.17.6.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Garcia-Gimeno MA, Struhl K. Aca1, and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon source utilization but not the response to stress. Mol Cell Biol. 2000;20:4340–4349. doi: 10.1128/mcb.20.12.4340-4349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin IM, Simpson RT. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Goffeau A, Halachmi D, Eilam Y. Calcium homeostasis and transport are affected by disruption of cta3, a novel gene encoding Ca2+-ATPase in Schizosaccharomyces pombe J. Biol Chem. 1990;265:18400–18407. [PubMed] [Google Scholar]
- Halachmi D, Ghislain M, Eilam Y. An intracellular ATP-dependent calcium pump within the yeast Schizosaccharomyces pombe, encoded by the gene cta3. Eur J Biochem. 1992;207:1003–1008. doi: 10.1111/j.1432-1033.1992.tb17136.x. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 1991;5:561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- Jabet C, Sprague ER, van Denmark AP, Wolberger C. Characterization of the N-terminal domain of the yeast transcriptional repressor Tup1. Proposal for an association model of the repressor complex Tup1 x Ssn6. J Biol Chem. 2000;275:9011–9018. doi: 10.1074/jbc.275.12.9011. [DOI] [PubMed] [Google Scholar]
- Janoo RTK, Neely LA, Braun BR, Whitehall SK, Hoffman CS. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors, and the CCAAT binding factor activation complex. Genetics. 2001;157:1205–1215. doi: 10.1093/genetics/157.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells. 1996;1:391–408. doi: 10.1046/j.1365-2443.1996.d01-247.x. [DOI] [PubMed] [Google Scholar]
- Kon N, Schroeder SC, Krawchuk MD, Wahls WP. Regulation of the Mts1-Mts2-dependent ade6–M26 meiotic recombination hot spot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol Cell Biol. 1998;18:7575–7583. doi: 10.1128/mcb.18.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez JA, Pascual-Ahuir A, Proft M, Serrano R. The Ssn6-Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J. 1998;17:2543–2553. doi: 10.1093/emboj/17.9.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Millar JBA, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. The meiotic recombination hot spot created by the single-base substitution ade6–M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mukai Y, Matsuo E, Roth SY, Harashima S. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol Cell Biol. 1999;19:8461–8468. doi: 10.1128/mcb.19.12.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely LA, Hoffman CS. Protein kinase A, and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol Cell Biol. 2000;20:6426–6434. doi: 10.1128/mcb.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AN, Lee A, Place W, Shiozaki K. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol Biol Cell. 2000;11:1169–1181. doi: 10.1091/mbc.11.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Aiba H, Mizuno T. The cta3+ gene that encodes a cation-transporting P-type ATPase is induced by salt stress under control of the Wis-Sty1 MAPKK-MAPK cascade in fission yeast. FEBS Lett. 1999;455:183–187. doi: 10.1016/s0014-5793(99)00876-5. [DOI] [PubMed] [Google Scholar]
- Ohmiya R, Kato C, Yamada H, Aiba H, Mizuno T. A fission yeast gene (prr1+) that encodes a response regulator implicated in oxidative stress response. J Biochem. 1999a;125:1061–1066. doi: 10.1093/oxfordjournals.jbchem.a022387. [DOI] [PubMed] [Google Scholar]
- Ohmiya R, Kato C, Yamada H, Aiba H, Mizuno T. Isolation of multicopy suppressors of the calcium sensitivity of a mutant lacking the bZIP transcription factor Atf1 in fission yeast. Mol Gen Genet. 1999b;261:297–306. doi: 10.1007/s004380050970. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Conlan RS, Gounalaki N, Copf T, Tzamarias D. Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J Biol Chem. 2000;275:8397–8403. doi: 10.1074/jbc.275.12.8397. [DOI] [PubMed] [Google Scholar]
- Pascual-Ahuir A, Serrano R, Proft M. The Sko1 repressor, and Gcn4 activator antagonistically modulate stress-regulated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:16–25. doi: 10.1128/MCB.21.1.16-25.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R, Posas F. Regulation of the Sko1 transcription repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001;20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Serrano R. Repressors and upstream repressing sequences of the stress regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J, Findlay VJ, Dawson K, Millar JBA, Jones N, Morgan BA, Toone WM. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in the fission yeast S. pombe. Mol Biol Cell. 2002;13:804–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd MJ, Arnaud MB, Johnson AD. A complex composed of Tup1 and Ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol. 2001;40:1067–1083. doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- Stettler S, Warbrick E, Prochnik S, Mackie S, Fantes P. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J Cell Sci. 1996;109:1927–1935. doi: 10.1242/jcs.109.7.1927. [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Cell cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Smith R, Johnson AD. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-liketranscription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- Toone WM, Jones N. Stress-activated signaling pathways in yeast. Genes Cells. 1998;3:485–498. doi: 10.1046/j.1365-2443.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase. StyI/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varanasi US, Klis M, Mikesell PB, Trumbly RJ. The Cyc8 (Ssn6)-Tup1 co-repressor is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol. 1996;16:6707–6714. doi: 10.1128/mcb.16.12.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol. 1996;16:704–711. doi: 10.1128/mcb.16.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AD, Edmondson DG, Bone JR, Mukai Y, Yu Y, Stillman DJ, Roth SY. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 2000;14:2737–2744. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JH, Barker DG, Nurse P, Johnston LH. Periodic transcription as a means of regulatory gene expression during the cell cycle, contrasting modes of expression of DNA ligase in budding and fission yeast. EMBO J. 1986;5:1705–1709. doi: 10.1002/j.1460-2075.1986.tb04414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehall S, Stacey P, Dawson K, Jones N. Cell cycle regulated transcription in fission yeast: Cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol Biol Cell. 1999;10:3705–3715. doi: 10.1091/mbc.10.11.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh J-C, Toda T, Millar JBA, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- Zaman Z, Ansari AZ, Koh SS, Young R, Ptashne M. Interaction of a transcriptional repressor with the RNA polymerase II holoenzyme plays a crucial role in repression. Proc Natl Acad Sci USA. 2001;98:2550–2554. doi: 10.1073/pnas.041611198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Takeda T, Nasmyth K, Jones N. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8:885–898. doi: 10.1101/gad.8.8.885. [DOI] [PubMed] [Google Scholar]