Abstract
The Saccharomyces cerevisiae genome contains five genes encoding P-type ATPases that are potential aminophospholipid translocases (APTs): DRS2, NEO1, and three uncharacterized open reading frames that we have named DNF1, DNF2, and DNF3 for DRS2/NEO1 family. NEO1 is the only essential gene in APT family and seems to be functionally distinct from the DRS2/DNF genes. The drs2Δ dnf1Δ dnf2Δ dnf3Δ quadruple mutant is inviable, although any one member of this group can maintain viability, indicating that there is a substantial functional overlap between the encoded proteins. We have previously implicated Drs2p in clathrin function at the trans-Golgi network. In this study, we constructed strains carrying all possible viable combinations of null alleles from this group and analyzed them for defects in protein transport. The drs2Δ dnf1Δ mutant grows slowly, massively accumulates intracellular membranes, and exhibits a substantial defect in the transport of alkaline phosphatase to the vacuole. Transport of carboxypeptidase Y to the vacuole is also perturbed, but to a lesser extent. In addition, the dnf1Δ dnf2Δ dnf3Δ mutant exhibits a defect in recycling of GFP-Snc1p in the early endocytic-late secretory pathways. Drs2p and Dnf3p colocalize with the trans-Golgi network marker Kex2p, whereas Dnf1p and Dnf2p seem to localize to the plasma membrane and late exocytic or early endocytic membranes. We propose that eukaryotes express multiple APT subfamily members to facilitate protein transport in multiple pathways.
INTRODUCTION
P-type ATPases are a large family of multitransmembrane domain, ATP-dependent transporters. The P-type designation derives from a covalent aspartyl-phosphate catalytic intermediate formed during ATP hydrolysis, which is coupled to conformational changes that can drive ion transport across membranes against an electrochemical gradient. Based on the substrate transported, P-type ATPases are classified as heavy metal ion ATPases, such as the copper transporters that are mutated in patients with Menkes or Wilson disease; and nonheavy metal ion ATPases, such as Ca2+-, H+-, and Na+,K+-ATPases (Moller et al., 1996). From sequence analyses, members of a third subfamily of P-type ATPases have been distinguished from the above-mentioned two types (Catty et al., 1997; Halleck et al., 1998) and are proposed to be aminophospholipid translocases (APTs) or flippases (Zachowski et al., 1989; Auland et al., 1994; Tang et al., 1996; Gomes et al., 2000; Ujhazy et al., 2001). APTs translocate phosphatidylserine (PS) and/or phosphatidylethanolamine (PE) from one leaflet of the membrane bilayer to the other, and seem to be responsible for concentrating PS and PE on the cytosolic leaflet of biological membranes.
Genome sequencing projects have identified a large number of potential APT subfamily genes in eukaryotic organisms, although no prokaryotic member has been identified yet. There are five members of this subfamily in both Saccharomyces cerevisiae and Schizosaccharomyces pombe (Costanzo et al., 2001). The Caenorhabditis elegans genome contains six members, Arabidopsis has 11 (Gomes et al., 2000), and there seem to be at least 21 members in humans. Little is known about the cellular function of the APT subfamily of proteins or why eukaryotes maintain so many of these genes. However, a few biological disorders have been linked or attributed to genes from this subfamily. FIC1 mutations cause familial intrahepatic cholestasis in humans, which is a defect in bile secretion (Thompson and Jansen, 2000; Ujhazy et al., 2001). The ATP10C gene has been linked to Angelman syndrome and autism in some patients (Herzing et al., 2001; Meguro et al., 2001), and the mouse ATP10C homolog pfatp is a candidate gene for increased body fat (Dhar et al., 2000). In addition, the PDE1 gene is a pathogenic factor for the rice blast fungus, Magnaporthe grisea (Balhadere and Talbot, 2001), and the ALA1 gene in Arabidopsis is required for growth at low temperature (Gomes et al., 2000).
The first member of the APT subfamily to be identified genetically and cloned was DRS2 from S. cerevisiae (yeast) (Ripmaster et al., 1993). Drs2p is 47% identical in amino acid sequence to the mammalian chromaffin granule ATPase II (Tang et al., 1996), which is proposed to be the PS translocase characterized with intact chromaffin granules (Zachowski et al., 1989). DRS2 was initially identified in a genetic screen for mutants that exhibit a cold-sensitive defect in ribosome synthesis and was shown to be a nonessential gene, although disruption of DRS2 causes a strong cold-sensitive growth defect (Ripmaster et al., 1993). Subsequently, we identified DRS2 in another genetic screen for mutations that are synthetically lethal with arf1 (Chen et al., 1999).
ADP-ribosylation factor (ARF) is a small GTP-binding protein whose functions in initiating COPI and clathrin-coated vesicle (CCV) budding have been well studied (Donaldson and Jackson, 2000). The drs2Δ null allele is also synthetically lethal with clathrin heavy chain temperature-sensitive (ts) alleles but not with ts alleles of COPI subunits (Chen et al., 1999). These genetic interactions between DRS2, ARF, and clathrin suggested a protein transport function for Drs2p at the trans-Golgi network (TGN). In fact, Drs2p localizes to the TGN and the drs2Δ mutant exhibits TGN defects comparable with those exhibited by clathrin mutants. These include 1) the mislocalization of TGN proteins, resulting in a defect in the proteolytic processing of pro-α-factor; 2) the accumulation of aberrant Golgi cisternae; and 3) a deficiency of CCVs in subcellular fractions (Chen et al., 1999). These phenotypes are much more severe at the nonpermissive growth temperature. Recently, however, we have found that the drs2Δ mutant exhibits a defect at permissive growth temperatures in generating a specific class of CCVs carrying invertase and acid phosphatase to the plasma membrane (Gall et al., 2002).
The ribosome synthesis defect observed originally in drs2Δ is likely a secondary effect of its protein transport defect. Many secretory mutants globally down-regulate ribosomal protein synthesis through a protein kinase C signal transduction pathway (Nierras and Warner, 1999; Li et al., 2000). In addition, the severe protein transport defects are observed immediately after shifting cells to the nonpermissive temperature, whereas several hours are required before a protein synthesis defect is observed (Chen et al., 1999). The drs2Δ mutant has also been reported to exhibit a defect in APT activity at the plasma membrane (Tang et al., 1996; Gomes et al., 2000), although others report no defect in this activity (Siegmund et al., 1998; Marx et al., 1999). Moreover, the localization of Drs2p suggests a primary function for this protein in the TGN rather than the plasma membrane (Chen et al., 1999). Although PS and/or PE translocation is the most likely biochemical activity for Drs2p, direct evidence for this remains elusive. In fact, only an erythrocyte plasma membrane APT activity has been reconstituted in vitro with purified enzyme (Auland et al., 1994).
The drs2Δ strain grows as well as wild type at 24°C or above, but fails to grow at 23°C or below. One possible explanation for the cold sensitivity of drs2Δ null strain is that other proteins could replace the essential function of Drs2p at higher temperatures, with the best candidates being the Drs2p homologs encoded within the yeast genome. A phylogenetic sequence comparison of the 16 P-type ATPases encoded in the yeast genome suggested that five of these proteins belong to the APT subfamily (Catty et al., 1997). Two of these genes (DRS2 and NEO1) have been identified genetically, whereas the remaining three genes (YER166W, YDR093W, and YMR162C) have not been characterized. We have designated the three uncharacterized genes DNF1 (YER166W), DNF2 (YDR093W), and DNF3 (YMR162C) for DRS2/NEO1 family. The similarity of these proteins range from 39% for DNF3 and NEO1 to 83% for DNF1 and DNF2 (Table 1). We have constructed strains carrying all possible combinations of drs2Δ and dnfΔ null alleles and have found that DRS2 and the DNF genes constitute an essential gene family that exert overlapping functions in protein transport. Drs2p and Dnf1p have redundant functions in protein transport from the TGN to the vacuole, and Dnf1p, Dnf2p, and Dnf3p have redundant functions in recycling the v-SNARE Snc1p from endosomes to the TGN. The potential role of these proteins in generating membrane asymmetry will be the focus of another report.
Table 1.
Similarity and identity of DRS2/NEO1 genes
| DRS2 | DNF1 | DNF2 | DNF3 | NEO1 | |
|---|---|---|---|---|---|
| DRS2 | 48 | 48 | 45 | 48 | |
| DNF1 | 31 | 83 | 45 | 43 | |
| DNF2 | 31 | 69 | 45 | 43 | |
| DNF3 | 29 | 28 | 27 | 39 | |
| NEO1 | 30 | 26 | 26 | 23 |
Upper right lists the similarity of DRS2/NEO1 genes (%), and lower left lists their identity (%). (Adapted from Halleck et al., 1998).
MATERIALS AND METHODS
Media, Strains, and Plasmids
Yeast were grown in standard rich medium (YPD) or SD minimal media containing required supplements (Sherman, 1991). The calcofluor white (CW) sensitivity test was done on YPD 2% agar plates containing 10, 30, or 50 μg/ml calcofluor white (F6259; Sigma-Aldrich, St. Louis, MO).
Yeast strains used in this study are listed in Table 2. The original DRS2/NEO1 family gene deletion strains were made by Saccharomyces genome deletion project (Winzeler et al., 1999) and were purchased from American Type Culture Collection (Manassas, VA). The drs2/dnf combination deletion strains were generated by standard genetic crosses or gene disruptions. The genotype of each spore was determined by a polymerase chain reaction (PCR) method as described by the Saccharomyces genome deletion project (http://sequence-www.stanford.edu/group/yeast_deletion_project/deletions3.html). ZHY3271G was generated by mating two dnf1,2,3Δ strains generated from tetrad dissection. The other dnf deletion diploid strains were generated by transforming cells with pHO (Russell et al., 1986), encoding the HO endonuclease. The strains that harbor Myc-tagged proteins were generated by PCR-based gene targeting into the BY4742 strain and selected on G418 plates. The pFA6a-13Myc-kanMX6 was used as PCR template (Bahler et al., 1998), and the Myc integrated strains were confirmed by PCR. The epitope-tagged Drs2 and Neo1 proteins are functional because they support cell growth at all temperatures tested. We assume that the epitope-tagged Dnf proteins are functional although this has not been tested directly.
Table 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3 leu2 ura3 met15 | ATCC |
| BY4742 | MATα his3 leu2 ura3 lys2 | ATCC |
| BY4743 | MATa/α his3/his3 leu2/leu2 ura3/ura3 met15/MET15 lys2/LYS2 | ATCC |
| BY4739 YAL026C | MATα leu2 ura3 lys2 drs2Δ | ATCC |
| BY4741 YER166W | MATa his3 leu2 ura3 met15 dnf1Δ | ATCC |
| BY4742 YER166W | MATα his3 leu2 ura3 lys2 dnf1Δ | ATCC |
| BY4741 YDR093W | MATa his3 leu2 ura3 met15 dnf2Δ | ATCC |
| BY4742 YDR093W | MATα his3 leu2 ura3 lys2 dnf2Δ | ATCC |
| BY4741 YMR162C | MATa his3 leu2 ura3 met15 dnf3Δ | ATCC |
| BY4742 YMR162C | MATα his3 leu2 ura3 lys2 dnf3Δ | ATCC |
| BY4743 YIL048W | MATa/α his3/his3 leu2/leu2 ura3/ura3 neo1Δ/NEO1 | ATCC |
| ZHY9072B | MATa his3 leu2 ura3 met15 neo1Δ pBM743-NEO1 | This study |
| ZHY615D1C | MATa his3 leu2 ura3 lys2 drs2Δ | This study |
| ZHY615M2D | MATα his3 leu2 ura3 lys2 drs2Δ | This study |
| ZHY2149D | MATα his3 leu2 ura3 lys2 drs2Δ dnf1Δ | This study |
| ZHY615D2A | MATα leu2 ura3 drs2Δ dnf2Δ | This study |
| ZHY615M2A | MATα leu2 ura3 met15 drs2Δ dnf3Δ | This study |
| PFY3275F | MATa his3 leu2 ura3 met15 dnf1Δ dnf2Δ | This study |
| PFY3272G | MATa his3 leu2 ura3 met15 dnf1Δ dnf3Δ | This study |
| ZHY5057B | MATα his3 leu2 ura3 met15 dnf2Δ dnf3Δ | This study |
| PFY3273A | MATα his3 leu2 ura3 met15 dnf1Δ dnf2Δ dnf3Δ | This study |
| PFY3272C | MATa his3 leu2 ura3 met15 dnf1Δ dnf2Δ dnf3Δ | This study |
| ZHY7282C | MATa leu2 ura3 drs2Δ dnf2Δ dnf3Δ | This study |
| ZHY708 | MATa his3 leu2 ura3 met15 dnf1Δ dnf3Δ drs2∷LEU2 | This study |
| ZHY709 | MATa his3 leu2 ura3 met15 dnf1Δ dnf2Δ drs2∷LEU2 | This study |
| ZHY704 | MATα his3 leu2 ura3 met15 dnf1Δ dnf2Δ dnf3Δ drs2∷LEU2 pRS416-DRS2 | This study |
| ZHY3271G | MATa/α his3/his3 leu2/leu2 ura3/ura3 met15/met15 dnf1Δ/dnf1Δ dnf2Δ/dnf2Δ dnf3Δ/dnf3Δ | This study |
| ZHY3272G1 | MATa/α his3/his3 leu2/leu2 ura3/ura3 met15/met15 dnf1Δ/dnf1Δ dnf3Δ/dnf3Δ | This study |
| ZHY3275F1 | MATa/α his3/his3 leu2/leu2 ura3/ura3 met15/met15 dnf1Δ/dnf1Δ dnf2Δ/dnf2Δ | This study |
| ZHYDPdnf23 | MATa/α his3/his3 leu2/leu2 ura3/ura3 met15/met15 dnf2Δ/dnf2Δ dnf3Δ/dnf3Δ | This study |
| ZHYDPdnf1 | MATa/α his3/his3 leu2/leu2 ura3/ura3 lys2/lys2 dnf1Δ/dnf1Δ | This study |
| ZHYNEO1-MYC | MATα his3 leu2 ura3 lys2 NEO1∷13XMYC | This study |
| ZHYDRS2-MYC | MATα his3 leu2 ura3 lys2 DRS2∷13XMYC | This study |
| ZHYDNF1-MYC | MATα his3 leu2 ura3 lys2 DNF1∷13XMYC | This study |
| ZHYDNF2-MYC | MATα his3 leu2 ura3 lys2 DNF2∷13XMYC | This study |
| ZHYDNF3-MYC | MATα his3 leu2 ura3 lys2 DNF3∷13XMYC | This study |
| 6210 vps35Δ | MATα his3 leu2 ura3 lys2 ade2 trp1 vps35∷HIS3 | Chen, et al., 1999 |
| 6210 ap15Δ | MATα his3 leu2 ura3 lys2 ade2 trp1 apl5∷HIS3 | Cowles, et al., 1997 |
ATCC, American Type Culture Collection.
pRS416-DRS2 was produced by subcloning of a SpeI-SalI fragment from pRS315-DRS2 (Chen et al., 1999) into SpeI-SalI digested pRS416. pZH523 (DRS2 disruption plasmid) was constructed by replacing a BamHI-SnaBI fragment from pPR10 with a ∼2.1-kb BamHI-PvuII fragment containing LEU2 from pJJ283 (Jones and Prakash, 1990). The plasmid pZH523 was linearized with SacI and HpaI and transformed into different strains to delete DRS2. The full-length NEO1 gene was PCR amplified using primers (forward 5′-AAGGATATGTCTTTTCGTGTACGTG and reverse 5′-GCCGCTTGATATGTAAGTTCGTCA) and cloned into pYGW1 by using the GeneWeaver I kit from Research Genetics (Huntsville, AL). The EcoRI-SalI fragment of the NEO1 gene from pYGW1-NEO1 was subcloned into EcoRI-SalI site of pBM743 so that NEO1 is under the control of GAL1 promoter and can be suppressed in glucose media (Johnston and Davis, 1984). GFP-ALP (Cowles et al., 1997), pRS416 SNC1-GFP (Lewis et al., 2000), pRS426 STE2-GFP (Odorizzi et al., 1998), and p426 KH (2 μ Kex2-HA in pRS426) (Chen et al., 1999) were transformed into strains as indicated.
Sporulation Test
To test for sporulation efficiency, cells were grown overnight in YPD medium, diluted into PSP2 (1% KOAc, 0.1% yeast extract, and 0.67% yeast nitrogen base) presporulation medium, and cultured for 18–24 h to late log phase. Cells were then washed once in distilled H2O and resuspended in 1% KOAc sporulation medium at 1–2 × 107 cells/ml. After shaking for 3–7 d at 30°C, the sporulating cells were examined under the microscope for visible tetrads. In addition, cells that were in sporulation medium for 5–7 d were treated with glusulase and ether to kill the nonsporulated cells and spread on YPD plates to germinate. Strains scored as “+” in Table 4 showed no visible or viable spores by these tests.
Table 4.
Sporulation defect and CW sensitivity of drs2/dnf deletion mutants
| Strains | Sporulation defect | Calcofluor white sensitivity |
|---|---|---|
| Wild type | − | − |
| drs2Δ | + | ++ |
| dnf1Δ | − | − |
| dnf2Δ | − | − |
| dnf3Δ | − | − |
| dnf1Δ dnf2Δ | + | ++ |
| dnf1Δ dnf3Δ | +/− | − |
| dnf2Δ dnf3Δ | − | − |
| dnf1Δ dnf2Δ dnf3Δ | + | +++ |
Sporulation was assessed in homozygous diploids, whereas the Calcofluor white sensitivity was examined in haploid strains as described in MATERIALS AND METHODS. Sporulation defect: −, no significant sporulation defect; +, complete sporulation defect; +/−, sporulation efficiency is ∼0.5% of wild type. Calcofluor white sensitivity: growth on 50 μg/ml CW (−); no growth on 30 μg/ml CW plates (++); no growth on 10 μg/ml CW plates (+++).
Immunological Methods
Cell labeling, immunoprecipitation (Gaynor and Emr, 1997), and immunoblotting (Chen et al., 1999) were performed as described previously. Anti-α-factor (Graham and Emr, 1991), anti-carboxypeptidase Y (CPY) (Klionsky et al., 1988), and anti-ALP (Seeger and Payne, 1992) serum were used (1 μl/OD of cells) for immunoprecipitations. The 9E10 mouse monoclonal c-Myc antibody (Oncogene Research Products, Darmstadt, Germany) was used at 1:2000 for Western blot and 1:100 for immunofluorescence. Polyclonal affinity-purified rabbit anti-hemagglutinin (HA) antibody (Zymed Laboratories, South San Francisco, CA) was used at 1:100 dilution to detect Kex2-HA by immunofluorescence. Alexa-488 goat anti-rabbit IgG and Alexa-594 goat anti-mouse IgG (Molecular Probes, Eugene, OR) were used at 1:200 as secondary antibodies for immunofluorescence. Monoclonal CPY and ALP antibody (Molecular Probes) were used at 1:2000 and 1:125 in Western blot experiments. NIH Image 1.62 was used to quantify the band intensities from scanned images of Western blots. Immunofluorescence and green fluorescent protein (GFP) fluorescence were observed using an Axioplan microscope (Carl Zeiss, Thornwood, NY), and fluorescent images were quantified and processed using MetaMorph 4.5 software (Universal Imaging, Downingtown, PA). Samples for electron microscopy were prepared as described previously (Rieder et al., 1996). Sections (50–60 nm) were viewed on a CM12 electron microscope (Philips, Eindhoven, Netherlands).
Invertase Secretion Assay
Cells were grown overnight in YP medium with 5% glucose at 30°C. Cells were diluted to 0.1–0.2 OD600/ml in YP medium with 5% glucose and were grown at 30°C to log phase (0.6–1.0 OD600/ml). Cells were pelleted and resuspended in 0.1% glucose and incubated at 30°C. Aliquots of cells were collected at 0, 15, 30, and 45 min after induction and processed for invertase activity assay (Novick et al., 1981). External invertase was assayed at 30°C as described by Goldstein and Lampen (1975). Total invertase activity was determined by assaying whole cell lysate prepared by freeze-thaw method. Invertase secretion ratio is calculated by dividing total invertase activity by external activity. The small amount of invertase activity at time 0 was subtracted from all time points.
RESULTS
NEO1 Is an Essential Gene at All Temperatures
NEO1 was identified in a screen for genes that confer resistance to the aminoglycoside neomycin upon overexpression and was reported to be an essential gene at 30°C (Prezant et al., 1996). Because disruption of DRS2 causes a temperature-conditional growth phenotype, we tested the neo1Δ strain for viability at several different temperatures. NEO1 was placed under transcriptional regulation of the GAL1 promoter, which is tightly repressed when cells are grown on glucose and expressed strongly on galactose (Johnston and Davis, 1984). The neo1Δ GAL:NEO1 strain grew well on galactose but failed to grow when plated on media containing glucose at any temperature tested (Table 3, neo1Δ). Thus, NEO1 provides an essential function that cannot be performed by DRS2, DNF1, 2, or 3. We also found that overexpression of NEO1 could not suppress the cold-sensitive growth defect of drs2Δ strains, nor could overexpression of DRS2 suppress the lethality of neo1Δ, suggesting that there is no significant genetic overlap between DRS2 and NEO1.
Table 3.
Growth profile of DRS2/NEO1 family deletion mutants
| Strains | 15°C | 20°C | 24°C | 30°C | 37°C |
|---|---|---|---|---|---|
| Wild type | +++ | +++ | +++ | +++ | +++ |
| neo1Δ | − | − | − | − | − |
| drs2Δ | − | − | ++ | +++ | +++ |
| dnf1Δ | +++ | +++ | +++ | +++ | +++ |
| dnf2Δ | +++ | +++ | +++ | +++ | +++ |
| dnf3Δ | +++ | +++ | +++ | +++ | +++ |
| drs2Δ dnf1Δ | − | − | + | + | ++ |
| drs2Δ dnf2Δ | − | − | ++ | +++ | +++ |
| drs2Δ dnf3Δ | − | − | ++ | +++ | +++ |
| dnf1Δ dnf2Δ | +++ | +++ | +++ | +++ | +++ |
| dnf1Δ dnf3Δ | +++ | +++ | +++ | +++ | +++ |
| dnf2Δ dnf3Δ | +++ | +++ | +++ | +++ | +++ |
| drs2Δ dnf1Δ dnf2Δ | − | − | +/− | + | + |
| drs2Δ dnf1Δ dnf3Δ | − | − | +/− | ++ | ++ |
| drs2Δ dnf2Δ dnf3Δ | − | − | + | +++ | +++ |
| dnf1Δ dnf2Δ dnf3Δ | +++ | +++ | +++ | +++ | +++ |
| drs2Δ dnf1Δ dnf2Δ dnf3Δ | − | − | − | − | − |
Growth was examined on YPD plates incubated at the temperatures indicated. Symbols indicate relative growth rate from wild type (+++) to no growth (−).
DRS2 and DNF Genes Constitute an Essential Family
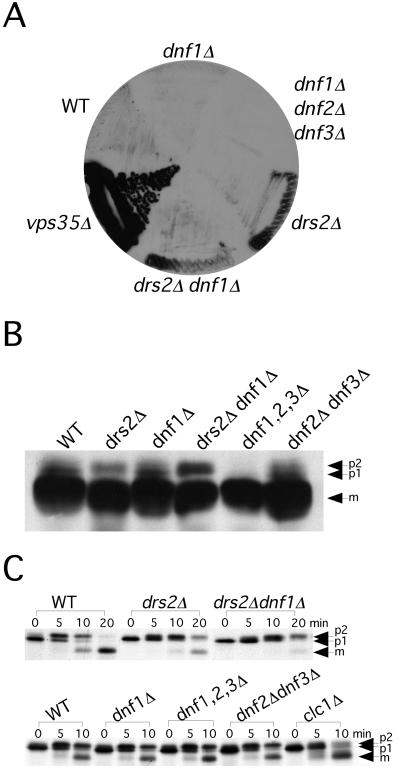
Except for NEO1, none of the other members of the DRS2/NEO1 family are essential (Winzeler et al., 1999). To test whether there is any functional redundancy between DRS2 and the DNF genes, we generated strains carrying all possible combinations of the drs2 and dnf null alleles (Tables 2 and 3 and Figure 1A). All possible double and triple mutants were recovered among the progeny of standard crosses, although this technique failed to yield a viable quadruple mutant. To further test whether the quadruple mutant is viable, the dnf1,2,3Δ strain was transformed with a second copy of the DRS2 gene carried on URA3 marked plasmid (pRS416-DRS2), and the chromosomal copy of DRS2 was disrupted. Serial dilutions of these strains (dnf1,2,3Δ pRS416-DRS2 and drs2Δ dnf1,2,3Δ pRS416-DRS2) were replica-plated to minimal medium containing 5-fluoroorotic acid (5-FOA) to select against cells that retained the pRS416-DRS2 plasmid. As shown in Figure 1B, the dnf1,2,3Δ strain could lose the DRS2 plasmid, whereas the drs2Δ dnf1,2,3Δ strain could not. In fact, the drs2Δ dnf1,2,3Δ pRS416-DRS2 strain failed to yield colonies on 5-FOA plates at any temperature tested (15, 20, 24, 30, and 37°C), indicating that the drs2Δ dnf1,2,3Δ strain is not viable. The fact that any one member of the four genes can maintain the viability of the cells (Table 3 and Figure 1) and that the quadruple mutant is not viable indicates that DRS2 and the DNF genes constitute an essential gene family with substantial functional overlap.
Figure 1.
DRS2 and DNFs constitute an essential gene family. (A) BY4742 (WT), ZHY615D1C (drs2Δ), BY4742 YER166W (dnf1Δ), ZHY2143A (drs2Δ dnf1Δ), ZHY709 (drs2Δ dnf1Δ dnf2Δ), ZHY708 (drs2Δ dnf1Δ dnf3Δ), ZHY7282C (drs2Δ dnf2Δ dnf3Δ), and PFY3273A (dnf1Δ dnf2Δ dnf3Δ) strains were grown on YPD plates at 30, 37, or 20°C as indicated in the lower right panel. (B) The dnf1,2,3Δ drs2Δ quadruple mutant is inviable. Serial dilutions of strains PFY3273A pRS416-DRS2 (dnf1,2,3Δ pRS416-DRS2) and ZHY704 (dnf1,2,3Δ drs2Δ pRS416-DRS2) were spotted on YPD or minimal 5-FOA plates, and incubated at 30°C (shown), and 37, 24, 20, or 15°C (not shown).
Growth Profiles of Double and Triple Mutants
Most of the double mutants grow as well as their parental single mutants except for drs2Δ dnf1Δ, which exhibits a strong growth defect at 30°C (Figure 1A). Interestingly, this double mutant grows much better at 37°C (Figure 1A) and can be propagated at this temperature. The remaining double mutants carrying drs2Δ exhibit the same cold-sensitive growth defect as the drs2Δ single mutant, whereas the dnf double mutants grow as well as the wild-type strain (Table 3). These data indicate that DRS2 and DNF1 show the greatest overlap in function, whereas DNF2 or DNF3 contribute little to viability in the absence of DRS2, which was surprising considering the sequence similarity between DNF1 and DNF2.
The drs2Δ dnf1,2Δ and drs2Δ dnf1,3Δ triple deletion strains exhibit a severe growth defect as predicted, because drs2Δ dnf1Δ exhibits a strong growth defect. Interestingly, the drs2Δ dnf1,2Δ and drs2Δ dnf1,3Δ strains grew slightly better than drs2Δ dnf1Δ at 30°C but fared worse at 37°C (Figure 1A). The drs2Δ dnf2,3Δ strain showed a slight temperature-sensitive growth phenotype compared with drs2Δ single mutant and the dnf1,2,3Δ strain grew nearly as well as the wild-type strain at all temperatures (Figure 1A), indicating that DRS2 by itself can fully maintain cell growth.
Sporulation Defect and Calcofluor White Hypersensitivity of drs2/dnf Mutants
During the course of generating the strains shown in Table 2, we noticed that some diploid deletion mutants could not sporulate. To further define this defect, we examined several diploid strains homozygous for the null alleles indicated in Table 4 for their ability to sporulate. Unexpectedly, the homozygous dnf1,2,3Δ diploid exhibited a severe sporulation defect even though it does not show an obvious growth defect. The drs2Δ, dnf1,2Δ, and dnf1,3Δ homozygous strains also exhibited a sporulation defect. However, dnf2,3Δ sporulated normally, as did all the single deletion dnf strains (Table 4).
The sporulation defect of homozygous diploids correlated to an increased sensitivity of the haploid mutants to CW (Table 4). Strains deficient for cell wall chitin are resistant to CW, whereas other mutants that increase chitin deposition or perturb cell wall structure are hypersensitive to this agent. The dnf1,2,3Δ strain exhibited the greatest sensitivity to CW, whereas the single mutants were similar to the wild-type strain. The drs2Δ and dnf1,2Δ strains showed an intermediate sensitivity (Table 4). We also noticed that the dnf1,2,3Δ cells are elongated when grown at lower temperature or in minimal media (our unpublished observation), which may be caused by a weakened cell wall at the bud tip. Although it is not clear whether the sporulation defect and CW hypersensitivity of these strains are caused by a common defect, these data do indicate that Dnf1p, Dnf2p, and Dnf3p overlap to exert an important cellular function that cannot be replaced by Drs2p or Neo1p.
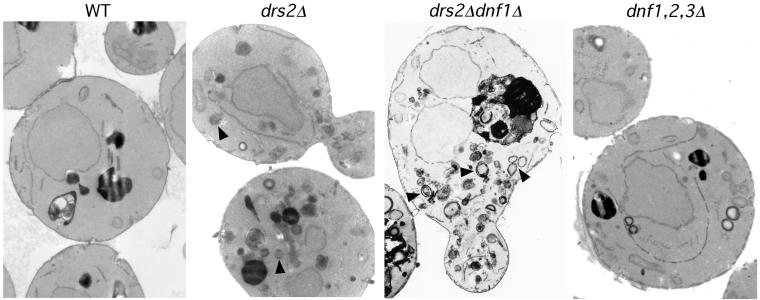
dnf1Δ Exacerbates Membrane Accumulation Defect of drs2Δ
Mutants that perturb protein transport in the secretory pathway typically accumulate abnormal membrane structures. The drs2Δ mutant accumulates abnormal membrane-bound structures containing Golgi enzymes that are similar to Berkeley bodies in morphology (Chen et al., 1999). Examination of drs2Δ dnf1Δ cells by electron microscopy (EM) revealed a massive accumulation of membrane-bound structures resembling Berkeley bodies (Figure 2), a more severe phenotype than observed in the drs2Δ single deletion cells. This is consistent with the severe growth defect of drs2Δ dnf1Δ cells and suggested that the drs2Δ dnf1Δ might exhibit a stronger defect in protein transport from the Golgi complex than either single mutant. In contrast, the dnf1,2,3Δ cells did not accumulate abnormal membrane structures and was similar in appearance to wild-type cells (Figure 2). Strains constructed in the BY4741/BY4742 background exhibit more internal membranes than other wild-type strains (e.g., SEY6210), so the appearance of the dnf1,2,3Δ cells shown is not significantly different from wild-type.
Figure 2.
drs2Δ dnf1Δ mutant massively accumulates abnormal membrane bound structures resembling Berkeley bodies. BY4742 (WT), ZHY2149D (drs2Δ dnf1Δ), and PFY3273A (dnf1,2,3Δ) cells were prepared for electron microscopy as described previously (Chen et al., 1999). Numerous double-membrane ring structures resembling Berkeley bodies (indicated with arrows) accumulate in drs2Δ dnf1Δ cells, whereas the dnf1,2,3Δ cells were similar to wild-type cells.
drs2 dnf1 Exhibits a Defect in Protein Transport to Vacuole
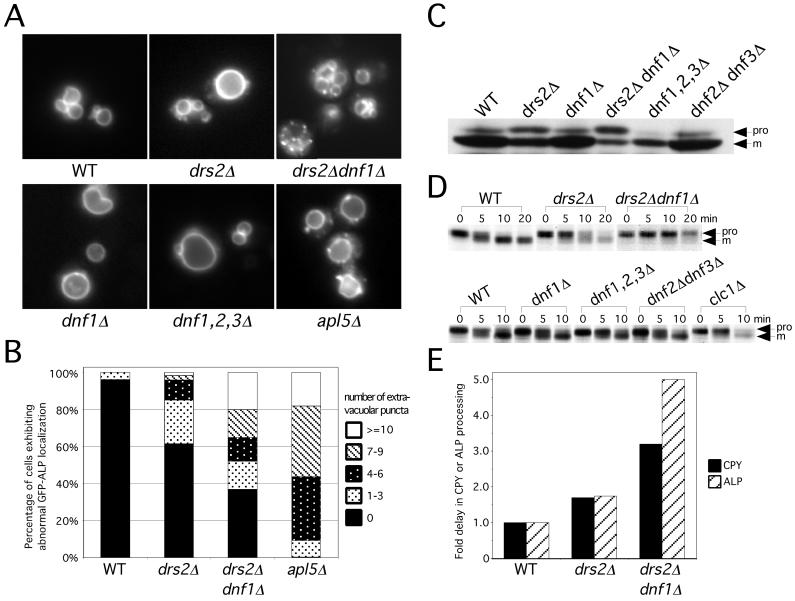
The functional redundancy between DRS2 and the DNF genes suggested that the Dnf proteins might be involved in protein transport in the secretory and/or endocytic pathways. To test this, we examined the transport of ALP and CPY to the vacuole. ALP is transported directly from the TGN to the vacuole in adaptor protein (AP)-3 coated vesicles (Cowles et al., 1997) and is processed from a high molecular weight precursor form to the mature form in the vacuole (Klionsky and Emr, 1989). To determine whether the Dnf proteins are involved in the ALP pathway, we first examined the localization of a GFP-ALP fusion protein in the different mutant strains. In wild-type cells, GFP-ALP was primarily localized to the vacuole membrane, and <4% of the cells contained one to three punctate structures outside of the vacuole (Figure 3, A and B). In drs2Δ cells, most of the GFP-ALP localized to the vacuole membrane, but an increase in the percentage of cells exhibiting extravacuolar puncta was observed. This defect in GFP-ALP localization was strongly exacerbated in the drs2Δ dnf1Δ cells, approaching the defect observed in the apl5Δ (AP-3) mutant (Figure 3, A and B). Close to 50% of the drs2Δ dnf1Δ cells contained more than four puncta, with many of the cells also containing a normal-appearing vacuole(s). This suggests that the primary defect in these cells is GFP-ALP mislocalization rather than vacuole fragmentation. This defect is specific to the drs2Δ dnf1Δ mutant cells, because neither dnf1Δ nor the dnf1,2,3Δ mutant exhibited a defect in ALP localization (Figure 3A).
Figure 3.
Drs2p and Dnf1p are required for ALP transport to the vacuole. (A) Localization of GFP-ALP. BY4742 (WT), ZHY615D1C (drs2Δ), ZHY2149D (drs2Δ dnf1Δ), BY4742 YER166W (dnf1Δ), PFY3273A (dnf1,2,3Δ), and 6210 apl5Δ harboring pGO41 (GFP-ALP) were grown at 30°C to log phase and examined by fluorescence microscope. (B) Percentage of cells exhibiting abnormal GFP-ALP localization. Cells containing 0, 1–3, 4–6, 7–9, or ≥10 punctate structures bearing GFP-ALP were counted and presented as the percentage of the total number of cells counted. Data shown is the average from counting >100 cells from three independent transformants of each strain. (C) Steady-state distribution of ALP forms. Cells were grown in YPD media at 30°C to log phase. Whole cell lysates were prepared and immunoblotted to detect precursor (pro) and mature (m) forms of ALP. (D) Kinetics of ALP transport to the vacuole. Cells were grown to log phase at 30°C, labeled with [35S]methione/cysteine for 5 min and chased at 0, 5, 10, and 20 min at 30°C. ALP was recovered from each sample by immunoprecipitation and subjected to SDS-PAGE. (E) CPY and ALP processing rate assessed from pulse-chase experiments were plotted relative to WT cells. Data shown is the average of at least three experiments.
Western blots indicated that the drs2Δ dnf1Δ cells accumulated more ALP precursor (49 ± 29% of total ALP, n = 7) than the wild-type cells (4 ± 0%). The drs2Δ dnf1Δ strains tended to pick up faster growing suppressors, which probably contributed to the wide range of error for this mutant. There was also an intermediate level of proALP accumulation in the drs2Δ cells (18 ± 8%), whereas dnf1Δ was indistinguishable from wild type (Figure 3C). To follow the transport kinetics of ALP in these mutant strains, cells were pulse-labeled and chased for the times indicated in Figure 3D. Consistent with the previous results, ALP processing is significantly delayed (∼5-fold) in the drs2Δ dnf1Δ mutant compared with the wild-type cells (Figure 3E). The drs2Δ mutant again showed an intermediate phenotype, whereas the dnf1,2,3Δ cells exhibited normal kinetics of ALP processing (Figure 3D). We conclude from these data that Drs2p and Dnf1p are required for the ALP transport pathway, whereas dnf1,2,3Δ does not affect this pathway.
We next examined the CPY transport pathway in these mutants. CPY is synthesized in the endoplasmic reticulum (ER) as the p1 precursor form and is modified on N-linked oligosaccharides by Golgi mannosyltransferases to form the p2 precursor. p2 CPY is sorted from secreted proteins in the TGN and is transported through the late endosome to the vacuole where it is processed to the mature form (mCPY) (Stevens et al., 1982). The CPY pathway is independent of AP-3 but is perturbed in vps mutants, which secrete the p2 form of CPY (Conibear and Stevens, 1998). For example, vps35 secretes p2 CPY and produces a dark signal on a colony blot probed for CPY (Figure 4A). The drs2Δ strain exhibits a very modest CPY secretion phenotype, which does not seem to be exacerbated in the drs2Δ dnf1Δ double mutant. Neither dnf1Δ nor the dnf1,2,3Δ triple mutant secreted CPY (Figure 4A). None of the drs2/dnf deletion strains tested accumulated a significant amount of p2 CPY as detected by Western Blot (Figure 4B), although a modest increase was observed in the drs2Δ dnf1Δ mutant (Figure 4B). To further explore the kinetics of CPY processing, these strains were subjected to a pulse-chase analysis at 30°C. The drs2Δ mutant exhibited a 1.7 fold kinetic delay in CPY transport, whereas the drs2Δ dnf1Δ mutant exhibited a 3.2-fold kinetic delay (Figures 3E and 4C). We had previously noted a partial, Golgi-specific glycosylation defect in drs2Δ that prevents a clear separation of p1 and p2 CPY by SDS-PAGE (Chen et al., 1999). This is also evident in the drs2Δ dnf1Δ cells (Figure 4C), making it difficult to assess the kinetics of ER-to-Golgi transport for CPY. However, analysis of α-factor transport (described below) indicated that protein transport from the ER to the TGN is unaffected in the drs2Δ dnf1Δ mutant. Thus, we see exacerbation of the CPY transport defect in the drs2Δ dnf1Δ double mutant, although not as substantial as observed for the ALP pathway. In addition, the Western blot seemed to show that there was less CPY precursor in the dnf1,2,3Δ mutant. However, the pulse-chase result indicated that the kinetics of CPY transport was normal in this mutant (Figure 4C). Similarly, the other mutants displayed wild-type kinetics of CPY transport (Figure 4C).
Figure 4.
CPY pathway is partially affected in drs2/dnf deletion strains at 30°C. (A) Colony-blot analysis of CPY secretion. Freshly streaked BY4742 (WT), BY4742 YER166W (dnf1Δ), PFY3273A (dnf1,2,3Δ), ZHY615D1C (drs2Δ), ZHY2149D (drs2Δ dnf1Δ) and 6210 vps35Δ cells were grown on YPD plates at 30°C (shown), and 37 or 20°C (data not shown) for 24 h. Nitrocellulose membranes were overlaid onto the colonies and incubated for another 24 h. Membranes were washed and probed with a monoclonal CPY antibody. (B) Steady-state distribution of CPY precursor forms. Cells were grown in YPD medium at 30°C to log phase. Whole cell lysates were prepared and subjected to SDS-PAGE and Western blotting to detect p1, p2 and mature forms of CPY. (C) Kinetics of CPY transport. Cells were grown to log phase at 30°C, labeled with [35S]methionine/cysteine for 5 min, and chased at 0, 5, and 10 min at 30°C. CPY was recovered from each sample by immunoprecipitation and subjected to SDS-PAGE.
dnf1Δ Does Not Exacerbate Pro-α-Factor Processing and Invertase Secretion Defects of drs2Δ
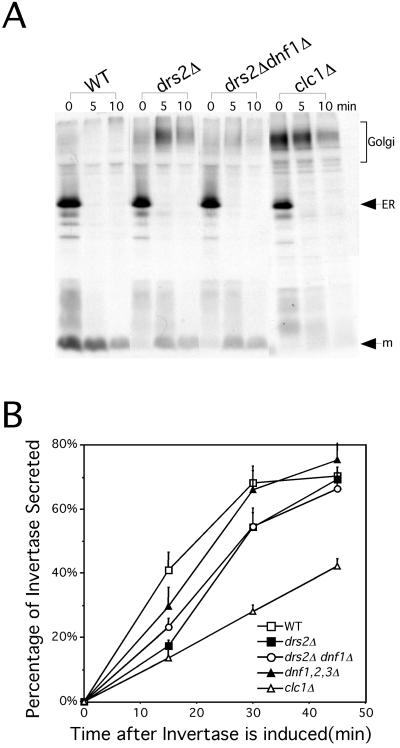
The yeast α-factor mating pheromone is synthesized in the ER as a high molecular weight precursor and further modified in the Golgi complex to produce the heterogeneously glycosylated pro-α-factor form, which migrates as a high molecular mass smear by SDS-PAGE. This precursor is further processed in the TGN through a series of proteolytic events, initiated by Kex2p, to produce the mature α-factor peptide, which is secreted (Fuller et al., 1988). Kex2p normally cycles between the TGN and the endosomal system (Conibear and Stevens, 1998). Mutants that mislocalize Kex2p and other TGN proteases required for pro-α-factor processing secrete the high molecular weight Golgi form of pro-α-factor (Payne and Schekman, 1989). The drs2Δ mutant exhibited a defect in pro-α-factor processing at 30°C (Figure 5A), although this phenotype is more severe at lower temperatures (Chen et al., 1999). The drs2Δ dnf1Δ cells also exhibited an incomplete processing of pro-α-factor, but this defect was actually less severe than that observed in the drs2Δ mutant (Figure 5A). The rapid disappearance of the ER form of pro-α-factor in the mutant cells indicates that ER-to-Golgi transport was not perturbed. As shown before, the clathrin light chain mutant (clc1Δ) exhibited a substantial defect with very little mature α-factor being produced. Furthermore, the dnf1Δ, dnf1,2,3Δ, and dnf2,3Δ strains processed pro-α-factor normally (our unpublished observation). Thus, dnf1Δ does not exacerbate the Kex2p mislocalization phenotype of drs2Δ.
Figure 5.
dnf1Δ does not exacerbate the pro-α-factor processing and invertase secretion defects of drs2Δ. (A) Kinetics of pro-α-factor processing. Cells were grown to log phase at 30°C, labeled with [35]methione/cysteine for 5 min, and chased at 0, 5, and 10 min at 30°C. Proα-factor was recovered from each sample by immunoprecipitation and subjected to SDS-PAGE. (B) Invertase secretion assay. Cells were grown in 5% YP glucose media at 30°C to log phase and shifted to 0.1% YP glucose media to start the induction of invertase. Aliquots of cells were collected at 0, 15, 30, and 45 min after invertase is induced. Secreted and total invertase activities were measured and expressed as the percentage of invertase secreted.
There is evidence for two classes of late secretory vesicles in yeast: a dense class carrying invertase and a lighter class carrying Pma1p (Harsay and Bretscher, 1995). In addition, clathrin is required to sort invertase into the dense vesicles, and in the absence of clathrin, invertase is shunted to the Pma1p vesicle pathway (Gurunathan et al., 2002; Harsay and Schekman, 2002). Thus, secretion of invertase is not blocked, although a kinetic defect is observed. To test whether the Dnf proteins are involved in exocytosis, we measured the kinetics of invertase secretion in the different dnf deletion strains. Cells were grown in 5% YP glucose media to log phase and then shifted to 0.1% YP glucose media to induce invertase synthesis. The rate of induction was equivalent for all strains tested (our unpublished observation), so the initial rate of invertase appearance outside of the cell reflected the rate of secretion. In wild-type cells, 40% of newly synthesized invertase was secreted within 15 min after induction and started to plateau (70%) at ∼30 min (Figure 5B). The dnf1,2,3Δ cells exhibited wild-type kinetics of invertase secretion while the drs2Δ and clathrin mutants exhibited a significant delay in this process. Although the drs2Δ dnf1Δ mutant exhibits severe growth and membrane accumulation defects, the invertase secretion defect is no more severe than that of the drs2Δ cells (Figure 5B). These results suggest that Drs2p is the only family member that contributes significantly to the invertase secretion pathway, and that the severe growth and membrane accumulation defects of the drs2Δ dnf1Δ cells, compared with drs2Δ cells, is not due to a more severe defect in secretion.
dnf1,2,3Δ Exhibits a Defect in Recycling and Polarized Localization of GFP-Snc1p
Other than the involvement of Dnf1p in the vacuolar protein transport pathways with Drs2p, we have not found a requirement for the other Dnf proteins in the secretory pathway. We examined the endocytic pathway in these mutants by labeling and chasing the cells with FM4-64, a fluorescent endocytic marker (Vida and Emr, 1995). All the dnf mutants, including the double and triple deletion mutants, were able to endocytose the FM4-64 and deliver the dye to the vacuole at 30°C as well as the wild-type strain (our unpublished observations). Interestingly, at 15°C the dnf1,2,3Δ cells seemed to internalize FM4-64 from the plasma membrane more rapidly than wild-type cells, but exhibited a delay in transport from endosomes to the vacuole (Figure 6). Surprisingly, the dnf1,2Δ and dnf1,3Δ exhibited a delay in FM4-64 internalization from plasma membrane at 15°C, but without accumulation of internal punctate structures. The reason for these differences between the dnf1,2,3Δ triple mutant and the double mutants is unclear. However, all these defects are cold sensitive and reversible, because when cells were shifted up to 22°C, the FM4-64 moved quickly to the vacuole (our unpublished observations).
Figure 6.
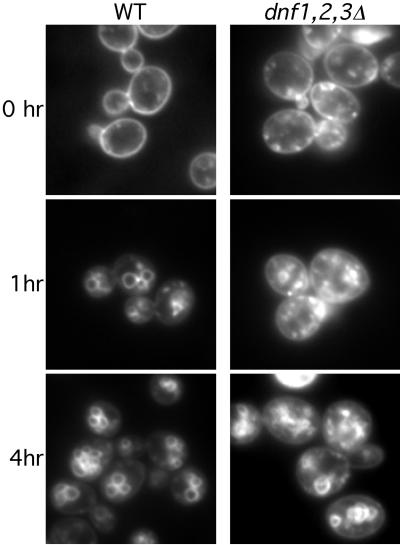
dnf1,2,3Δ mutant exhibits a cold-sensitive defect in FM4-64 transport to the vacuole. Cells were stained with FM4-64 on ice and shifted to 15°C for 0, 1, or 4 h and then viewed by fluorescence microscopy.
During the FM4-64 labeling experiments, we found that the drs2Δ and dnf1,2,3Δ cells always seemed to internalize FM4-64 faster than wild-type cells at either 15 or 30°C. Others have proposed that after internalization of FM4-64 to an early endosome, a portion of the dye follows a recycling pathway to the TGN and then back to the plasma membrane (Lewis et al., 2000). In this case, a defect in the recycling pathway will cause an apparent increase in the rate of FM4-64 internalization. To directly test whether the recycling pathway is perturbed, we examined the localization of a GFP-Snc1p fusion protein as a marker to follow this pathway. Snc1p is an exocytic v-SNARE protein (Gerst, 1999) that has been shown to recycle by endocytosis from the plasma membrane to an early endosome, and from there it is sorted to the TGN. From the TGN, Snc1p can be packaged into exocytic vesicles to target them to the plasma membrane (Lewis et al., 2000).
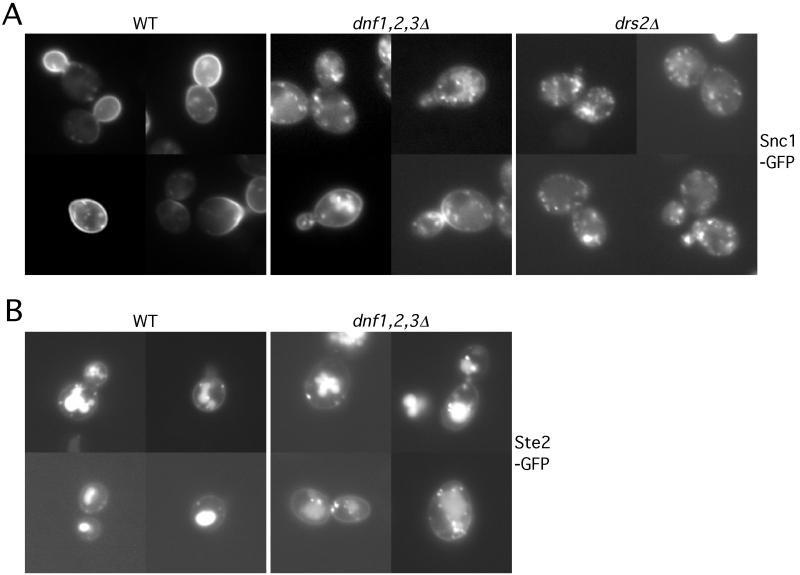
In wild-type cells, GFP-Snc1p was primarily localized to the plasma membrane, concentrating in the bud or the regions of polarized growth. Only a small fraction of the GFP-Snc1p was observed in punctate structures within the cell (Figure 7A). In contrast, there was a substantial decrease of GFP-Snc1p localized to the plasma membrane in dnf1,2,3Δ cells, with a concomitant increase to internal punctate structures (Figure 7A), which could be early endosomes or TGN compartments. The GFP-Snc1p that remains on the plasma membrane was evenly distributed between the mother and daughter cells, suggesting a defect in the polarized delivery of Snc1p to the bud. The mislocalization of Snc1p in the dnf1,2,3Δ cells supports the hypothesis that recycling from the early endosome back to the plasma membrane is disturbed in this mutant. Because the kinetics of secretion seems normal in the dnf1,2,3Δ cells (Figure 5B), we propose that the dnf1,2,3Δ mutant is defective in the endosome to TGN branch of this pathway. The dnf single and dnf2,3Δ mutants traffic GFP-Snc1p normally; however, the dnf1,2Δ and dnf1,3Δ mutants do exhibit a noticeable defect, although less severe than in the dnf1,2,3Δ triple mutant (our unpublished observation). The drs2Δ cells exhibit a similar GFP-Snc1p mislocalization defect as shown in dnf1,2,3Δ cells (Figure 7A), which is also consistent with the more efficient internalization of FM4-64 observed in these cells. However, this could be explained by the exocytic defect of drs2Δ, and so it is unclear whether drs2Δ also has an endosome to TGN transport defect.
Figure 7.
dnf1,2,3Δ mutant exhibits a defect in Snc1p-GFP recycling. (A) pRS416 SNC1-GFP was introduced into BY4742 (WT), PFY3273A (dnf1,2,3Δ) and ZHY615D1C (drs2Δ) strains. Cells were grown at 30°C to log phase and examined by fluorescence microscopy. (B) Cells harboring pRS426 STE2-GFP were grown at 30°C to log phase and examined by fluorescence microscopy. MetaMorph 4.5 was used to process the image and measure the fluorescence intensity on cell surface and in the vacuoles.
To rule out the possibility that Snc1p is actually internalized faster in the dnf1,2,3Δ cells rather than a slowed recycling back to the plasma membrane, we examined the localization of GFP-Ste2p in these cells (Figure 7B). Ste2p, the α-factor receptor, is constitutively internalized from the plasma membrane and delivered through the endosomal system to the vacuole for degradation (Schandel and Jenness, 1994). If endocytosis is enhanced in the dnf1,2,3Δ cells, we would expect to see a decrease in plasma membrane fluorescence relative to internal signal as shown for Snc1p. However, in both wild-type and the dnf1,2,3Δ cells, the ratio of Ste2p-GFP fluorescence intensity on the plasma membrane to that in the vacuole was 0.2 (plasma membrane pixel value ÷ vacuolar pixel value). This result suggests that the mislocalization of Snc1p in the dnf1,2,3Δ cells is not caused by enhanced endocytosis, but is due to inefficient transport in the endosome → TGN → plasma membrane loop.
Dnf Protein Expression and Localization
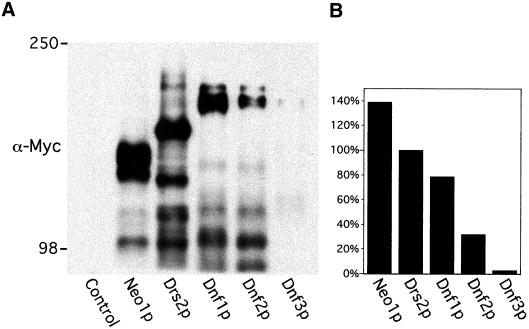
To study the expression pattern of the Dnf proteins, we epitope-tagged all five DRS2/NEO1 family members with c-Myc at the carboxyl-terminal end of these proteins. Immunoblotting of whole cell extracts indicated that all three previously uncharacterized genes (DNF1, 2, and 3) were expressed under normal growth conditions (Figure 8A). Neo1p and Drs2p are expressed at the highest level, whereas Dnf3p is expressed at a very low level compared with the other proteins (Figure 8B). This is consistent with the observation that Dnf3p seems to contribute the least to both cell viability and CW sensitivity. The protein level for each family member was greatly decreased when the cells were grown at 37°C and were moderately higher at 20°C as revealed by both Western blot and immunofluorescence analyses (our unpublished observations). Consistent with the growth profiles, this suggests that cells have a greater requirement for Drs2p and Dnf proteins at lower temperatures. The marked susceptibility to proteolysis, noted previously for Drs2p, is also the case for Neo1p and the Dnf proteins. We cannot distinguish at this time whether the proteolysis occurs in vivo or during preparation of the cell lysates.
Figure 8.
Expression levels of Drs2/Neo1 family proteins. (A) Strains BY4742 (control), ZHYNEO1-MYC, ZHYDRS2-MYC, ZHYDNF1-MYC, ZHYDNF2-MYC, and ZHYDNF3-MYC were grown in YPD media at 30°C. Equal amounts of whole cell lysates were subjected to SDS-PAGE and immunoblotting with a monoclonal c-Myc antibody to detect the fusion proteins. The migration of standard protein markers is labeled at the left. Predicted molecular mass of each full-length protein fused with 13XMyc tag is Neo1p, 146 kDa; Drs2p, 170 kDa; Dnf1p, 194 kDa; Dnf2p, 198 kDa; and Dnf3p, 204 kDa. Each lane contained an equivalent amount of CPY (not shown). (B) Full length and near full-length bands from A were analyzed by NIH Image 1.62. The signal intensity of each fusion protein was plotted as the percentage of Drs2p signal strength. Data shown in this figure is representative of three experiments.
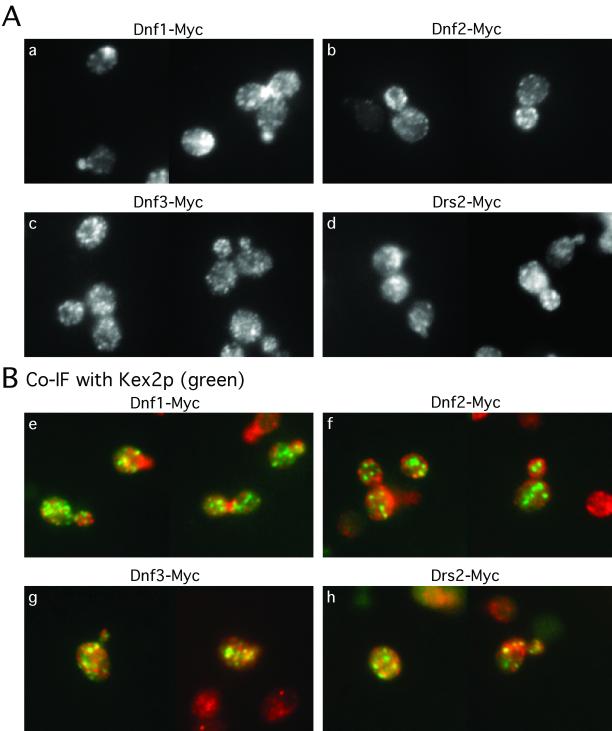
We then examined the localization of the epitope-tagged proteins by immunofluorescence microscopy and found that all the proteins exhibited a punctate staining pattern. However, the three Dnf proteins show different localization patterns (Figure 9A). Dnf1p is found in internal membranes and the plasma membrane, but it was more concentrated at the emerging bud site, small buds, and the mother-daughter neck of dividing cells (Figure 9, a and e), which is very similar to the localization of cortical actin patches. This suggests that Dnf1p may be concentrated in exocytic vesicles targeted to polarized sites of growth. Dnf2p is found in unique, smaller punctate structures that lie directly underneath the plasma membrane (Figure 9, b and f), but it does not exhibit a polarized distribution as Dnf1p does. The punctate structures might be endocytic vesicles or plasma membrane invaginations. In some cells, small regions of continuous plasma membrane staining could be seen for Dnf2p. Dnf3p staining reveals punctate structures evenly spread throughout the cell, which resembles the staining pattern for Drs2p (Figure 9, c, d, g, and h).
Figure 9.
Localization of Dnf proteins. Cells were grown in YPD media at 30°C and prepared for immunofluorescence. (A) Cells were visualized with mouse monoclonal c-Myc antibody (a–d). (B) Cells transformed with a 2 μ KEX2HA plasmid were stained with a mouse monoclonal c-Myc antibody and a rabbit polyclonal HA antibody to visualize Myc-tagged Drs2/Dnf proteins (red) and HA-tagged Kex2p (green) (e–h). The yellow punctate structures indicate the colocalization of Drs2/Dnf proteins with Kex2p.
Even though Dnf1p and Dnf2p exhibit unique localization patterns clustered on or near the plasma membrane, both were also found distributed in internal structures as well. Drs2p colocalizes substantially with the TGN marker Kex2p. We examined the colocalization of the Dnf proteins with Kex2p and found that all of them colocalized with Kex2p to some extent (Figure 9B). Dnf3p showed the greatest overlap with Kex2p (Figure 9B, g) comparable with the staining pattern observed for Drs2p (Figure 9B, h). Dnf1p and Dnf2p showed the least overlap with Kex2p, and further experiments will be required to determine whether a small percentage of Dnf1p and Dnf2p is localized to the TGN. The localization of the Dnf1 and Dnf2 proteins to plasma membrane and what seem to be transient exocytic and/or endocytic membranes suggests that these proteins might cycle in a TGN-plasma membrane–endosome–TGN pathway. This is consistent with the defect in GFP-Snc1 recycling in the dnf1,2,3Δ mutant.
DISCUSSION
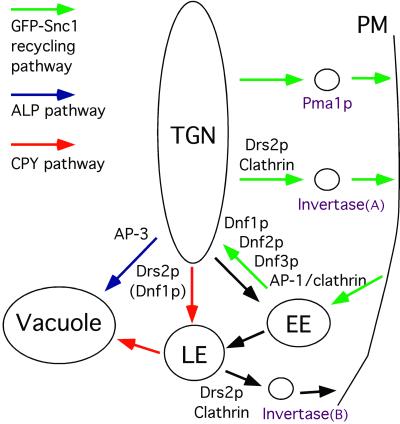
We have previously implicated Drs2p in clathrin function at the TGN (Chen et al., 1999), and this report focuses on the requirements for Drs2p and Dnfs in protein transport. We find that 1) the DRS2 and DNF genes constitute an essential gene family with overlapping functions. 2) Specific protein transport steps in the late secretory, endocytic, and vacuolar pathways have specific requirements for Drs2/Dnf proteins (Figure 10). 3) Drs2p, Dnf1p, and Dnf2p exhibit unique localization patterns, suggesting that each may have a primary function at different transport steps.
Figure 10.
Model for the roles of Drs2p and Dnf proteins in late secretory, vacuolar, and endocytic pathways. PM, plasma membrane; EE, early endosome; LE, late endosome.
Of the five DRS2/NEO1 family genes, NEO1 is the only essential gene and it does not seem to overlap genetically with the other four family members. Yeast can survive with multiple combinations of null alleles for DRS2, DNF1, 2, and 3 but the quadruple mutant is inviable. The fact that any single member of the Drs2/Dnf group can support cell growth demonstrates a functional interchangeability for these proteins, and suggests that each protein shares a common biochemical activity. A substantial body of work suggests that these proteins are APTs, although this activity has not yet been reconstituted with purified enzyme. Although it is possible that the Drs2 and Dnf proteins transport different substrates, it seems more likely that the existence of multiple members of this family reflects a requirement for these proteins at different subcellular organelles. This view is supported by the unique localization patterns observed for Drs2p, Dnf1p, and Dnf2p.
Role of Drs2p and Dnf1p in Protein Transport from TGN
Drs2p and Dnf1p seem to contribute most of the essential function of the Drs2/Dnf subgroup because the drs2Δ dnf1Δ double mutant grows very slowly. In addition, dnf1Δ strongly exacerbates the membrane accumulation phenotype of drs2Δ, suggesting an overlap in function for protein and membrane exit from the Golgi complex. Multiple exit pathways were examined and we found that the ALP pathway was most significantly perturbed in the drs2Δ dnf1Δ double mutant. A less severe synthetic defect in the CPY pathway was also observed in this double mutant, although the defects in the invertase secretion kinetics or pro-α-factor processing observed in the drs2Δ mutant were not exacerbated in the double mutant.
The redundant roles for Drs2p and Dnf1p in the ALP pathway are somewhat surprising because only a fraction of Dnf1p seems to localize to the TGN, where Drs2p is localized. Rather, Dnf1p is found in membranes concentrated near sites of polarized growth in wild-type cells. These include the incipient bud site, tips of small buds, and the mother/bud neck during cytokinesis, a distribution very similar to that of actin cortical patches in yeast. This dynamic localization pattern suggests that Dnf1p is cycling between the exocytic and endocytic pathways and could therefore be a transient occupant of the TGN. Because drs2Δ partially perturbs the exocytosis of invertase, it is possible that Dnf1p exocytosis is also inefficient, resulting in an increased residence in the TGN of drs2Δ cells. This could allow Dnf1p to partially compensate for the loss of Drs2p in the ALP pathway. We have observed modest defects in ALP transport in the drs2Δ single mutant but not in the dnf1Δ mutant. Therefore, we suggest that Drs2p play a primary role, and Dnf1p a compensatory role, in budding AP-3–coated vesicles carrying ALP to the vacuole (Figure 10).
Clathrin has recently been implicated in the invertase secretion pathway with the observation that clathrin mutants missort invertase into the less dense vesicle fractions containing Pma1p (Gurunathan et al., 2002; Harsay and Schekman, 2002). Mutations that perturb the actin cytoskeleton also affect invertase secretion (Novick and Botstein, 1985; Mulholland et al., 1997) and cause the accumulation of the invertase class of exocytic vesicles (Harsay and Bretscher, 1995). This is presumably because the targeted delivery of these vesicles to polarized sites of growth requires actin. We have found that Drs2p and clathrin are required to form vesicles that accumulate upon actin disruption and that these vesicles contain acid phosphatase, another cargo of the dense exocytic vesicles (Gall et al., submitted). Because Drs2p localizes to the TGN and this seems to be site where invertase and Pma1p are segregated into separate exocytic pathways, we prefer a model in which the invertase vesicles bud directly from the TGN for targeting to polarized sites of growth [Figure 10, Invertase (A)].
Surprisingly, the sorting of invertase and Pma1p into two separate exocytic pathways at the TGN is also perturbed by several mutations that disrupt protein trafficking through the late endosome. These observations have led others to propose that invertase is sorted first to a late endosome and is then packaged into clathrin-coated vesicles for exocytosis (Gurunathan et al., 2002; Harsay and Schekman, 2002) [Figure 10, Invertase (B)]. However, other interpretations are possible. For example, late Golgi proteins continuously cycle between the TGN and endosomes (Conibear and Stevens, 1995) and so a strong block in endosome trafficking could rapidly deplete the TGN resident proteins required for sorting invertase into CCVs. In addition, there is no direct evidence that invertase passes through the late endosome (or prevacuolar compartment) of wild-type cells before it is secreted. Further work will be required to clearly define the donor compartment that buds the invertase vesicles.
A function for Drs2p in vesicle budding was first suggested by the discovery of a synthetic lethal interaction between drs2Δ and arf1 mutations (Chen et al., 1999). ARF has been implicated in recruiting tetrameric adaptins (AP-1, AP-3, and AP-4) and clathrin to the TGN and endosomes (Boehm et al., 2001; Ooi et al., 1998; Zhu et al., 1999). The CPY pathway may use clathrin and the GGA adaptins, also ARF-dependent for membrane association (Boman et al., 2000; Hirst et al., 2000; Dell'Angelica et al., 2000; Deloche et al., 2001; Costaguta et al., 2001; Mullins and Bonifacino, 2001), for exit from the TGN. The requirements for Drs2p and Dnf proteins in these different ARF-dependent pathways are very interesting. Drs2p seems to be the only family member capable of supporting the clathrin-dependent invertase pathway because the drs2Δ single mutant exhibits a defect in this pathway (Figure 5B; Gall et al., 2002). However, both Drs2p and Dnf1p seem to be capable of contributing to the AP-3/ALP pathway. The CPY transport defect observed in the drs2Δ dnf1Δ mutant suggests that the Drs2/Dnf proteins may also contribute to a GGA-dependent pathway as well (Figure 10). Unfortunately, we have not yet successfully isolated conditional alleles of the DRS2/DNF genes that would allow us to score the immediate consequences of a complete loss of function for all four proteins.
Requirement for Dnf1, 2, and 3p in Snc1p Recycling Pathway
The dnf1,2,3Δ mutant grows well and does not exhibit a defect in the ALP, CPY, or invertase pathways. However, this mutant does exhibit a striking defect in GFP-Snc1p recycling. Snc1p is a v-SNARE that is thought to function in exocytic and endocytic vesicles, but at steady-state most of the GFP-Snc1p fluorescence is concentrated on the plasma membrane of wild-type cells (Gerst, 1999; Lewis et al., 2000). This localization pattern suggests that internalization of GFP-Snc1p from the plasma membrane is rate limiting in the recycling pathway shown in Figure 10. This localization pattern is altered in the dnf1,2,3Δ mutant with most of the GFP-Snc1p concentrated in internal structures. Because exocytosis does not seem to be perturbed, we suggest that endosome to TGN transport is partially defective in this mutant. The recycling defect could also explain the CW hypersensitivity and sporulation defects of the dnf1,2,3Δ mutant. Chitin synthase III (Chs3p) is known to be dynamically recycled through endocytic intermediates for targeting to polarized sites of growth (Chuang and Schekman, 1996; Santos and Snyder, 1997). Recent work strongly suggests that ARF, AP-1, and clathrin are required for transport of Chs3p from an early endosome to the TGN. Mutations in these clathrin coat components results in an unregulated transport of Chs3p to the cell surface, and increased sensitivity of cells to CW (Valdivia et al., 2002). Clathrin and AP-1 have also been implicated in endosome → TGN as well as TGN → plasma membrane routes in mammalian cells (Folsch et al., 1999; Meyer et al., 2000; Boehm and Bonifacino, 2001). The formation of spore membranes and cell walls may also require a dynamic recycling of materials. In fact, Chs3p is delivered to the prospore membranes for synthesis of the spore cell wall (Pammer et al., 1992), and homozygous clathrin mutants also fail to sporulate (Lemmon et al., 1990).
The localization of Dnf2p to both internal structures as well as the plasma membrane suggests that this protein may also be cycling between the endocytic and exocytic pathways. Thus, Dnf1p and Dnf2p would be positioned to directly contribute to the clathrin/AP-1–dependent endosome to TGN route. Remarkably, our studies position a Drs2-related P-type ATPase in each pathway where an ARF/AP/clathrin-dependent vesicle budding event has been implicated. In fact, we have observed a dramatic defect in the ability of drs2Δ mutants to produce CCVs at 15°C, whereas normal-appearing CCV can be isolated at 30°C. We postulate that Dnf1, 2, and 3 proteins facilitate CCV budding in the drs2Δ mutant at 30°C but fail to do so at lower temperatures. This would explain why the clathrin deficiency phenotypes of drs2Δ are much more severe at 20°C or below.
APT Genes in Other Organisms
The DRS2/NEO1 family of potential APT genes exists ubiquitously in eukaryotes, and a few other members of this large family have been identified genetically. The PDE1 gene from rice blast fungus was identified in a screen for pathogenicity factors and is most similar to DNF3. Mutation in PDE1 perturbs formation of penetration hyphae, a narrow outgrowth of the cell surface used to puncture the plant cuticle. The Talbot group has suggested that Pde1p could play a role in the massive recycling of membrane and cell wall biosynthetic enzymes required for the formation of penetration hyphae (Balhadere and Talbot, 2001). The defect in GFP-Snc1p recycling and polarized delivery we have observed in the dnf1,2,3Δ mutant is consistent with this interpretation.
The ALA1 gene from Arabidopsis is most similar to DRS2 and remarkably, decreased expression of ALA1 causes cold-sensitive growth of Arabidopsis. In addition, the ALA1 gene can complement the cold-sensitive growth defect of the drs2Δ mutant underscoring the conservation of Drs2p/Ala1 function across phylogenetic boundaries. The ALA1 gene also complements a cold-sensitive defect in APT activity at the plasma membrane of drs2Δ cells (Gomes et al., 2000). Because Drs2p is not localized to the plasma membrane but is required for protein transport in the late secretory pathway, this defect may be caused by mislocalization of other APTs. Our localization data suggest that Dnf1 and Dnf2 are the best candidates for the plasma membrane APT in yeast, and we are currently testing this possibility.
The roles of the Drs2/Dnf family in protein traffic in yeast could also shed light on the function of their homologs implicated in human disease. Both FIC1 and BSEP mutations cause familial intrahepatic cholestasis in humans. FIC1 is most homologous to the DNF1 and DNF2 genes in yeast and BSEP is a bile salts export pump (in the ABC ATPase family) localized to the bile canalicular plasma membrane of hepatocytes (Thompson and Jansen, 2000; Ujhazy et al., 2001). How FIC1 is involved in bile production is not clear, but based on the phenotype of the drs2/dnf yeast mutants, it is possible that FIC1 might be required for the polarized delivery of BSEP to the canalicular membrane.
ATP10C, another member of the APT family in humans, has been linked to the neurological disorders Angelman syndrome and autism (Herzing et al., 2001; Meguro et al., 2001). Recycling of membrane from synapses plays a critical role in synaptic vesicle production, a process where ARF and AP-3 have been implicated (Faundez et al., 1998; Kantheti et al., 1998). In addition, the recycling or polarized delivery of AMPA receptors to specific sites on the plasma membrane is essential for proper function of neurons and may contribute to long-term potentiation (Luscher and Frerking, 2001). Interestingly, mutations in the mouse ATP10C homolog pfatp are linked to increased body fat (Dhar et al., 2000). This phenotype could have a neurological basis or could reflect perturbations in the trafficking of glucose transporters or insulin receptors in adipocytes.
The precise mechanism by which Drs2p and the Dnf proteins contribute to protein trafficking remains to be elucidated. In the case of Drs2p, mutation of aspartic acid critical for ATPase activity strongly perturbs the formation of clathrin-coated vesicles targeted to the plasma membrane (Gall at al., 2002). This suggests that lipid translocation is required to either generate a membrane environment permissive for vesicle budding or perhaps to help deform the membrane during vesicle budding (see Chen et al., 1999, for further discussion on the potential roles of lipid translocases in vesicle budding). Further biochemical analysis of these proteins will be required to better assess their role in protein trafficking.
ACKNOWLEDGMENTS
We thank Scott Emr, Hugh Pelham, Jeff Flick, and Steve Nothwehr for antibodies, strains, or plasmids. We thank Howard Wynder for assistance with electron microscopy. We also thank Walter Gall, Sophie Chen, Diane Hopkins, and the other members of the Graham laboratory for help during the course of these experiments. This work was supported by National Institutes of Health Grant GM-62367) (to T.R.G.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0172. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0172.
REFERENCES
- Auland ME, Roufogalis BD, Devaux PF, Zachowski A. Reconstitution of ATP-dependent aminophospholipid translocation in proteoliposomes. Proc Natl Acad Sci USA. 1994;91:10938–10942. doi: 10.1073/pnas.91.23.10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balhadere PV, Talbot NJ. PDE1 Encodes a P-type ATPase involved in appressorium-mediated plant infection by the rice blast fungus Magnaporthe grisea. Plant Cell. 2001;13:1987–2004. doi: 10.1105/TPC.010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Aguilar RC, Bonifacino JS. Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs) EMBO J. 2001;20:6265–6276. doi: 10.1093/emboj/20.22.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS. Adaptins: the final recount. Mol Biol Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Zhang C, Zhu X, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catty P, de Kerchove d'Exaerde A, Goffeau A. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 1997;409:325–332. doi: 10.1016/s0014-5793(97)00446-8. [DOI] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p [published erratum appears in J. Cell Biol. (1996) 135, 1925] J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vacuolar biogenesis in yeast: sorting out the sorting proteins. Cell. 1995;83:513–516. doi: 10.1016/0092-8674(95)90088-8. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS. Yeast GGA coat proteins function with clathrin in Golgi to endosome transport. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, et al. YPD, PombePD, and WormPD. Model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloche O, Yeung BG, Payne GS, Schekman R. Vps10p transport from the trans-Golgi network to the endosome is mediated by clathrin-coated vesicles. Mol Biol Cell. 2001;12:475–485. doi: 10.1091/mbc.12.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar M, Webb LS, Smith L, Hauser L, Johnson D, West DB. A novel ATPase on mouse chromosome 7 is a candidate gene for increased body fat. Physiol Genomics. 2000;4:93–100. doi: 10.1152/physiolgenomics.2000.4.1.93. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- Faundez V, Horng JT, Kelly RB. A function for the AP3 coat complex in synaptic vesicle formation from endosomes. Cell. 1998;93:423–432. doi: 10.1016/s0092-8674(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Folsch H, Ohno H, Bonifacino JS, Mellman I. A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell. 1999;99:189–198. doi: 10.1016/s0092-8674(00)81650-5. [DOI] [PubMed] [Google Scholar]
- Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Gall WE, Geething NC, Hua Z, Ingram MF, Liu K, Chen S, Graham TR. Drs2p-dependent budding of clathrin-coated vesicles in vivo. 2002. (in press). [DOI] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst JE. SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell Mol Life Sci. 1999;55:707–734. doi: 10.1007/s000180050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Lampen JO. β-d-Fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Gomes E, Jakobsen MK, Axelsen KB, Geisler M, Palmgren MG. Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell. 2000;12:2441–2454. [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S, David D, Gerst JE. Dynamin and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 2002;21:602–614. doi: 10.1093/emboj/21.4.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halleck MS, Pradhan D, Blackman C, Berkes C, Williamson P, Schlegel RA. Multiple members of a third subfamily of P-type ATPases identified by genomic sequences and ESTs. Genome Res. 1998;8:354–361. doi: 10.1101/gr.8.4.354. [DOI] [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E, Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–286. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing LB, Kim SJ, Cook EH, Jr, Ledbetter DH. The human aminophospholipid-transporting ATPase gene ATP10C maps adjacent to UBE3A and exhibits similar imprinted expression. Am J Hum Genet. 2001;68:1501–1505. doi: 10.1086/320616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lui WW, Bright NA, Totty N, Seaman MN, Robinson MS. A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, Carskadon SL, Kapfhamer D, Sufalko D, Robinson MS, Noebels JL, Burmeister M. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon SK, Freund C, Conley K, Jones EW. Genetic instability of clathrin-deficient strains of Saccharomyces cerevisiae. Genetics. 1990;124:27–38. doi: 10.1093/genetics/124.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Moir RD, Sethy-Coraci IK, Warner JR, Willis IM. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol Cell Biol. 2000;20:3843–3851. doi: 10.1128/mcb.20.11.3843-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher C, Frerking M. Restless AMPA receptors. implications for synaptic transmission and plasticity. Trends Neurosci. 2001;24:665–670. doi: 10.1016/s0166-2236(00)01959-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx U, Polakowski T, Pomorski T, Lang C, Nelson H, Nelson N, Herrmann A. Rapid transbilayer movement of fluorescent phospholipid analogues in the plasma membrane of endocytosis-deficient yeast cells does not require the Drs2 protein. Eur J Biochem. 1999;263:254–263. doi: 10.1046/j.1432-1327.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- Meguro M, Kashiwagi A, Mitsuya K, Nakao M, Kondo I, Saitoh S, Oshimura M. A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat Genet. 2001;28:19–20. doi: 10.1038/ng0501-19. [DOI] [PubMed] [Google Scholar]
- Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P. μ1A-adaptin-deficient mice, lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller JV, Juul B, le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol Biol Cell. 1997;8:1481–199. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins C, Bonifacino JS. Structural requirements for function of yeast GGAs in vacuolar protein sorting, alpha-factor maturation, and interactions with clathrin. Mol Cell Biol. 2001;21:7981–7994. doi: 10.1128/MCB.21.23.7981-7994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierras CR, Warner JR. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:13235–13241. doi: 10.1074/jbc.274.19.13235. [DOI] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–9. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–58. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-Ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammer M, Briza P, Ellinger A, Schuster T, Stucka R, Feldmann H, Breitenbach M. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast. 1992;8:1089–1099. doi: 10.1002/yea.320081211. [DOI] [PubMed] [Google Scholar]
- Payne GS, Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Prezant TR, Chaltraw WE, Jr, Fischel-Ghodsian N. Identification of an overexpressed yeast gene which prevents aminoglycoside toxicity. Microbiology. 1996;142:3407–3414. doi: 10.1099/13500872-142-12-3407. [DOI] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripmaster TL, Vaughn GP, Woolford JL., Jr DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7901–7912. doi: 10.1128/mcb.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Jensen R, Zoller MJ, Burke J, Errede B, Smith M, Herskowitz I. Structure of the Saccharomyces cerevisiae HO gene and analysis of its upstream regulatory region. Mol Cell Biol. 1986;6:4281–4294. doi: 10.1128/mcb.6.12.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Siegmund A, Grant A, Angeletti C, Malone L, Nichols JW, Rudolph HK. Loss of Drs2p does not abolish transfer of fluorescence-labeled phospholipids across the plasma membrane of Saccharomyces cerevisiae. J Biol Chem. 1998;273:34399–34405. doi: 10.1074/jbc.273.51.34399. [DOI] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity [published erratum in Science (1996) 274, 1597] Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- Thompson R, Jansen PL. Genetic defects in hepatocanalicular transport. Semin Liver Dis. 2000;20:365–372. doi: 10.1055/s-2000-9384. [DOI] [PubMed] [Google Scholar]
- Ujhazy P, Ortiz D, Misra S, Li S, Moseley J, Jones H, Arias IM. Familial intrahepatic cholestasis 1: Studies of localization and function. Hepatology. 2001;34:768–775. doi: 10.1053/jhep.2001.27663. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zachowski A, Henry JP, Devaux PF. Control of transmembrane lipid asymmetry in chromaffin granules by an ATP-dependent protein. Nature. 1989;340:75–76. doi: 10.1038/340075a0. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Drake MT, Kornfeld S. ADP-ribosylation factor 1 dependent clathrin-coat assembly on synthetic liposomes. Proc Natl Acad Sci USA. 1999;96:5013–5018. doi: 10.1073/pnas.96.9.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]