Abstract
Phosphatidylcholine and phosphatidylethanolamine are the two main phospholipids in eukaryotic cells comprising ∼50 and 25% of phospholipid mass, respectively. Phosphatidylcholine is synthesized almost exclusively through the CDP-choline pathway in essentially all mammalian cells. Phosphatidylethanolamine is synthesized through either the CDP-ethanolamine pathway or by the decarboxylation of phosphatidylserine, with the contribution of each pathway being cell type dependent. Two human genes, CEPT1 and CPT1, code for the total compliment of activities that directly synthesize phosphatidylcholine and phosphatidylethanolamine through the CDP-alcohol pathways. CEPT1 transfers a phosphobase from either CDP-choline or CDP-ethanolamine to diacylglycerol to synthesize both phosphatidylcholine and phosphatidylethanolamine, whereas CPT1 synthesizes phosphatidylcholine exclusively. We show through immunofluorescence that brefeldin A treatment relocalizes CPT1, but not CEPT1, implying CPT1 is found in the Golgi. A combination of coimmunofluorescence and subcellular fractionation experiments with various endoplasmic reticulum, Golgi, and nuclear markers confirmed that CPT1 was found in the Golgi and CEPT1 was found in both the endoplasmic reticulum and nuclear membranes. The rate-limiting step for phosphatidylcholine synthesis is catalyzed by the amphitropic CTP:phosphocholine cytidylyltransferase α, which is found in the nucleus in most cell types. CTP:phosphocholine cytidylyltransferase α is found immediately upstream cholinephosphotransferase, and it translocates from a soluble nuclear location to the nuclear membrane in response to activators of the CDP-choline pathway. Thus, substrate channeling of the CDP-choline produced by CTP:phosphocholine cytidylyltransferase α to nuclear located CEPT1 is the mechanism by which upregulation of the CDP-choline pathway increases de novo phosphatidylcholine biosynthesis. In addition, a series of CEPT1 site-directed mutants was generated that allowed for the assignment of specific amino acid residues as structural requirements that directly alter either phospholipid head group or fatty acyl composition. This pinpointed glycine 156 within the catalytic motif as being responsible for the dual CDP-alcohol specificity of CEPT1, whereas mutations within helix 214–228 allowed for the orientation of transmembrane helices surrounding the catalytic site to be definitively positioned.
INTRODUCTION
Phospholipids are the major components of cellular membranes. Phosphatidylcholine (PtdCho) and phosphatidylethanolamine (PtdEtn) are the two most abundant phospholipids present in eukaryotic cell membranes, comprising ∼50 and 25% of phospholipid mass, respectively (Raetz, 1986; Zinser et al., 1991; Schneiter et al., 1999). Both PtdCho and PtdEtn are actively metabolized through both agonist stimulated and constitutive processes to release a plethora of biologically active molecules, including arachidonic acid for the synthesis of the inflammatory mediators prostaglandins and leukotrienes, and diacylglycerol for the activation of signaling molecules, which include members of the protein kinase C family (Hodgkin et al., 1998; Parekh et al., 2000). In the face of this complex metabolic regulation the levels of PtdCho and PtdEtn remain essentially unchanged, because the cell is capable of responding to alterations in lipid catabolism through increases in synthesis and subsequent transport of lipids from their site of synthesis to the intracellular destination at which the lipid has been catabolized. Indeed, in any model of intracellular lipid homeostasis one must couple lipid metabolism at a particular site within the cell to increased de novo lipid synthesis and targeted transport of the lipid to the site of its catabolism. The regulation of lipid transport may overlap with vesicle trafficking processes; indeed, the major cellular phospholipid PtdCho is believed to be inhibitory to fission of vesicles from the Golgi (Xie et al., 2001), whereas its metabolites phosphatidic acid and diacylglycerol appear to be positive regulators of vesicle transport (Bi et al., 1997; Kearns et al., 1997; Schmidt et al., 1999; Weigert et al., 1999; Siddhanta et al., 2000; Henneberry et al., 2001; Bankaitis, 2002; Baron and Malhotra, 2002). Research into the transport of the major membrane lipids PtdCho and PtdEtn has been hampered by a lack of knowledge with regard to precise sites of synthesis of these lipids within the cell.
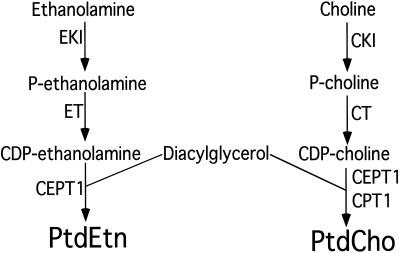
PtdCho is synthesized almost exclusively through the CDP-choline pathway in all mammalian cell types (except the liver; Figure 1; Cui et al., 1993; Kent, 1995; Walkey et al., 1998). The first step in the synthesis of PtdCho through the CDP-choline pathway is the phosphorylation of choline by a cytoplasmic choline kinase to form phosphocholine. In the second step, CMP is transferred from CTP to phosphocholine to form CDP-choline by the rate-limiting CTP:phosphocholine cytidylyltransferase (CT). There are two isoforms of CT found in mammalian cells, with CTα being ubiquitous and likely the major contributor to PtdCho synthesis in most cell types, whereas the second CT isoform, CTβ, is found in a much more restricted tissue distribution. (Wang et al., 1993; Kent, 1995; Wang et al., 1995; Lykidis et al., 1998; Northwood et al., 1999; Cornell and Northwood, 2000; DeLong et al., 2000; Ridsdale et al., 2001). CTα is directed to the nucleus in most mammalian cell types including the Chinese hamster ovary cells (CHO) cells used in this study by a basic motif found in its N-terminal region, whereas CTβ is extranuclear in all cell types thus far examined (Wang et al., 1993, 1995; Lykidis et al., 1998; Northwood et al., 1999; DeLong et al., 2000). The final step in the synthesis of PtdCho is catalyzed by a cholinephosphotransferase activity that transfers phosphocholine from CDP-choline to diacylglycerol to form PtdCho (Weiss et al., 1958; Hjelmstad and Bell, 1987, 1988; McMaster and Bell, 1994a; Williams and McMaster, 1998; Henneberry and McMaster, 1999; Mancini et al., 1999; Henneberry et al., 2000). The intracellular location of the cholinephosphotransferase reaction defines the site of PtdCho synthesis.
Figure 1.
Synthesis of PtdCho and PtdEtn by the CDP-alcohol pathways. The gene names of the enzymes that catalyze each step are indicated. EKI, ethanolamine kinase; ET CTP:phosphocholine cytidylyltransferase; CEPT1, choline/ethanolaminephosphotransferase; CKI, choline kinase; CT, CTP:phosphocholine cytidylyltransferase; CPT1, cholinephosphotransferase.
PtdEtn is synthesized through an analogous pathway using a soluble ethanolamine kinase to produce phosphoethanolamine, which is converted by an endoplasmic reticulum bound CTP:phosphoethanolamine cytidylyltransferase to CDP-ethanolamine. Phosphoethanolamine is transferred from CDP-ethanolamine to diacylglycerol by an ethanolaminephosphotransferase activity of unknown intracellular location to produce PtdEtn. (Hjelmstad and Bell, 1990, 1991a, 1991b; Kent, 1995; Henneberry and McMaster, 1999; Birner et al., 2001). PtdEtn can also be synthesized by the decarboxylation of phosphatidylserine with the contribution of the CDP-ethanolamine versus phosphatidylserine decarboxylation pathways being cell type dependent (Voelker, 1984; McMaster and Choy 1992; Shiao et al., 1995). Our laboratory recently identified the first mammalian cholinephosphotransferase and ethanolaminephosphotransferase encoding cDNAs (Henneberry and McMaster, 1999; Henneberry et al., 2000). CPT1 codes for a CDP-choline specific cholinephosphotransferase for the exclusive synthesis of PtdCho, whereas CEPT1 codes for a dual specificity choline/ethanolaminephosphotransferase that can use both CDP-choline and CDP-ethanolamine as substrates to synthesize both PtdCho and PtdEtn. An analysis of the human genome and those of other eukaryotic cells indicates that these two genes likely code for the entire set of cholinephosphotransferases and ethanolaminephosphotransferases in mammalian cells. Genetic inactivation of the analogous genes in yeast resulted in complete loss of both enzyme activities in vitro and an inability to metabolically reconstitute lipid synthesis by either route in vivo (McMaster and Bell, 1994a, 1994b; Hjelmstad and Bell, 1991a; Hjelmstad et al.; 1994). Human CEPT1 and CPT1, by virtue of their choice in diacylglycerol fatty acyl and CDP-alcohol species, affect the form and function of PtdCho and PtdEtn (Samborski et al., 1990; DeLong et al., 1999; Hunt et al., 2001; Jansen et al., 2001). In this study we have pinpointed the sites of PtdCho and PtdEtn synthesis in mammalian cells. We have also taken advantage of the dual CDP-alcohol specificity of CEPT1 to identify amino acid residues that determine its ability to synthesize PtdCho versus PtdEtn and have identified amino acid residues that alter its diacylglycerol fatty acyl species preference.
MATERIALS AND METHODS
Materials
[Methyl-14C]Cytidine 5′-diphosphocholine and [ethanolamine 1,2-14C]cytidine 5′-diphosphoethanolamine were purchased from American Radiolabeled Chemicals (St. Louis, MO). [Methyl-14C]-choline chloride was purchased from NEN Life Science Products (Boston, MA). [Ethanolamine 1,2-14C]-ethanolamine hydrochloride was purchased from ICN (Costa Mesa, CA). All materials used in the preparation of bacterial and yeast media were purchased from Difco Laboratories (Detroit, MI). Lipids were purchased from Avanti Polar Lipids (Birmingham, AL). The T7 mouse mAb and T7 mouse mAb conjugated to horseradish peroxidase were purchased from Novagen (Madison, WI). The anticalnexin rabbit polyclonal antibody was a product of StressGen Biotechnologies Corp (Victoria, British Columbia, Canada). The Lap-2 mouse mAb was purchased from Transduction Laboratories (Lexington, KY). The CTα rabbit polyclonal antibody was a gift from Dr. Martin Post (Hospital for Sick Children, Toronto, Canada). FITC-labeled Lens culinaris (LcH) lectin was purchased from Sigma (St. Louis, MO). Goat anti-mouse Texas Red, goat anti-rabbit Texas Red, goat anti-mouse FITC, and goat anti-rabbit FITC secondary antibodies were products of Molecular Probes (Eugene, OR).
Generation of Constructs for Expression in Mammalian Cells
Human CEPT1 was amplified by PCR from our original cDNA using the oligonucleotide primers, 5′-GCGGGATCCATGAGTGGGCATCGATCAACA-3′ (forward) and 5′-GCGGTCGACTTAATGATTAGAATGAGCTGT-3′ (reverse), which contain BamHI and SalI restriction sites, respectively. The PCR product was subcloned into the pCR2.1-Topo vector (Invitrogen, Carlsbad, CA), excised with BamHI and SalI and subcloned into the Escherichia coli expression vector pET23a (Novagen), resulting in the addition of an 11-residue, T7 epitope tag to the N-terminus of the protein. The T7-tagged version of CEPT1 was excised using BglII and SalI and subcloned into the BamHI and SalI sites of the mammalian expression vector pcDNA3 (Invitrogen). GFP was added to the C terminus of T7-tagged CEPT1 by amplification of CEPT1 by PCR using the oligonucleotide primers, 5′-GCAAGATCTATGGCTAGCATGACTGGTGGA-3′ (forward) and 5′-GCGCAGAATTCGATGATGATTAGAATGAGC-3′ (reverse), which have BglII and EcoRI restriction sites, respectively, built into the primers, and subcloned into the BglII and EcoRI sites of the mammalian expression vector pEGFP-N1 (Clontech, Palo Alto, CA). Expression of this construct in mammalian cells results in the production of a CEPT1 protein with an N-terminal T7 epitope tag and a C-terminal green fluorescent protein (GFP).
The open reading frame of human CPT1 was amplified by PCR using the oligonucleotide primers 5′-GCCAGATCTATGGCGGCGGCGCCGGGGCC-3′ (forward) and 5′-GCCGTCGACTCAATCCATGTTATTCTGATG-3′ (reverse), which have BglII and SalI restriction sites, respectively, and subcloning into the TA cloning into the pCR2.1-TOPO (Invitrogen). The BglII/SalI fragment was excised and subcloned into the BamHI and SalI sites of the pET23a vector, resulting in the addition of a T7 epitope tag to the N-terminus of CPT1. The T7-tagged version of CPT1 was digested with BglII and NotI and subcloned into the BamHI and NotI restriction sites of pcDNA3 (Invitrogen). The DNA sequence of all PCR-amplified constructs was confirmed by sequencing both DNA strands.
Site-directed mutants were made using the MORPH site-directed mutagenesis kit (5 Prime 3 Prime, Inc., West Chester, PA). Mutagenic oligonucleotides were 5′-phosphorylated and purchased from Life Technologies (Rockville, MD). Mutations were confirmed by manual dideoxy sequencing using the T7sequencing kit (Amersham Pharmacia Biotech, Piscataway, NJ).
Cell Culture
CHO-K1 cells were transiently transfected using LIPOFECTAMINE Reagent (Life Technologies) in DMEM containing 5% (vol/vol) fetal calf serum and 33 μg/ml proline and maintained at 37°C in an atmosphere of 5% CO2.
The calcium chloride method of cell transfection was used for the preparation of stable cell lines in 100-mm dishes. Transfected cells were incubated overnight at 37°C in DMEM containing 5% (vol/vol) fetal calf serum and 33 μg/ml proline. Medium was replaced with 8 ml of CHO-K1 media containing 500 μg/ml G418 to start selection of stably transfected clones. Subsequently, the medium was replaced every 2–3 d with 8 ml of fresh CHO-K1 medium containing 500 μg/ml G418 to select for clones that were stably transfected with the desired plasmid. Single colonies of stably transfected CHO-K1 cells were selected by dilution cloning. Positive colonies were initially identified by Western blotting with the T7-horseradish peroxidase–conjugated antibody and their homogeneity confirmed by immunofluorescence.
Immunofluorescence
All cells used for immunofluorescence were grown on glass coverslips in 60-mm dishes at a density of 2 × 105 cells per dish. Cells were fixed in 3% (vol/vol) formaldehyde in 10 mM sodium phosphate (pH 7.4), 225 mM NaCl, 2 mM MgCl2 (PBS-B) for 15 min at room temperature. The coverslips were washed twice at room temperature with PBS-B containing 5 mM ammonium chloride for 5 min each wash, with PBS-B for 5 min, and then permeabilized by adding PBS-B containing 0.05% Triton X-100 and incubating at 40°C for 15 min and then room temperature for 15 min. The coverslips were washed twice with PBS-B containing 1% fatty acid–free BSA (PBS/BSA) for 5 min per wash, once for 15 min, and treated with primary antibody in PBS/BSA for 1 h at room temperature. The coverslips were washed twice with PBS/BSA for 5 min each. LcH-lectin (which was conjugated directly to FITC) and secondary antibody was added in PBS/BSA, and cells were incubated for 1 h at room temperature and washed twice with PBS/BSA for 5 min each, and the coverslips were mounted on slides with 2.5% (wt/vol) 1,4-diazadicyclo-[2,2,2]-octane in 50 mM Tris-HCl (pH 9.0) and 90% (vol/vol) glycerol. T7 antibodies were used at 1:1000 dilution, CTα antibodies at 1:4000, and calnexin antibodies at 1:200. Secondary antibodies and FITC-coupled LcH-lectin were used at a 1:4000 dilution. Very faint intranuclear staining was observed with the T7 mAb in mock-transfected cells.
To stain the mitochondria, cells were grown on glass coverslips as described above. Before fixing, the medium was aspirated from the cells and replaced with fresh CHO-K1 medium containing 200 nM MitoTracker Red CMXRos (Molecular Probes). The cells were incubated with the dye for 45 min at 37°C and washed twice with warm PBS. Cells were fixed and mounted as described above.
Cells transiently transfected with GFP-fusion and YFP-fusion proteins were grown on glass coverslips, washed three times with 58 mM Na2HPO4, 17 mM NaH2PO4, 68 mM NaCl (PBS-C, pH 7.4), and either viewed directly or fixed by incubating with PBS-C/4% formaldehyde for 30 min at room temperature, washed twice with PBS-C, and mounted as described above. The YFP-Golgi marker is a fusion protein consisting of enhanced yellow fluorescent protein and the sequence encoding the N-terminal 81 amino acids of human β-1,4-galactosyltransferase (Clontech).
Fluorescence microscopy was performed using a Zeiss axiophot microscope (Thornwood, NY). Green and yellow fluorescent proteins and FITC-coupled antibodies were visualized with Zeiss filter number 10, which excites at 450/490 and emits at 550/565. Texas Red and MitoTracker were visualized with Zeiss filter number 15, which excites at 546/560 and emits at 590.
Nuclear Fraction Isolation
NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Pierce, Rockford, IL) was used to isolate nuclei. We seeded 5 × 105 CHO cells in a 100-mm plate containing DMEM with 5% (vol/vol) fetal calf serum and 33 μg/ml proline and maintained at 37°C. The cells were maintained in an atmosphere of 5% CO2 for 4 d. The plates were placed on ice, and the medium was aspirated. The cells were scraped into 1 ml ice-cold PBS and spun at 500 × g at 4°C for 3 min. The PBS was aspirated, and the cell pellet was resuspended in 200 μl cytoplasmic extraction reagent I. The sample was vortexed at the highest setting for 15 s to fully resuspend the cell pellet. The sample was incubated on ice for 10 min, and 11 μl cytoplasmic extraction reagent II was added. The sample was vortexed 5 s and incubated on ice for 1 min. The sample was vortexed 5 s on the highest setting and then spun at 16,000 × g and 4°C for 5 min. The supernatant was transferred to a new, precooled tube and mixed with an equal volume of SDS-loading buffer. The pellet was resuspended in 100 μl nuclear extraction reagent. The sample was vortexed for 15 s and incubated on ice for 10 min. This was repeated for a total of 40 min. The sample was spun at 16,000 × g and 4°C for 10 min. The supernatant was transferred to a new, precooled tube and then mixed with an equal volume of SDS-loading buffer. Samples were stored at −20°C. A 20-μl sample of nuclear and extranuclear extracts was loaded and proteins separated by SDS-PAGE and identified by Western blot.
Western Blot Analysis
HJ091 Saccharomyces cerevisiae cells (a his3-Δ1 leu2-3 leu2-112 ura3-52 trp1–289 cpt1::LEU2 ept1−) were grown to midlog phase in synthetic dextrose medium containing the appropriate nutrients to ensure plasmid maintenance (Kaiser et al., 1994), and microsomal membranes were prepared as described (Williams and McMaster, 1998). Protein extracts were incubated with SDS-PAGE sample buffer at 37°C for 30 min, separated on a 12% SDS-polyacrylamide gel, and transferred to polyvinylidene difluoride membranes. Blots were probed with a T7 epitope tag-specific mAb coupled directly to horseradish peroxidase (1:5000, Novagen) for subsequent detection using the ECL system (Amersham Pharmacia Biotech).
Enzyme Assays
To determine in vitro choline- and ethanolaminephosphotransferase enzyme activities of wild-type and site-directed CEPT1 mutants the microsomal membranes were isolated from HJ091 yeast (cpt1::LEU2 ept1−) grown to midlog phase in synthetic dextrose media containing the appropriate nutrients to ensure plasmid maintenance of wild-type or mutant versions of CEPT1 from the constitutive glyceraldehyde-3-phosphate dehydrogenase promoter in the expression vector p416GPD (Munberg et al., 1995). A mixed micelle assay was used as previously described (Henneberry et al., 2001; Wright et al., 2001). All assays were performed at least three times in duplicate and the indicated values represent their mean. SEs were <15% of the mean for each experiment.
Cholinephosphotransferase and ethanolaminephosphotransferase activities in CHO-K1 cell lines and CHO-K1 cells transiently transfected with CEPT1-GFP were determined after placing the cells on ice and washing them twice with ice-cold PBS. The cells were scraped into a microfuge and centrifuged at 15,000 × g for 5 min at 4°C. The cell pellet was resuspended in 0.5 ml of 10 mM HEPES-HCl (pH 7.4), 50 mM KCl, 1 mM EDTA, and Complete protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN)) and passed through a 23-gauge needle 20 times to lyse the cells. The mixture was centrifuged at 15,000 × g for 30 s at 4°C, and the supernatant was centrifuged at 450,000 × g for 15 min at 4°C to pellet cellular membranes. Membranes were resuspended in 10 mM HEPES-HCl (pH 7.4), 50 mM KCl, 1 mM EDTA, and Complete protease inhibitor cocktail (Roche Molecular Biochemicals) by using a Teflon pestle and stored at −70°C. The mixed micelle assay for cholinephosphotransferase activity was used as previously described (Henneberry et al., 2001; Wright et al., 2001).
Metabolic Labeling
S. cerevisiae HJ091 cells (cpt1::LEU2 ept1−) transformed with either wild-type or mutant versions of CEPT1 in the constitutive expression vector p416GPD were grown to midlog phase in synthetic dextrose media containing the appropriate nutrients to ensure plasmid maintenance (Kaiser et al., 1994). [14C]Choline (10 μM, 1 × 105 dpm/nmol) or [14C]ethanolamine (6.7 μM, 2.2 × 105 dpm/nmol) was added to the cultures for 1 h at 30°C. Cells were then concentrated by centrifugation and incorporation of radiolabel into lipids was performed as described (Henneberry et al., 2001). All labeling experiments were performed at least three times in duplicate and the values indicated represent their mean. SEs were <15% of the mean for each experiment.
Protein and Lipid Determination
Protein was determined by the method of Lowry et al. (1951) using bovine serum albumin as standard. Phospholipid phosphorus was determined by the method of Ames and Dubin (1960).
RESULTS
Sites of PtdCho and PtdEtn Synthesis
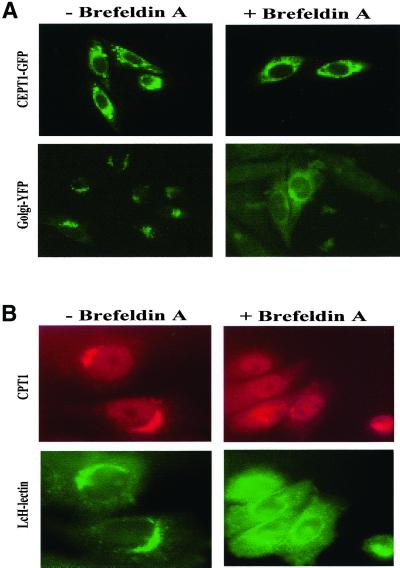
The CEPT1 open reading frame was fused with that of the GFP, and this construct was transiently transfected into Chinese hamster ovary (CHO-K1) cells. The CEPT1-GFP chimeric protein was functional as cells transiently transfected with CEPT1-GFP at 20% efficiency had a fourfold increase in cholinephosphotransferase activity. Indeed, transfection efficiency of CEPT1-GFP essentially mirrored the increase in cholinephosphotransferase activity, implying that all of the CEPT1-GFP fusion protein was enzymatically active (our unpublished results). As an initial step in determining the intracellular site of CEPT1 we treated the cells with brefeldin A, which essentially collapses the Golgi apparatus into the endoplasmic reticulum, resulting in a dramatic relocalization of most Golgi resident proteins. Brefeldin A treatment did not result in the relocalization of CEPT1, whereas the Golgi marker was effectively relocalized (Figure 2A). Similar results were observed whether live cells were viewed or if cells were fixed before microscopy (our unpublished results).
Figure 2.
Effect of brefeldin A on CEPT1 and CPT1 subcellular localization. (A) CHO-K1 cells transiently transfected with T7-CEPT1-GFP or the first 81 amino of the Golgi resident β-1,4-galactosyltransferase fused to yellow fluorescent protein were treated with 2 μg/ml brefeldin A for 30 min. Live and fixed cells resulted in identical images. (B) CHO-K1 cells stably expressing T7-CPT1 were treated with brefeldin A and T7 monoclonal antibodies followed by Texas Red–conjugated secondary antibodies were used to determine the location of CPT1. The location of the Golgi was determined by the addition of FITC-coupled L. culinaris lectin.
To further define the intracellular site of CEPT1 and CPT1, stable CHO-K1 cell lines expressing T7-epitope–tagged CEPT1 and CPT1 were constructed. These same T7-tagged CEPT1 and CPT1 proteins expressed in yeast devoid of their endogenous cholinephosphotransferase and ethanolaminephosphotransferase activities were previously used to characterize the substrate specificity of the CEPT1 and CPT1 enzymes in vitro by enzyme assay and in vivo through metabolic labeling experiments (Henneberry and McMaster, 1999; Henneberry et al., 2000). In addition, increased expression of T7-tagged CEPT1 was demonstrated to prevent farnesol induced inhibition of cholinephosphotransferase activity in CHO-K1 cells, once again demonstrating that the tagged versions of these enzymes are active and can reconstitute the CDP-alcohol pathways in a variety of eukaryotic cell types, indicating they are properly localized (Wright et al., 2001). We isolated five CHO-K1 cell lines expressing T7-tagged CEPT1 or CPT1 that possessed barely perceptible increases in CEPT1 and CPT1 enzyme activity compared with those with up to fivefold increased expression of CPT1 or CEPT1. We found that the level of expression of CPT1 and CEPT1 did not alter their intracellular locations (our unpublished results), and representative images of cell lines with an estimated twofold increase in CEPT1 and CPT1 expression are presented (Figures 2 and 3).
Figure 3.
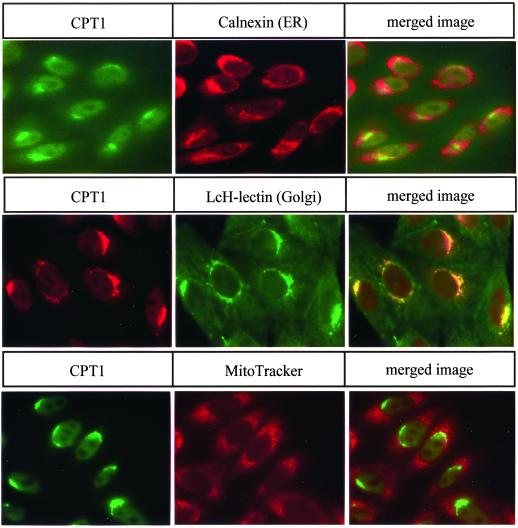
Intracellular localization of CPT1. The intracellular location of CPT1 was determined in CHO-K1 cells stably expressing T7-CPT1 using T7 monoclonal primary antibodies. The location of the endoplasmic reticulum was assessed using primary antibodies to calnexin. Golgi location was determined by the addition of FITC-coupled L. culinaris lectin. MitoTracker dye was used to determine the location of the mitochondria. Secondary antibodies were coupled to either FITC or Texas Red. The yellow color in merged images indicates overlap between CPT1 and the organelle marker.
Brefeldin A treatment of CHO cells expressing T7-CPT1 resulted in a redistribution of CPT1 from a large punctate region to a more diffuse region (Figure 2B). This implies that CPT1 synthesizes PtdCho in the Golgi apparatus.
Coimmunofluorescence of CPT1 and CEPT1 with organelle-specific markers was performed. CPT1 colocalized with the Golgi-specific L. culinaris lectin (Ridgway et al., 1992) and was independent of the endoplasmic reticulum and mitochondrial markers (Figure 3). This was consistent with the redistribution of CPT1 in response to brefeldin A treatment. The CEPT1 protein colocalized with the endoplasmic reticulum marker calnexin and was not found colocalized with the Golgi or mitochondrial markers (Figure 4). The CEPT1-GFP chimera was also found to colocalize with the endoplasmic reticulum resident protein disulfide isomerase (our unpublished results).
Figure 4.
Intracellular localization of CEPT1. The intracellular location of CEPT1 was determined in CHO-K1 cells stably expressing T7-CEPT1 using T7 monoclonal primary antibodies. The location of the endoplasmic reticulum was assessed using primary antibodies to calnexin. Golgi location was determined by the addition of FITC coupled L. culinaris lectin. MitoTracker dye was used to determine the location of the mitochondria. Secondary antibodies were coupled to either FITC or Texas Red. The yellow color in merged images indicates overlap between CEPT1 and the organelle marker.
Reconstitution of the PtdCho Biosynthetic Pathway
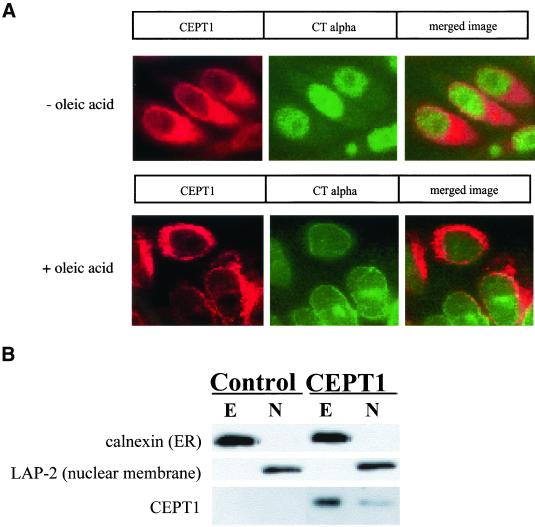
Closer observation of the merged CEPT1/calnexin image resulted in our observance of a small region around the nuclear membrane that did not appear to completely merge with the endoplasmic reticulum marker. The synthesis of CDP-choline for consumption by cholinephosphotransferase is the rate-limiting step in the synthesis of PtdCho and is catalyzed by a pair of CTP:phosphocholine cytidylyltransferase (CT) enzymes. CTα is the main isoform present in most cell types including CHO cells (Wang et al., 1995; Lykidis et al., 1998; Attard et al., 2000; Cornell and Northwood, 2000), and upon activation of PtdCho synthesis (e.g., through fatty acid supplementation) CTα translocates to the nuclear membrane (Watkins and Kent, 1992; Wang et al., 1993, 1995; Northwood et al., 1999; Cornell and Northwood, 2000; DeLong et al., 2000; Ridsdale et al., 2001). In our hands, activation of CTα also resulted in its colocalization to the nuclear membrane, and this overlapped with the portion of CEPT1 that did not colocalize with the endoplasmic reticulum (Figure 5A). To ensure that a portion of CEPT1 was indeed associated with the nuclear membrane, we separated CHO-K1 cellular nuclei from endoplasmic reticulum by subcellular fractionation. The nuclei are clearly separated from the endoplasmic reticulum because the endoplasmic reticulum marker calnexin is found exclusively in the extranuclear fraction, whereas the nuclear membrane marker Lap-2 is found exclusively in the nuclear fraction. Similar to our coimmunofluorescence experiments, the bulk of CEPT1 was associated with the extranuclear fraction, whereas a smaller proportion was found in the nucleus. These experiments demonstrate that CEPT1 resides in both the endoplasmic reticulum and nuclear membranes, and increasing PtdCho synthesis via CTα translocation is by redistribution of this rate-limiting enzyme to nuclear located CEPT1. However, whether CEPT1 and CTα reside within the same nuclear membrane and/or directly interact will require further experimentation.
Figure 5.
PtdCho synthesis pathway reconstitution at the nuclear membrane. (A) CHO-K1 stably expressing T7-CEPT1 were treated with oleic acid (500 μM in 0.5% bovine serum albumin) for 24 h to translocate CTα from its inactive soluble intranuclear location to its active nuclear membrane location before immunofluorescence. (B) Nuclei (N) were separated from extranuclear structures including the endoplasmic reticulum (E) by subcellular fractionation as described in MATERIALS AND METHODS, and the distribution of nuclear and endoplasmic reticulum markers are compared with CEPT1.
Structure/Function Analysis of CEPT1
The first genes identified to code for cholinephosphotransferase and ethanolaminephosphotransferase activities were isolated from the yeast S. cerevisiae (Hjelmstad and Bell 1987, 1988, 1990, 1991a). The yeast CPT1 gene product utilizes CDP-choline in vitro and in vivo, whereas the EPT1 gene product can use both CDP-choline and CDP-ethanolamine (Hjelmstad and Bell 1990, 1991a, 1991b; Hjelmstad et al., 1994; McGee et al., 1994a, 1994b; McMaster and Bell, 1994a, 1994b; Henneberry et al., 2001). Genetic inactivation of the CPT1 and EPT1 genes resulted in the complete loss of measurable cholinephosphotransferase and ethanolaminephosphotransferase in vitro enzyme activity and an inability to incorporate specific radiolabeled precursors for each pathway into either PtdCho or PtdEtn (Hjelmstad and Bell, 1991b; Hjelmstad et al., 1994; McMaster and Bell, 1994a, 1994b; Henneberry et al., 2001). Inactivation of the both CDP-alcohol pathways in most cell types would be lethal; however, yeast cells are still viable because they can de novo synthesize phosphatidylserine, which can be decarboxylated to PtdEtn and methylated to PtdCho (Paltauf et al., 1992; Birner et al., 2001). The ability of yeast to survive in the absence of functional CDP-choline and CDP-ethanolamine pathways for PtdCho and PtdEtn synthesis allowed for their use as a null expression system for structure/function analysis of human CEPT1.
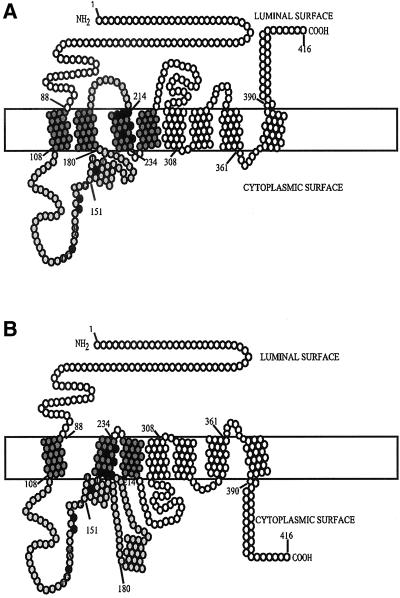
Extrapolation of previous chimeric enzyme analysis of the yeast Cpt1p and Ept1p enzymes to human CEPT1 positions the outside limit of the CDP-alcohol–binding region to amino acid residues 88–213, whereas the diacylglycerol binding site is located to the transmembrane helices spanning residues 88–258 (Figure 6; Hjelmstad et al., 1994; McMaster and Bell, 1994a). Within this region is a CDP-alcohol phosphotransferase motif, DG(x)2AR(x)8G(x)3D(x)3D, that runs from residues 136–158 in human CEPT1. The final two aspartates within this motif are required for catalysis with the remainder of the conserved residues responsible for substrate affinity or steric stability (Williams and McMaster, 1998). Secondary structure predictions and hydropathy plots weave either three of four membrane-spanning domains through the substrate binding region of the yeast and human cholinephosphotransferases and choline/ethanolaminephosphotransferases (Figure 6). Amino acids residues within the CDP-alcohol phosphotransferase motif were changed from those found in the dual specificity human CEPT1 and yeast Ept1p to those found in CDP-choline–restricted human CPT1 and yeast Cpt1p (Figure 7A) to determine if the divergent residues within this region impart CDP-alcohol specificity.
Figure 6.
Predicted secondary structures for human CEPT1. Seven or eight membrane spans are strongly predicted with helix 181–199 positioned either in (A) or outside (B) of the membrane using the SMART or TmPred algorithms. The numbers denote amino acid residues with the black filled circles representing those mutated in the current study. Based on comparisons to studies on the yeast Cpt1p and Ept1p enzymes, the dark gray circles represent the maximum region required for diacylglycerol binding, and the light gray the maximum region required for CDP-alcohol binding. The conserved residues within the CDP-alcohol phosphotransferase motif, DG(x)2AR(x)8G(x)3D(x)3D, are also indicated.
Figure 7.
CEPT1 CDP-alcohol phosphotransferase motif mutants. (A) The site-directed mutations made in CEPT1 are indicated. The numbers denote CEPT1 amino acid residues. (B) Western blot of wild-type and mutant versions of CEPT1.
CEPT1 CDP-Alcohol Site-directed Mutants
Plasmids carrying wild-type and site-directed mutants of CEPT1 were transformed into S. cerevisiae HJ091, a yeast strain that is devoid of cholinephosphotransferase and ethanolaminephosphotransferase activities due to inactivating mutations within its endogenous CPT1 and EPT1 genes (cpt1::LEU2 ept1−; Hjelmstad and Bell, 1991a; Hjelmstad et al., 1994; McMaster and Bell, 1994a). This expression system ensures that any cholinephosphotransferase or ethanolaminephosphotransferase activity detected would be plasmid encoded. Western blot analysis of HJ091 membrane fractions expressing the wild-type and mutant versions of CEPT1 established that similar amounts of full-length enzyme were being expressed (Figure 7B).
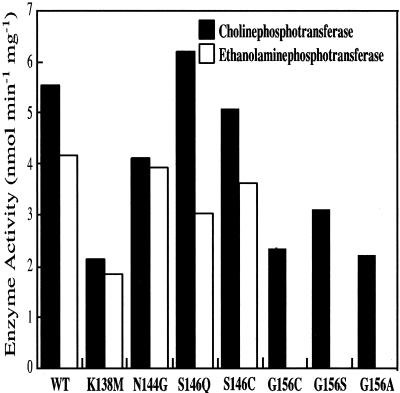
The CDP-alcohol specificities of CEPT1 and the CDP-alcohol phosphotransferase motif mutants were determined by in vitro enzyme assay. There were very small differences in enzyme activity or CDP-alcohol specificity with the CEPT1 N144G, S146Q, or S146C mutations compared with the wild-type enzyme (Figure 8). A decrease in both cholinephosphotransferase and ethanolaminephosphotransferase activity to ∼50% wild-type activity was seen in the K138 M mutant; however, this demonstrates an overall effect on activity but not substrate specificity because use of both CDP-choline and CDP-ethanolamine as substrates was equally affected. Mutation of glycine 156 to either alanine, serine, or cysteine also decreased cholinephosphotransferase activity to 50% wild-type, but more importantly abolished the ability of CEPT1 to utilize CDP-ethanolamine as a substrate (Figure 8). Increasing the specific radioactivity of the CDP-ethanolamine substrate to values 10-fold normally used still did not allow for detectable enzyme activity. This indicates a specific role for glycine 156 in CDP-ethanolamine binding by CEPT1.
Figure 8.
Enzyme activity of CEPT1 CDP-alcohol phosphotransferase motif mutants. Enzyme activities were determined from microsomal membrane preparations of S. cerevisiae cells (cpt1:: LEU2 ept1−) constitutively expressing CEPT1 or the indicated site-directed mutants. The ability to use either CDP-choline (black) or CDP-ethanolamine (white) as a substrate is indicated.
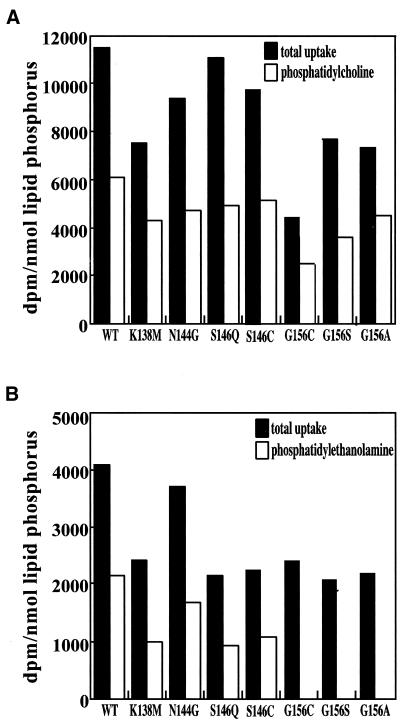
To confirm the in vitro CDP-alcohol substrate specificity results, the ability of wild-type and mutant versions of CEPT1 to reconstitute the synthesis of PtdCho or PtdEtn from radiolabeled choline or ethanolamine, respectively, was tested in S. cerevisiae HJ091 cells (cpt1::LEU2 ept1−). All of the yeast expressing mutant CEPT1 proteins were able to take up choline and synthesize PtdCho at levels similar to yeast expressing parental CEPT1 (Figure 9A). All of the yeast expressing mutant CEPT1 proteins were also capable of taking up ethanolamine at levels near to those expressing parental CEPT1; however, none of the CEPT1 proteins containing a substitution for glycine 156 were able to reconstitute de novo PtdEtn synthesis (Figure 9B). Hence, the in vivo metabolic pathway reconstitution results for the CEPT1 mutants were consistent with the in vitro assessment of their CDP-alcohol specificities and further support a role for glycine 156 in CEPT1 CDP-alcohol specificity for human CEPT1.
Figure 9.
Metabolic reconstitution of PtdCho and PtdEtn synthesis by CEPT1 CDP-alcohol phosphotransferase mutants. Exponentially growing S. cerevisiae cells (cpt1:: LEU2 ept1−) constitutively expressing CEPT1 or the indicated site-directed mutants were radiolabeled with (A) [14C]choline to label PtdCho or (B) [14C]ethanolamine to label PtdEtn, for 1 h. Radiolabel incorporated into each phospholipid was determined by scintillation counting.
Identification of Diacylglycerol Fatty Acyl Specificity Residues
The 214–234 membrane spanning helix of CEPT1 lies within the predicted diacylglycerol binding region of CEPT1 (Hjelmstad et al., 1994; McMaster and Bell, 1994a). N-terminal to residues 214–234 is a helix that spans residues 181–199 that is weakly predictive to span the membrane. If helix 181–199 does indeed span the membrane, then the orientation of helix 214–234 within the membrane would be flipped (Figure 6). Because the incoming phosphobase is transferred directly from the CDP-alcohol onto diacylglycerol without going through a membrane-bound intermediate (Hirabayashi et al., 1976; Bae-Lee and Carman, 1984; Pontoni et al., 1985; Raetz et al., 1987; Williams and McMaster, 1998), then the relevant amino acids within helix 214–234 need to be in close proximity to the soluble CDP-alcohol phosphotransferase motif. Because diacylglycerol only spans half of the membrane bilayer, amino acid residues that encompass the half of the membrane that interact with diacylglycerol would have an effect on enzymatic activity and/or diacylglycerol specificity, whereas residues mutated on the opposite side of the helix would have little effect. A number of residues spanning the entire membrane spanning 214–234 helix were targeted for site-directed mutagenesis (Figure 10A) to determine if residues within this helix interact with diacylglycerol and to allow positioning of this helix in the active site of CEPT1 relative to the CDP-alcohol phosphotransferase motif.
Figure 10.
CEPT1 diacylglycerol specificity mutants. (A) The site-directed mutations made in CEPT1 are indicated. The same region in human CPT1 is provided for comparison. The numbers denote CEPT1 amino acid residues. (B) Western blot of wild-type and mutant versions of CEPT1.
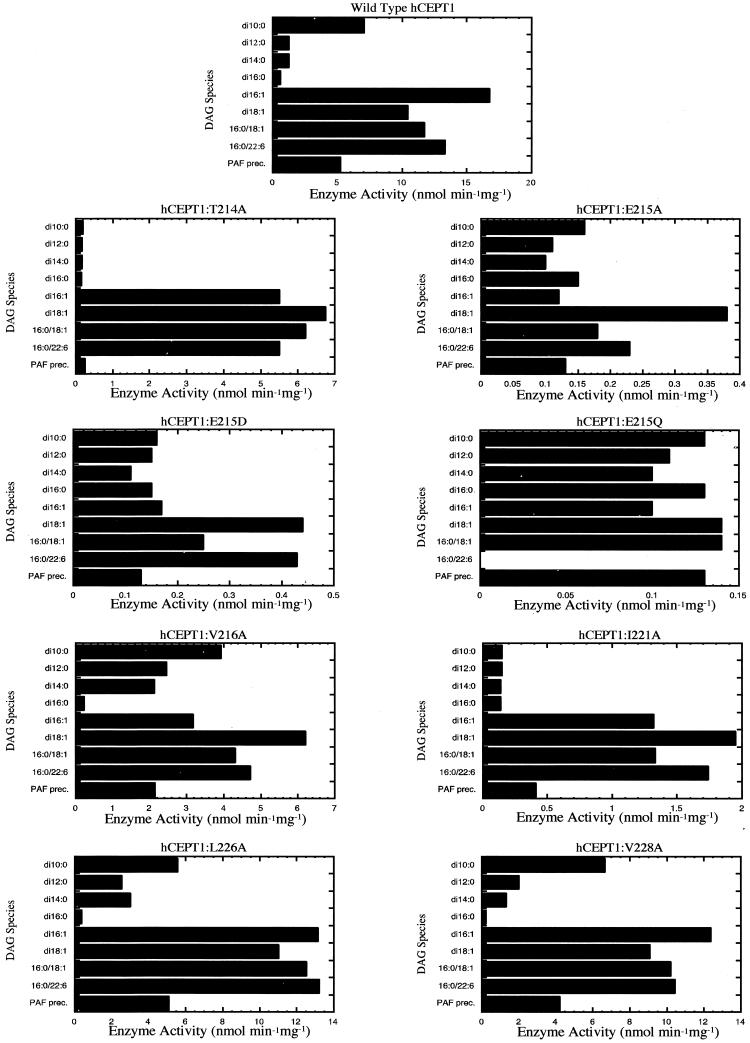
Plasmids carrying wild-type CEPT1 and various helix 214–234 mutations were transformed into S. cerevisiae strain HJ091 (cpt1::LEU2 ept1−). Western blot analysis of HJ091 membrane fractions established that similar levels of full-length CEPT1 protein were expressed (Figure 10B). The cholinephosphotransferase activities of CEPT1 mutants were determined by in vitro enzyme assay. The wild-type CEPT1 enzyme had a broad diacylglycerol substrate specificity with di16:1 ≥ 16:0/22:6 ≥ 16:0/18:1 ≥ di18:1 > di10:0 ≥ 16:0(O):20:4 (platelet activating factor [PAF] precursor). There was very little detectable activity toward di16:0, di14:0, or di12:0 diacylglycerols (Figure 10), and no activity was detected using 16:0(O):2:0 as substrate for the direct synthesis of PAF (our unpublished results). Each of the CEPT1 mutants was also analyzed for cholinephosphotransferase activity using a wide variety of defined diacylglycerols (Figure 11). A number of the mutations, namely T214A, V216A, and I221A, altered the profile of diacylglycerol utilization when compared with that of wild-type CEPT1 and also resulted in modest reductions in enzyme activity. The E215A, E215D, and E215Q mutations resulted in a much more dramatic reduction in CEPT1 enzyme activity. E215A and E215D did not alter diacylglycerol specificity, whereas the E215Q demonstrated altered diacylglycerol specificity. In contrast, the mutations L226A and V228A did not alter cholinephosphotransferase activity or diacylglycerol specificity of CEPT1. These results indicate that residues 226 and 228 are located on the opposite side of the helix from the catalytic site, whereas residues 214, 215, 216, and 221 are in close proximity to the catalytic site. Based on these results, it is predicted that the model proposed in Figure 5B is correct with regards to the orientation of membrane spanning helix 214–234 to the CDP-alcohol phosphotransferase motif.
Figure 11.
Enzyme activity of CEPT1 diacylglycerol specificity motif mutants. Enzyme activities were determined from microsomal membrane preparations of S. cerevisiae cells (cpt1:: LEU2 ept1−) constitutively expressing CEPT1 or the indicated site-directed mutants.
DISCUSSION
PtdCho and PtdEtn comprise 50–75% of cellular phospholipid mass. In this study we have identified the sites at which CEPT1 and CPT1, the enzymes that directly synthesize these two lipids, reside and provided molecular insights into specific amino acid determinants for both the fatty acid and lipid head group specificity of CEPT1. CEPT1 is a dual specificity enzyme that can synthesizes both PtdCho and PtdEtn, and glycine 156 within the catalytic CDP-alcohol phosphotransferase motif of CEPT1 was required for dual CDP-alcohol specificity. Any other amino acid at this position resulted in a loss of ethanolaminephosphotransferase activity by CEPT1. Mutagenesis of amino acid residues within the predicted transmembrane helix 214–234 of CEPT1 altered diacylglycerol substrate specificity, and enzyme activity confirming this helix spans the membrane. Only mutations in residues 214–221 of this helix affected diacylglycerol substrate specificity and enzyme activity, and because the phosphobase is transferred directly from the CDP-alcohol onto diacylglycerol without passing through an enzyme-bound intermediate (Hirabayashi et al., 1976; Bae-Lee and Carman, 1984; Pontoni et al., 1985; Raetz et al., 1987; Williams and McMaster, 1998), the first half of helix 214–234 must juxtapose the CDP-alcohol–binding site. This enabled the orientation of this diacylglycerol binding helix within the membrane span to be accurately positioned.
Our study also provides formal proof that PtdCho and PtdEtn are de novo synthesized at specific sites within the cell. CPT1 synthesizes PtdCho exclusively and was found in the Golgi, whereas the dual specificity CEPT1, which synthesizes both PtdCho and PtdEtn, was found in both endoplasmic reticulum and nuclear membranes. The rate-limiting step in the synthesis of PtdCho is catalyzed by CTα in most cell types and produces the CDP-choline that is used by cholinephosphotransferase activities to produce PtdCho. In most cell types, CTα is found in the nucleus as an amphitropic protein that is stored as an inactive soluble protein that translocates to the nuclear membrane to upregulate PtdCho synthesis (Watkins and Kent, 1992; Wang et al., 1993, 1995; Northwood et al., 1999; Cornell and Northwood, 2000; DeLong et al., 2000; Ridsdale et al., 2001). We demonstrated that activation of CTα through translocation to the nuclear membrane brings it into proximity with the nuclear membrane portion of CEPT1. This demonstrates that the activation of the CDP-choline pathway is likely through redistribution of the rate-limiting penultimate enzyme to the site of the ultimate step within the pathway. Whether CTα and CEPT1 reside in the same nuclear bilayer and/or physically interact remains to be determined, as does their colocalization with other upstream enzymes including CTβ and CTP:phosphoethanolamine cytidylyltransferase.
The role of diacylglycerol in the regulation of vesicle transport from the Golgi is well established in yeast and mammalian cells. In mammalian cells diacylglycerol production is required for recruitment of the Golgi vesicle biogenesis factor protein kinase D to the trans-Golgi network (Bankaitis, 2002; Baron and Malhotra, 2002). The regulation of Golgi diacylglycerol levels thus appears to be an important regulatory event in Golgi derived vesicle transport in mammalian cells. Naturally, the production of diacylglycerol in the Golgi for Golgi-derived vesicle transport must be balanced by diacylglycerol clearance. Our identification of a Golgi specific localization for mammalian CPT1 implies its consumption of diacylglycerol could play a role in the regulation of Golgi derived vesicle transport. A role for diacylglycerol consumption by the CDP-choline pathway during de novo PtdCho biosynthesis is supported by data obtained in yeast.
The yeast PtdCho/phosphatidylinositol transfer protein Sec14p is a soluble protein that translocates to Golgi membranes, and ablation of Sec14p function results in decreased Golgi-derived vesicle transport that eventually results in cell death (Bankaitis et al., 1989; Bankaitis et al., 1990; Cleves et al., 1991; Sha et al., 1998). Genetic inactivation of the CPT1-catalyzed step in the synthesis of PtdCho results in bypass of the essential function of Sec14p and allows cells to now live because of their ability to regain Golgi-derived vesicle transport. Decreasing PtdCho synthesis and thus increasing diacylglycerol levels by preventing its consumption through the CDP-choline pathway suppresses the need for Sec14p in Golgi transport (McGee et al., 1994a, 1994b; Xie et al., 2001), implying increased Golgi diacylglycerol promotes Golgi-derived vesicle transport (Cleves et al., 1991; Skinner et al., 1995; Kearns et al., 1997; Sreenivas et al., 1998; Xie et al., 1998; Phillips et al., 1999; Rivas et al., 1999; Henneberry et al., 2001; Xu et al., 2001). However, inactivation of the EPT1 pathway for PtdCho synthesis does not bypass the ability of cells to live in the absence of Sec14p, even if Ept1p is contributing to 50% of net CDP-choline–derived PtdCho synthesis (Henneberry et al., 2001). Our determination that human CEPT1 resides in the endoplasmic reticulum and nuclear membrane, whereas human CPT1 is in the Golgi, may provide an explanation for the ability of loss of function of yeast CPT1, but not EPT1, to bypass the essential function of Sec14p. Although speculative, if yeast Cpt1p and Ept1p localize to similar intracellular locations as their mammalian counterparts, then inactivation of yeast CPT1 would increase diacylglycerol and decrease PtdCho levels in the Golgi and thus provide a favorable shift in Golgi lipid levels for bypass of Sec14p function. Inactivation of EPT1 would primarily alter endoplasmic reticulum and nuclear lipid levels and would not have dramatic affects on Golgi diacylglycerol or PtdCho levels. Increased expression of EPT1 (and human CEPT1) in yeast was able to restore PtdCho synthesis to cells lacking their CPT1 gene, and this also prevented bypass of loss of Sec14p function in yeast carrying an inactivated CPT1 gene. We predict that increased expression of Ept1p and human CEPT1 results in either the mislocalization of a portion of these enzymes to the Golgi or drives increased synthesis of endoplasmic reticulum PtdCho, which is then transported to the Golgi. Our attempts to express human CPT1 in yeast were at levels too low to allow for complete restoration of PtdCho synthesis (Henneberry et al., 2000), and this is likely why human CPT1 was unable to bypass of the essential function of Sec14 due to inactivation of yeast CPT1. The precise site of yeast Cpt1p, Ept1p, and their human counterparts expressed in yeast is currently under investigation.
In this study we have addressed a fundamental issue of cell biology, where are phospholipids made? We have also demarcated molecular determinants that define product formation. The sites of synthesis of PtdCho and PtdEtn is requisite knowledge for any model attempting to describe how these lipids are transported to other organelles for restoration of PtdCho and PtdEtn levels subsequent to their signal transduction–mediated catabolism or for providing new membrane for cell growth. The data have also added credence to the model that interprets how Golgi-derived vesicle transport may be regulated by the consumption of diacylglycerol during the formation of PtdCho.
ACKNOWLEDGMENTS
We thank David Byers, Neale Ridgway, Harold Cook, and Vanina Zaremberg for helpful comments during the course of these studies. This work was supported by Operating and Scholarship grants from the Canadian Institutes of Health Research (to C.R.M.), a Canadian Institutes of Health Research Doctoral Award (to A.L.H.), and a graduate studentship from Cancer Care Nova Scotia and the Canadian Cancer Society (to M.M.W.).
Abbreviations used:
- PtdCho
phosphatidylcholine
- PtdEtn
phosphatidylethanolamine
- CPT1
cholinephosphotransferase of S. cerevisiae
- EPT1
choline/ethanolaminephosphotransferase of S. cerevisiae
- CEPT1
human choline/ethanolaminephosphotransferase
- CPT1
human cholinephosphotransferase
- CT
CTP:phosphocholine cytidylyltransferase
Footnotes
DOI: 10.1091/mbc.01–11–0540.
REFERENCES
- Ames BN, Dubin DT. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J Biol Chem. 1960;235:769–775. [PubMed] [Google Scholar]
- Attard GS, Templer RH, Smith WS, Hunt AN, Jackowski S. Modulation of CTP. phosphocholine cytidylyltransferase by membrane curvature elastic stress. Proc Natl Acad Sci USA. 2000;97:9032–9036. doi: 10.1073/pnas.160260697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae-Lee MS, Carman GM. Phosphatidylserine synthesis in Saccharomyces cerevisiae. Purification and characterization of membrane-associated phosphatidylserine synthase. J Biol Chem. 1984;259:10857–10862. [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Aitken JF, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA. Slick recruitment to the Golgi. Science. 2002;295:290–291. doi: 10.1126/science.1068446. [DOI] [PubMed] [Google Scholar]
- Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN, and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- Bi K, Roth MG, Ktistakis NT. Phosphatidic acid formation by phospholipase D is required for transport from the endoplasmic reticulum to the Golgi complex. Curr Biol. 1997;7:301–307. doi: 10.1016/s0960-9822(06)00153-9. [DOI] [PubMed] [Google Scholar]
- Birner R, Bürgermeister M, Schneiter R, Daum G. Roles of phosphatidylethanolamine, and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:997–1007. doi: 10.1091/mbc.12.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RB, Northwood IC. Regulation of CTP. phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biol Sci. 2000;25:441–447. doi: 10.1016/s0968-0004(00)01625-x. [DOI] [PubMed] [Google Scholar]
- Cui Z, Vance JE, Chen MH, Voelker DE, Vance DE. Cloning and expression of a novel phosphatidylethanolamine N-methyltransferase. A specific biochemical and cytological marker for a unique membrane fraction in rat liver. J Biol Chem. 1993;268:16655–16663. [PubMed] [Google Scholar]
- DeLong CJ, Qin L, Cui Z. Nuclear localization of enzymatically active green fluorescent protein-CTP. phosphocholine cytidylyltransferase α fusion protein is independent of cell cycle conditions and cell types. J Biol Chem. 2000;275:32325–32330. doi: 10.1074/jbc.M004644200. [DOI] [PubMed] [Google Scholar]
- DeLong CJ, Shen Y-J, Thomas MJ, Cui Z. Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J Biol Chem. 1999;274:29683–29688. doi: 10.1074/jbc.274.42.29683. [DOI] [PubMed] [Google Scholar]
- Henneberry AL, McMaster CR. Cloning and expression of a human choline/ethanolaminephosphotransferase: synthesis of phosphatidylcholine and phosphatidylethanolamine. Biochem J. 1999;339:291–298. [PMC free article] [PubMed] [Google Scholar]
- Henneberry AL, Lagace TA, Ridgway ND, McMaster CR. Phosphatidylcholine synthesis influences the diacylglycerol homeostasis required for Sec14p-dependent Golgi function, and cell growth. Mol Biol Cell. 2001;12:511–520. doi: 10.1091/mbc.12.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberry AL, Wistow G, McMaster CR. Cloning, genomic organization, and characterization of a human cholinephosphotransferase. J Biol Chem. 2000;275:29808–29815. doi: 10.1074/jbc.M005786200. [DOI] [PubMed] [Google Scholar]
- Hirabayashi T, Larson TJ, Dowhan W. Membrane-associated phosphatidylglycerophosphate synthetase from Escherichia coli: purification by substrate affinity chromatography on cytidine 5′-diphospho-1,2-diacyl-sn-glycerol sepharose. Biochemistry. 1976;15:5205–5211. doi: 10.1021/bi00669a002. [DOI] [PubMed] [Google Scholar]
- Hjelmstad RH, Bell RM. Mutants of Saccharomyces cerevisiae defective in sn-1,2-diacylglycerol cholinephosphotransferase. Isolation, characterization, and cloning of the CPT1 gene J. Biol Chem. 1987;262:3909–3917. [PubMed] [Google Scholar]
- Hjelmstad RH, Bell RM. The sn-1,2-diacylglycerol ethanolaminephosphotransferase activity of Saccharomyces cerevisiae. Isolation of mutants and cloning of the EPT1 gene J. Biol Chem. 1988;263:19748–19757. [PubMed] [Google Scholar]
- Hjelmstad RH, Morash SA, McMaster CR, Bell RM. Chimeric enzymes. structure-function analysis of segments of sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases. J Biol Chem. 1994;269:20095–21002. [PubMed] [Google Scholar]
- Hjelmstad RH, Bell RM. The sn-1,2-diacylglycerol cholinephosphotransferase of Saccharomyces cerevisiae. Nucleotide sequence, transcriptional mapping, and gene product analysis of the CPT1 gene. J Biol Chem. 1990;265:1755–1764. [PubMed] [Google Scholar]
- Hjelmstad RH, Bell RM. sn-1,2-Diacylglycerol choline- and ethanolaminephosphotransferases in Saccharomyces cerevisiae. Mixed micellar analysis of the CPT1 and EPT1 gene products. J Biol Chem. 1991a;266:4357–4365. [PubMed] [Google Scholar]
- Hjelmstad RH, Bell RM. sn-1,2-Diacylglycerol choline- and ethanolaminephosphotransferases in Saccharomyces cerevisiae. Nucleotide sequence of the EPT1 gene and comparison of the CPT1 and EPT1 gene products. J Biol Chem. 1991b;266:5094–5103. [PubMed] [Google Scholar]
- Hodgkin MN, Pettitt TR, Martin A, Michell RH, Pemberton AJ, Wakelam MJ. Diacylglycerols and phosphatidates: which molecular species are intracellular messengers? Trends Biochem Sci. 1998;23:200–204. doi: 10.1016/s0968-0004(98)01200-6. [DOI] [PubMed] [Google Scholar]
- Hunt AN, Clark GT, Attard GS, Postle AD. Highly saturated endonuclear phosphatidylcholine is synthesized in situ, and colocated with CDP-choline pathway enzymes. J Biol Chem. 2001;276:8492–8499. doi: 10.1074/jbc.M009878200. [DOI] [PubMed] [Google Scholar]
- Jansen SM, Groener JEM, Bax W, Suter A, Saftig P, Somerharju P, Poorthuis JHM. Biosynthesis of phosphatidylcholine from a phosphocholine precursor pool derived from the late endosomal/lysosomal degradation of sphingomyelin. J Biol Chem. 2001;276:18722–18727. doi: 10.1074/jbc.M101817200. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent C. Eukaryotic phospholipid biosynthesis. Annu Rev Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lykidis A, Murti KG, Jackowski S. Cloning and characterization of a second human CTP:phosphocholine cytidylyltransferase. J Biol Chem. 1998;273:14022–14029. doi: 10.1074/jbc.273.22.14022. [DOI] [PubMed] [Google Scholar]
- Mancini A, Del Rosso F, Roberti R, Orvietani P, Coletti L, Binaglia L. Purification of ethanolaminephosphotransferase from bovine liver microsomes. Biochim Biophys Acta. 1999;1437:80–92. doi: 10.1016/s1388-1981(98)00011-0. [DOI] [PubMed] [Google Scholar]
- McGee TP, Skinner HB, Bankaitis VA. Functional redundancy of the CDP-ethanolamine and CDP-choline pathway enzymes in phospholipid biosynthesis: ethanolamine-dependent effects on steady-state membrane phospholipid composition in Saccharomyces cerevisiae. J Bacteriol. 1994a;176:6861–6868. doi: 10.1128/jb.176.22.6861-6868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee TP, Skinner HB, Whitters EA, Henry SA, Bankaitis VA. A phosphatidylinositol transfer protein controls the phosphatidylcholine content of yeast Golgi membranes, J. Cell Biol. 1994b;124:273–287. doi: 10.1083/jcb.124.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster CR, Choy PC. Serine regulates phosphatidylethanolamine biosynthesis in the hamster heart. J Biol Chem. 1992;267:14586–14591. [PubMed] [Google Scholar]
- McMaster CR, Bell RM. Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2- diacylglycerol choline- and ethanolaminephosphotransferases. J Biol Chem. 1994a;269:28010–28016. [PubMed] [Google Scholar]
- McMaster CR, Bell RM. Phosphatidylcholine biosynthesis via the CDP-choline pathway in Saccharomyces cerevisiae. Multiple mechanisms of regulation. J Biol Chem. 1994b;269:14776–14783. [PubMed] [Google Scholar]
- Munberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Northwood IC, Tong AH, Crawford B, Drobnies AE, Cornell RB. Shuttling of CTP:Phosphocholine cytidylyltransferase between the nucleus and endoplasmic reticulum accompanies the wave of phosphatidylcholine synthesis during the G(0) → G(1) transition. J Biol Chem. 1999;274:26240–26248. doi: 10.1074/jbc.274.37.26240. [DOI] [PubMed] [Google Scholar]
- Paltauf F, Kohlwein SD, Henry SA. The Molecular and Cellular Biology of the Yeast Saccharomyces ed. E.W. Jones, J.R. Pringle, and J.R. Broach. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 415–500. [Google Scholar]
- Parekh DB, Ziegler W, Parker PJ. Multiple pathways control protein kinase C phosphorylation. EMBO J. 2000;19:496–503. doi: 10.1093/emboj/19.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SE, Sha B, Topalof L, et al. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol Cell. 1999;4:187–197. doi: 10.1016/s1097-2765(00)80366-4. [DOI] [PubMed] [Google Scholar]
- Pontoni G, Manna C, Salluzo A, del Piano L, Galletti P, De Rosa M, Zappia V. Studies on enzyme-substrate interactions of cholinephosphotransferase from rat liver. Biochim Biophys Acta. 1985;836:222–232. doi: 10.1016/0005-2760(85)90070-0. [DOI] [PubMed] [Google Scholar]
- Raetz CR. Molecular genetics of membrane phospholipid synthesis. Annu Rev Genet. 1986;20:253–295. doi: 10.1146/annurev.ge.20.120186.001345. [DOI] [PubMed] [Google Scholar]
- Raetz CRH, Carman GM, Dowhan W, Jiang R-T, Waszluc W, Loffredo W, Tsai M-D. Phospholipids chiral at phosphorus. Steric course of the reactions catalyzed by phosphatidylserine synthase from Escherichia coli and yeast. Biochemistry. 1987;26:4022–4027. doi: 10.1021/bi00387a042. [DOI] [PubMed] [Google Scholar]
- Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale R, Tseu I, Wang J, Post M. CTP. phosphocholine cytidylyltransferase α is a cytosolic protein in pulmonary epithelial cells and tissues. J Biol Chem. 2001;276:49148–49155. doi: 10.1074/jbc.M103566200. [DOI] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Guo S, Xie Z, Sekar C, Hosaka K, Kagiwada S, York JD, Bankaitis BA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p”and inositol auxotrophy, Mol. Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samborski RW, Ridgway ND, Vance DE. Metabolism of molecular species of phosphatidylethanolamine and phosphatidylcholine in rat hepatocytes during prolonged inhibition of phosphatidylethanolamine N-methyltransferase. J Biol Chem. 1990;265:18322–18329. [Google Scholar]
- Schmidt A, Wolde M, Thiele C, Fest W, Kratzin H, Podtelejnikov AV, Witke W, Huttner WB, Soling HD. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature. 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- Schneiter R, Brugger B, Sandhoff R, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha BD, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- Shiao YJ, Lupo G, Vance JE. Evidence that phosphatidylserine is imported into mitochondria via a mitochondria-associated membrane and that the majority of mitochondrial phosphatidylethanolamine is derived from decarboxylation of phosphatidylserine. J Biol Chem. 1995;270:11190–11198. doi: 10.1074/jbc.270.19.11190. [DOI] [PubMed] [Google Scholar]
- Siddhanta A, Backer JM, Shields D. Inhibition of phosphatidic acid synthesis alters the structure of the Golgi apparatus, and inhibits secretion in endocrine cells. J Biol Chem. 2000;275:12023–12031. doi: 10.1074/jbc.275.16.12023. [DOI] [PubMed] [Google Scholar]
- Skinner HB, McGee TP, McMaster CR, Fry MR, Bell RM, Bankaitis VA. The Saccharomyces cerevisiae phosphatidylinositol-transfer protein effects a ligand-dependent inhibition of choline-phosphate cytidylyltransferase activity. Proc Natl Acad Sci USA. 1995;92:112–116. doi: 10.1073/pnas.92.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast, J. Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- Voelker DE. Phosphatidylserine functions as the major precursor of phosphatidylethanolamine in cultured BHK-21 cells. Proc Natl Acad Sci USA. 1984;81:2669–2673. doi: 10.1073/pnas.81.9.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey CJ, Yu L, Agellon LB, Vance DE. Biochemical and evolutionary significance of phospholipid methylation. J Biol Chem. 1998;273:27043–27046. doi: 10.1074/jbc.273.42.27043. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sweitzer TD, Weinhold P, Kent C. Nuclear localization of soluble CTP:phosphocholine cytidylyltransferase. J Biol Chem. 1993;268:5899–5904. [PubMed] [Google Scholar]
- Wang Y, MacDonald JI, Kent C. Identification of the nuclear localization signal of rat liver CTP:phosphocholine cytidylyltransferase. J Biol Chem. 1995;270:354–360. doi: 10.1074/jbc.270.1.354. [DOI] [PubMed] [Google Scholar]
- Watkins JD, Kent C. Immunolocalization of membrane-associated CTP:phosphocholine cytidylyltransferase in phosphatidylcholine-deficient Chinese hamster ovary cells. J Biol Chem. 1992;267:5686–5692. [PubMed] [Google Scholar]
- Weigert R, Silletta MG, Spano S, et al. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402:429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- Weiss SB, Smith SW, Kennedy EP. The enzymatic formation of lecithin from cytidine diphosphate choline and D-1,2-diglyceride. J Biol Chem. 1958;233:53–64. [PubMed] [Google Scholar]
- Williams JG, McMaster CR. Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiae cholinephosphotransferase. J Biol Chem. 1998;273:13482. doi: 10.1074/jbc.273.22.13482. ,13487. [DOI] [PubMed] [Google Scholar]
- Wright MM, Henneberry AL, Lagace TA, Ridgway ND, McMaster CR. Uncoupling Farnesol-induced Apoptosis from Its Inhibition of Phosphatidylcholine Synthesis. J Biol Chem. 2001;276:25254–25261. doi: 10.1074/jbc.M011552200. [DOI] [PubMed] [Google Scholar]
- Xie Z, Fang M, Bankaitis VA. Evidence for an intrinsic toxicity of Phosphatidylcholine to Sec14p-dependent protein transport from the yeast Golgi complex. Mol Biol Cell. 2001;12:1117–1129. doi: 10.1091/mbc.12.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht J, Bankaitis VA. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects, Proc. Natl Acad Sci USA. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liu Y, Ridgway ND, McMaster CR. Novel members of the human oxysterol-binding protein family bind phospholipids, and regulate vesicle transport, J. Biol Chem. 2001;276:18407–18414. doi: 10.1074/jbc.M101204200. [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CDM, Fasch E-V, Kohlwein SP, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]