Abstract
Telomerase is a ribonucleoprotein (RNP) complex that is minimally composed of a protein catalytic subunit, the telomerase reverse transcriptase (TERT), and an RNA component, the telomerase RNA. The survival of motor neuron (SMN) gene codes for a protein involved in the biogenesis of certain RNPs. Here, we report that SMN is a telomerase-associated protein. Using in vitro binding assays and immunoprecipitation experiments, we demonstrate an association between SMN and the telomerase RNP in vitro and in human cells. The specific immunopurification of SMN from human 293 cells copurified telomerase activity, suggesting that SMN associates with a subset of the functional telomerase holoenzyme. Our results also indicate that the human telomerase RNA and the human (h) TERT are not associated with Sm proteins, in contrast to Saccharomyces cerevisiae telomerase. Immunofluorescence analysis showed that hTERT does not specifically colocalize with wild-type SMN in gems or Cajal bodies. However, a dominant-negative mutant of SMN (SMNΔN27) previously characterized to elicit the cellular reorganization of small nuclear RNPs caused the accumulation of hTERT in specific SMNΔN27-induced cellular bodies. Furthermore, coexpression of SMNΔN27 and hTERT in rabbit reticulocyte lysates decreased the efficiency of human telomerase reconstitution in vitro. Our results establish SMN as a novel telomerase-associated protein that is likely to function in human telomerase biogenesis.
INTRODUCTION
Telomere maintenance in most eukaryotic cells is established by the ribonucleoprotein (RNP) enzyme telomerase. The telomerase RNP minimally consists of an RNA molecule and a protein catalytic subunit, the telomerase reverse transcriptase (TERT). Using an internal template sequence in the telomerase RNA subunit, this specialized reverse transcriptase synthesizes simple guanine-rich sequences at the 3′-end of chromosomal DNA. The telomeric DNA repeats and associated telomere-binding proteins protect chromosomes from nuclease digestion, end-to-end fusions, and other DNA rearrangement events (Lundblad, 2000).
The lengths and nucleotide sequences of the telomerase RNA subunits are highly divergent (Nugent and Lundblad, 1998; Chen et al., 2000). Secondary structure prediction suggests the presence of a small nucleolar (sno) RNA H/ACA box motif in the 3′-end of vertebrate telomerase RNAs (Mitchell et al., 1999a; Chen et al., 2000). Mutations that perturb the folding of the H/ACA box of human and mouse telomerase RNAs prevent both their cellular accumulation (Mitchell et al., 1999a; Martin-Rivera and Blasco, 2001) and their ability to properly localize to the nucleolus of microinjected Xenopus oocytes (Narayanan et al., 1999). Notably, the human telomerase RNA (hTR) associates with the H/ACA box snoRNA-binding proteins dyskerin, hGAR1, NH2P, and NOP10 (Mitchell et al., 1999b; Pogacic et al., 2001). The role of these proteins in vertebrate telomerase function is unclear. The X-linked form of the disease dyskeratosis congenita is caused by mutations in the gene that encodes the protein dyskerin (Heiss et al., 1998). Cells from individuals affected by this disease have low levels of hTR and telomerase activity as well as short telomeres (Mitchell et al., 1999b), supporting an important role for snoRNP proteins in telomerase biogenesis.
Human telomerase activity is detected in >85% of cancers and transformed cell lines, whereas it is absent from most normal human cells (Oulton and Harrington, 2000). Inhibition of human telomerase from immortal and cancer cell lines results in telomere shortening and, in certain cell types, cell death or senescence (Damm et al., 2001; Harrington and Robinson, 2002). Consequently, a better understanding of the mechanisms involved in the assembly and regulation of the human telomerase RNP will be important for the rational design of telomerase inhibitors. In Saccharomyces cerevisiae, the telomerase RNA (TLC1) associates with the heptameric Sm protein complex and acquires a 5′-2,2,7-trimethylguanosine (TMG) cap structure (Seto et al., 1999); both of these events are hallmarks of small nuclear (sn) RNP assembly. Yet, little is known about the molecular machinery involved in the localization and assembly of vertebrate telomerase.
The product encoded by the survival of motor neuron (SMN) gene is present both in the cytoplasm and in the nucleus, where it localizes in different nuclear structures: gems, Cajal bodies (CBs), and the nucleolus (Liu and Dreyfuss, 1996; Hebert et al., 2001; Young et al., 2001). SMN and a set of associated proteins (Gemins) form a complex involved in the biogenesis of at least four uridine (U)-rich snRNPs, U1, U2, U4, and U5, all major constituents of the splicing machinery (Fischer et al., 1997; Liu et al., 1997; Meister et al., 2001). Biochemical data demonstrate that SMN interacts directly with the arginine-glycine–rich domain of a subset of Sm proteins (Pellizzoni et al., 1999; Brahms et al., 2001; Friesen et al., 2001). The Sm core complex contains seven proteins (B/B′, D1–D3, E, F, and G) predicted to form a closed ring structure around a conserved sequence motif within some of the UsnRNAs (Kambach et al., 1999; Mura et al., 2001; Will and Luhrmann, 2001). The precise function of the SMN complex is not completely characterized; however, evidence strongly suggests that the SMN complex facilitates or stabilizes the association of Sm proteins with U1, U2, U4, and U5 snRNAs and the functional maturation of UsnRNPs (Fischer et al., 1997; Meister et al., 2001; Will and Luhrmann, 2001). Furthermore, using antibody addition experiments and a dominant-negative version of SMN that lacks the first 27 amino acids (SMNΔN27), Pellizzoni et al. (1998) demonstrated that SMN might also have a more direct role in pre-mRNA splicing. As expected from the wide range of cellular pathways in which SMN is implicated (Terns and Terns, 2001), disruption of the gene encoding SMN in different organisms is lethal (Schrank et al., 1997; Miguel-Aliaga et al., 1999; Paushkin et al., 2000).
The role of the Sm protein complex in S. cerevisiae telomerase biogenesis and the recent observation that SMN associates with snoRNP proteins (Jones et al., 2001; Pellizzoni et al., 2001a), prompted us to examine whether SMN and/or Sm proteins are involved in human telomerase function. We report that SMN is a novel telomerase-associated protein. The human Sm protein complex does not interact with human (h) TERT, hTR, or catalytically active telomerase, suggesting that the association of SMN with human telomerase is independent of Sm proteins. A previously characterized dominant-negative SMN protein (SMNΔN27) has the ability to perturb the normal subcellular localization of hTERT and decrease the efficiency of in vitro reconstitution of telomerase in rabbit reticulocyte lysates (RRLs). On the basis of these and other recent results (Jones et al., 2001; Pellizzoni et al., 2001a), we suggest that SMN is involved in human telomerase biogenesis as an H/ACA snoRNP.
MATERIALS AND METHODS
Experimental Procedures
Construction of Plasmids. The cDNAs encoding human SMN and SMNΔN27 were amplified by RT-PCR using total cellular RNA extracted from HeLa cells. SmB, SmD1, and SmD3 cDNAs were amplified by PCR from IMAGE clones. Expression of the Myc-tagged version of these proteins in cultured human cells or rabbit reticulocyte lysates was performed by cloning the DNA fragments corresponding to the above-mentioned cDNAs into a modified pcDNA3.1 vector (InVitrogen, San Diego, CA) containing the sequence for the Myc-tag epitope (Chen and Richard, 1998). The expression construct for FLAG-hTERT was a gift from Dr. Lea Harrington (Amgen, University of Toronto). For subcellular localization experiments, hTERT cDNA was subcloned into the pEGFP-C1 vector (Clonetech, Cambridge, UK).
Antibodies. The antibodies used were as follows: mouse monoclonal FLAG antibody (Sigma, St. Louis, MO); affinity-purified goat anti-GST serum (Amersham Biosciences, Arlington Heights, IL); affinity-purified rabbit anti-hTERT serum (Moriarty et al., 2002); mouse anti-Sm (Lerner et al., 1981); affinity-purified rabbit anti-TEP1 serum (Harrington et al., 1997a); mouse monoclonal anti-SMN (clone 8; Transduction Laboratories, Lexington, KY); mouse monoclonal anti-Myc (9E10; ATCC hybridoma; American Type Culture Collection, Manassas, VA); and mouse monoclonal 2,2,7-TMG-specific antibody (Oncogene Research Products, San Diego, CA).
Cell Culture and Manipulations. Human embryonic kidney cells (293) and HeLa cells were grown in DMEM with 10% fetal bovine serum and antibiotics. Transient transfections of 293 and HeLa cells were performed using Lipofectamine 2000 (Invitrogen) with 1–2 μg expression constructs combined per 35-mm dish.
In Vitro Binding Assays. Reconstitution of active human telomerase by coexpression of GST-hTERT and the hTR in yeast was described previously (Bachand and Autexier, 1999). To investigate SMN and telomerase interaction in vitro, we generated [35S]methionine-labeled Myc-SMN and luciferase using an RRL kit as described per the manufacturer's instructions (Promega, Madison, WI). Equal amounts of labeled proteins were first precleared overnight in 1 ml of in vitro binding buffer (50 mM Tris, pH 7.5, 200 mM NaCl, 2 mM EDTA, 0.1% NP-40, and protease inhibitors). Immunopurified human telomerase RNP was prepared by incubating protein extracts from 50 ml yeast pellets with GST-specific antibody (Amersham Biosciences) and protein-A–Sepharose (Sigma) in yeast lysis buffer (10 mM Tris, pH 7.5, 2 mM MgCl2, 5.0 mM β-mercaptoethanol, 20% glycerol, 1% NP-40, 0.25 mM sodium deoxycholate, 1.0 mM EGTA, and 150 mM NaCl, plus protease and RNase inhibitors). After a 1-hour incubation at 4°C, beads were washed 5 times in yeast lysis buffer supplemented with 500 mM NaCl. The precleared RRL-synthesized labeled proteins were then incubated with the immunopurified human telomerase RNP for an additional 2 h at 4°C. After washing 5 times with 1 ml of in vitro binding buffer, bound proteins were eluted by boiling in SDS-PAGE loading dye and subjected to electrophoresis.
Immunoprecipitation, Telomerase Activity, and Northern Blotting. Twenty to 24 h after transfection, cells were washed two times with PBS and resuspended in 500 μl of lysis buffer (20 mM HEPES, pH 7.9, 2 mM MgCl2, 0.2 mM EGTA, 10% glycerol, 1 mM DTT, 150 mM NaCl, and 1.0% NP-40, plus protease and RNase inhibitors). After homogenization by forcing the cells five times through a 25-gauge needle, the cell suspension was left rotating at 4°C for 30 min before the lysate was cleared in a microcentrifuge for 15–20 min. For protein coimmunoprecipitation experiments, cell lysates were incubated with antibodies for 30 min before the addition of protein-A–Sepharose for an additional 1-hour incubation at 4°C. After the beads had been washed four times with 1 ml of lysis buffer, the bound proteins were eluted and subjected to SDS-PAGE and immunoblotting. Five to 10% of immunoprecipitates were assayed for telomerase activity by Telomeric repeat amplification protocol (TRAP) as previously described (Bachand and Autexier, 2001). The preparation of HeLa nucleolar-enriched nuclear extracts was based on a previously described protocol (Jordan et al., 1996).
To analyze immunoprecipitated RNAs, HeLa total cell lysates were prepared from 70–80% confluent cells in a 10-cm dish in lysis buffer (20 mM HEPES, pH 7.9, 300 mM KCl, 10% glycerol, 0.5 mM DTT, 1 mM EDTA, 2 mM MgCl2, and 1% NP-40, plus protease and RNase inhibitors) and subjected to immunoprecipitation as previously described (Bachand and Autexier, 2001). Probes used for Northern blotting were DNA oligonucleotides complementary to human U1, U2, and U6 snRNAs and hTR.
Indirect Immunofluorescence. HeLa cells were cultured on coverslips in 6-well dishes. Twenty hours after transfection, HeLa cells were fixed for 5 min in 1.0% paraformaldehyde in PBS, pH 7.5, and then permeabilized for 5 min in 0.5% Triton X-100 in PBS. Myc-tagged SMN and SMNΔN27 proteins were labeled with anti-Myc antibody (9E10; 1:400). Cells were then washed with 0.1% Triton X-100 in PBS, followed by PBS, and were then incubated with secondary antibody (goat anti-mouse Cy3 from Chemicon, Temecula, CA) for 30 min. Cells were rinsed with 0.1% Triton X-100 in PBS or in PBS alone and then mounted in 1 mg/ml para-phenylenediamine in PBS/90% glycerol that also contained DAPI at 1 μg/ml. Digital imaging was performed with a SPOT cooled CCD camera (Diagnostic Instruments, Inc., Burroughs, MI) mounted on a Zeiss Axioplan immunofluorescence microscope.
RESULTS
SMN Associates with the Human Telomerase RNP In Vitro and In Vivo
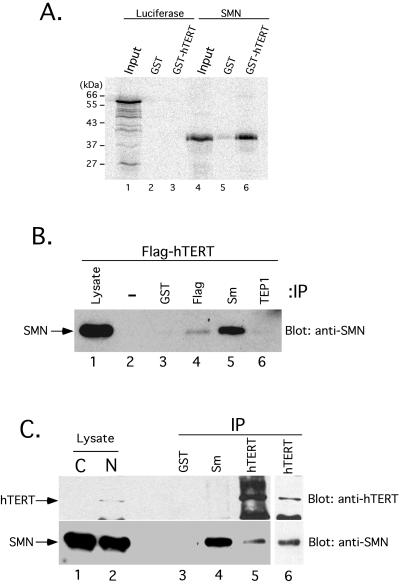
We used functional recombinant human telomerase expressed in S. cerevisiae (Bachand and Autexier, 1999) to investigate whether the SMN protein can form a complex with telomerase in vitro. First, the GST-hTERT/hTR telomerase complex was immunopurified from yeast extracts using an affinity-purified GST-specific antibody as described in Experimental Procedures. [35S]methionine-labeled luciferase and SMN proteins were generated in RRLs. The labeled proteins were incubated with the recombinant telomerase RNP previously immobilized on antibody-coated Sepharose beads. After a 2-h incubation, the complexes were washed extensively, eluted, and analyzed by SDS-PAGE. Figure 1A shows that SMN specifically bound to the GST-hTERT/hTR complex (lane 6), whereas SMN did not bind to GST alone (lane 5). Treatment of the GST-hTERT/hTR complex with a cocktail of RNases before addition of [35S]-labeled SMN did not affect or disrupt the SMN-telomerase interaction (data not shown). Although incomplete RNA digestion cannot be ruled out, the results suggest that the association of in vitro–synthesized SMN with recombinant hTERT is mediated via direct contact with hTERT or a yeast TERT-associated protein.
Figure 1.
SMN associates with hTERT in vitro and in vivo. (A) Human telomerase was reconstituted by coexpression of GST-hTERT and hTR in S. cerevisiae as previously described (Bachand and Autexier, 1999). GST (lanes 2 and 5) and GST-hTERT/hTR (lanes 3 and 6) were affinity-purified from equal volumes of yeast extracts and incubated with in vitro–translated [35S]methionine-labeled luciferase (lanes 1–3) and SMN (lanes 4–6). After extensive washing, bound proteins were analyzed by SDS-PAGE and autoradiography. The input lanes (1 and 4) show 5% of the RRL lysate used in the binding reaction. Molecular mass markers are indicated on the left (in kilodaltons, kDa). (B) 293 cells were transiently transfected with a DNA construct expressing FLAG-tagged hTERT. At 20 h after transfection, a total cell lysate was prepared and subjected to immunoprecipitation (IP) without antibody or using anti-GST, anti-FLAG, anti-Sm (Y12), or anti-TEP1. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting for endogenous SMN. The lysate lane corresponds to 5% of the total cell lysate used for the immunoprecipitation. (C) Nucleolar-enriched nuclear extracts were prepared from HeLa cells and subjected to immunoprecipitation (IP) using anti-GST, anti-Sm (Y12), and two different affinity-purified hTERT antibodies. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting for endogenous hTERT (top) and SMN (bottom). Five percent of the cytosolic (C) and nucleolar-enriched nuclear (N) extracts were also loaded.
We also used transient expression of a FLAG-tagged hTERT protein in telomerase-positive 293 cells to demonstrate the association between SMN and telomerase. Total cell extracts from 293 cells transiently transfected with a FLAG-hTERT construct were subjected to immunoprecipitation using different antibodies and analyzed by immunoblotting with a mouse monoclonal anti-SMN antibody. As previously demonstrated (Liu et al., 1997; Pellizzoni et al., 1999), an antibody specific to the Sm protein complex efficiently coimmunoprecipitated SMN from 293 cellular extracts (Figure 1B, lane 5). The precipitation of FLAG-tagged hTERT using anti-FLAG also coimmunoprecipitated SMN from 293 cell extracts (Figure 1B, lane 4). An antibody specific to a telomerase-associated protein (TEP1) (Harrington et al., 1997a), GST antibody, or protein-A–Sepharose alone did not precipitate the SMN protein (Figure 1B, lanes 6, 3, and 2, respectively).
Nucleolar-enriched nuclear extracts were prepared from HeLa cells to determine whether an endogenous SMN-hTERT complex exists in cells. The SMN protein was found in both the cytosolic and nuclear extracts (Figure 1C, lanes 1 and 2), as expected from its previously determined subcellular localization (Liu and Dreyfuss, 1996). Confirming the efficiency of our nuclear extract preparation, the hTERT protein was primarily nuclear (Figure 1C, lanes 1 and 2), in agreement with the previous immunofluorescence analysis of hTERT (Harrington et al., 1997b) and mTERT (Martin-Rivera et al., 1998). Proteins from the nuclear extracts were subjected to immunoprecipitation using the Sm protein–specific antibody (Y12), an affinity-purified hTERT antibody (Moriarty et al., 2002), and a GST-specific antibody as a negative control. After extensive washing of the antibody-coated beads, the immunoprecipitated proteins were analyzed for recovery of SMN and hTERT as determined by Western blotting with antibodies specific for the respective proteins. As demonstrated in Figure 1B, immunoprecipitation performed with the Y12 antibody coprecipitated SMN from HeLa cell nuclear extracts, but not the hTERT protein (Figure 1C, lane 4). hTERT-specific immunoprecipitation also recovered SMN from the HeLa nuclear extracts (lane 5). The coimmunopurification of SMN and hTERT was also confirmed using a different affinity-purified hTERT antibody (Harrington et al., 1997b) (Figure 1C, lane 6). As a control, neither SMN nor hTERT was present in anti-GST immunoprecipitates (lane 3). We conclude that hTERT and SMN can associate in vitro and in human cells.
SMN Is Associated with Catalytically Active Human Telomerase
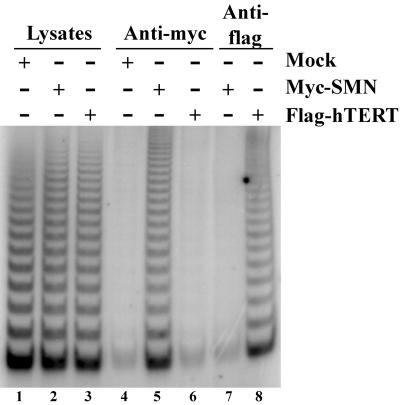
We used transient transfection experiments to investigate whether SMN is associated with active human telomerase. A Myc-tagged version of SMN and FLAG-hTERT were transiently expressed in human 293 cells, and total cell lysates were prepared for immunoprecipitations using either Myc or FLAG antibodies. As a control, a lysate from mock-transfected cells was used. TRAP assays demonstrated that equal levels of telomerase activity were present in the different total cell lysates before immunopurification (Figure 2, lanes 1–3). As previously demonstrated (Harrington et al., 1997a; Mitchell et al., 1999b), FLAG antibody resin precipitated the FLAG-hTERT protein (data not shown) and human telomerase activity from the FLAG-hTERT–containing extracts (Figure 2, lane 8). However, no telomerase activity was present on the FLAG-antibody resin incubated with Myc-SMN–containing extracts (lane 7). Telomerase activity (lane 5) was also recovered from Myc-antibody resin prepared from lysates containing the Myc-SMN protein, but not from anti-Myc immunoprecipitates prepared from lysates of mock-transfected and FLAG-hTERT–transfected cells (lanes 4 and 6, respectively). Similar results were observed using HeLa cells (data not shown). These results indicate that SMN associates with a fully assembled and catalytically active telomerase RNP.
Figure 2.
SMN associates with catalytically active telomerase. 293 cells were transiently transfected with DNA constructs expressing Myc-tagged SMN (lanes 2, 5, and 7), FLAG-tagged hTERT (lanes 3, 6, and 8), or with vector alone (mock; lanes 1 and 4). Total cell lysates were prepared and subjected to immunoprecipitation using either anti-Myc (lanes 4–6) or anti-FLAG (lanes 7 and 8). Immunoprecipitates were analyzed for telomerase activity by the TRAP assay. Of the total cell lysates (lanes 1–3), 0.5% were also assayed for telomerase activity.
The Sm Protein Complex Is Not Associated with Active Human Telomerase and hTR
The Sm proteins form an RNA-binding complex that interacts with a specific region of UsnRNAs (Will and Luhrmann, 2001). Direct interactions between SMN and specific members of the Sm proteins are thought to recruit the SMN complex to the Sm-snRNA complex (Liu et al., 1997; Pellizzoni et al., 1999). Interestingly, the S. cerevisiae telomerase RNA subunit, TLC1, contains an Sm protein–binding site, as determined by the coimmunoprecipitation of the TLC1 RNA and yeast telomerase activity with epitope-tagged versions of the SmD1 and SmD3 proteins (Seto et al., 1999).
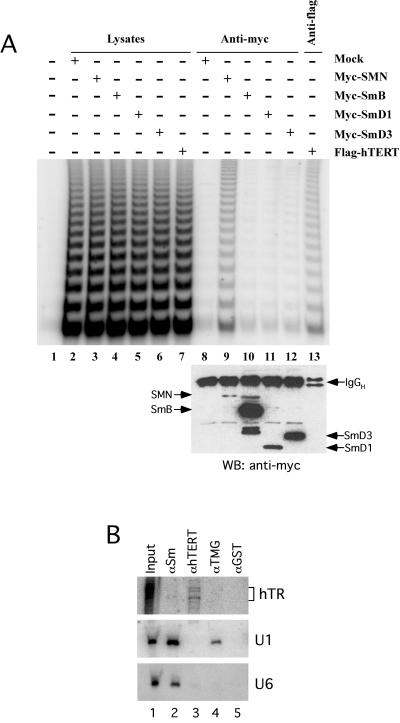
Myc-tagged versions of the human SmB, SmD1, and SmD3 proteins were used to examine whether the association of SMN with telomerase could be mediated by the involvement of the Sm protein complex in human telomerase biogenesis. Addition of the Myc epitope to the N-terminus of the SmB, D1, and D3 proteins did not affect their function, as determined by their ability to coimmunoprecipitate SMN and by immunofluorescence analysis (data not shown). Constructs expressing the Myc-SmB, Myc-SmD1, and Myc-SmD3 proteins were transfected into human 293 cells, and total cell extracts were prepared for immunopurification using the Myc antibody. As was previously shown (Figure 2), Myc-antibody resin incubated with Myc-SMN–containing extracts recovers telomerase activity (Figure 3, lane 9). However, immunoprecipitations performed using the Myc antibody and prepared from either the Myc-SmB–, Myc-SmD1–, or Myc-SmD3–containing extracts did not recover levels of human telomerase activity (Figure 3, lanes 10–12, respectively) significantly higher than background (lane 8). Western blot analysis of the immunoprecipitated proteins (Figure 3A, bottom) revealed that the myc-tagged Sm proteins were expressed and immunoprecipitated to considerably higher levels than myc-tagged SMN, yet only myc-SMN copurified human telomerase activity. As a control, anti-FLAG antibody efficiently precipitated telomerase activity from lysates prepared from FLAG-hTERT–transfected cells (lane 13). These results, in addition to the absence of hTERT in Y12 immunoprecipitates (Figure 1C), indicate that the Sm protein complex is not associated with the human telomerase RNP and suggest that Sm proteins do not mediate the SMN-telomerase association.
Figure 3.
The hTR and telomerase activity are not associated with the Sm protein complex. (A) 293 cells were transiently transfected with DNA constructs expressing Myc-SMN (lanes 3 and 9), Myc-SmB (lanes 4 and 10), Myc-SmD1 (lanes 5 and 11), Myc-SmD3 (lanes 6 and 12), FLAG-hTERT (lanes 7 and 13), or with vector alone (mock; lanes 2 and 8). At 20 h after transfection, total cell lysates were prepared and subjected to immunoprecipitation using either anti-Myc (lanes 8–12) or anti-FLAG (lane 13). Immunoprecipitates were analyzed for telomerase activity by the TRAP assay (top) and for protein content by Western blotting (WB) using anti-myc (bottom). Of the total cell lysates (lanes 2–7) or lysis buffer (lane 1), 1% were analyzed for telomerase activity. (B) A total cell lysate from HeLa cells was prepared and subjected to immunoprecipitation using anti-Sm (Y12-lane 2), affinity-purified anti-hTERT (lane 3), anti-TMG (lane 4), or anti-GST (lane 5). Immunoprecipitates were analyzed by denaturing PAGE and Northern blotting for endogenous hTR (top), U1 snRNA (middle), and U6 snRNA (bottom). The input lane (lane 1) was loaded with 2.5% of the total RNA extracted from the HeLa total cell lysate. For hTR, exposure time was three times longer than for U1 and U6.
To further investigate whether the Sm protein complex is involved in human telomerase biogenesis, we determined whether endogenous hTR from HeLa total cell extracts copurifies with the Sm complex using the Y12 antibody. As previously demonstrated (Lerner and Steitz, 1979), endogenous U1 and U6 snRNAs are coprecipitated with Sm proteins using the Y12 antibody (Figure 3B, lane 1). However, the Y12 antibody did not coimmunoprecipitate hTR (Figure 3B, lane 2). hTERT-associated hTR was coprecipitated using an affinity-purified hTERT antibody (Figure 3B, lane 3), whereas an anti-GST immunoprecipitation did not recover U1, U6, or hTR (Figure 3B, lane 5).
We also determined whether a subpopulation of the hTR contains a TMG cap structure as was previously shown for the yeast telomerase RNA (Seto et al., 1999). This type of hypermethylated 5′-cap structure is a well-known characteristic of some spliceosomal snRNAs, such as U1, U2, U4, and U5 (Will and Luhrmann, 2001). HeLa total cell lysates were subjected to immunoprecipitation using a TMG-specific monoclonal antibody, and the copurified RNAs were analyzed by Northern blotting. Both U1 and U2 snRNAs were recovered from the anti-TMG immunoprecipitate (Figure 3B, lane 4 and data not shown), whereas the presence of hTR in TMG-specific immunoprecipitates was undetectable (Figure 3B, lane 4). Similarly, U6 snRNA was not coimmunoprecipitated by the TMG-specific antibody (lane 4) and was used as a negative control because it does not have a 5′-TMG cap structure (Singh and Reddy, 1989). In conclusion, these results indicate that the Sm proteins, whether endogenous or transiently overexpressed, associate neither with hTR nor with the catalytically active human telomerase RNP. Our results further suggest that hTR does not acquire a 5′-TMG cap.
Expression of a Dominant-Negative Version of SMN (SMNΔN27) Perturbs the Subcellular Localization of hTERT
The SMN protein is present in the cytoplasm as well as in the nucleus of cells, where it is known to concentrate in nuclear structures such as gems, CBs, and nucleoli (Liu and Dreyfuss, 1996; Hebert et al., 2001; Young et al., 2001). A previous study characterizing a dominant-negative mutant of SMN lacking its first N-terminal 27 amino acids (SMNΔN27) revealed that this mutant protein causes the reorganization of snRNPs in the nucleus and in the cytoplasm and also negatively affects pre-mRNA splicing in vitro (Pellizzoni et al., 1998). This SMN mutant protein was also used to demonstrate the functional interaction between SMN and the RNA polymerase II complex (Pellizzoni et al., 2001b) as well as with the snoRNP proteins fibrillarin and hGAR1 (Pellizzoni et al., 2001a).
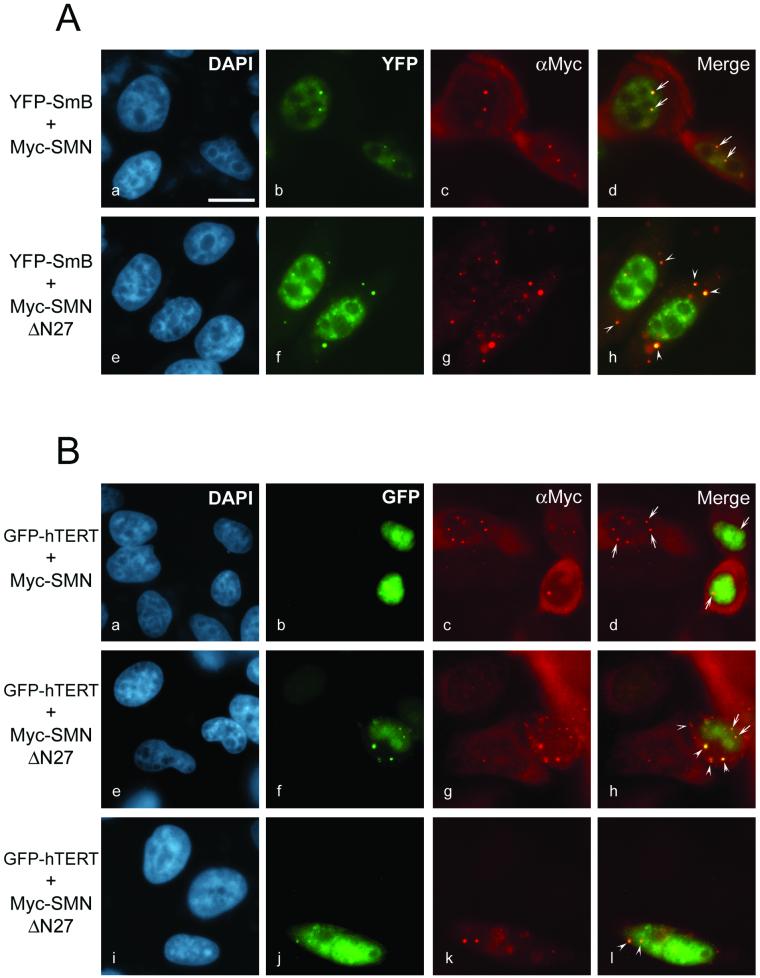
We generated SMNΔN27 to investigate the effect of this dominant-negative mutant SMN on hTERT cellular localization. Indirect immunofluorescence with the Myc antibody was used to detect the Myc-SMN and Myc-SMNΔN27 proteins in transfected HeLa cells. The Myc-SMN protein localized in the cytoplasm and in the nucleus, where it accumulated in gems and CBs (Figure 4, A and B, c). When the Myc-SMN construct was cotransfected with a plasmid expressing a yellow fluorescent protein (YFP)-SmB fusion, Myc-SMN and YFP-SmB colocalized in gems and CBs (Figure 4A, a–d), as previously reported (Liu et al., 1997; Pellizzoni et al., 1998). In cells transfected with Myc-SMNΔN27, the mutant protein accumulated in large cytoplasmic bodies that partially redistributed the YFP-SmB fusion (Figure 4A, e–h). In contrast, the YFP-SmB fusion protein was barely detectable in the cytoplasm of Myc-SMN transfected cells (Figure 4A, b) and untransfected cells (data not shown), consistent with the localization of endogenous Sm proteins as detected using the Sm-specific Y12 antibody (Liu et al., 1997; Pellizzoni et al., 1998). Similar results were obtained when a YFP-SmD1 fusion was used (data not shown), in agreement with the observations that a mutant of SMN lacking the first 27 amino acids causes a reorganization of Sm snRNPs proteins (Pellizzoni et al., 1998).
Figure 4.
The expression of a dominant-negative mutant of SMN (SMNΔN27) perturbs the nuclear localization of hTERT. (A) HeLa cells were transiently cotransfected with DNA constructs expressing either Myc-SMN and YFP-SmB (a–d) or Myc-SMNΔN27 and YFP-SmB (e–h). The fixed and permeabilized cells were stained for Myc-SMN (c) and Myc-SMNΔN27 (g) using anti-Myc. DNA stained with DAPI shows the nucleus of each cell (a and e). Images b and c, and f and g, are merged to form d and h, respectively. (B) HeLa cells were transiently cotransfected with DNA constructs expressing either Myc-SMN and GFP-hTERT (a–d) or Myc-SMNΔN27 and GFP-hTERT (e–l). The fixed and permeabilized cells were stained for Myc-SMN (c) and Myc-SMNΔN27 (g and k) using anti-Myc. DNA stained with DAPI shows the nucleus of each cell (a, e, and i). Images b and c, f and g, and j and k are merged to form d, h, and l, respectively. The arrows point to the nuclear gems and the arrowheads to the SMNΔN27-induced cytoplasmic accumulations. Bar, 12 μm.
We used GFP-tagged hTERT to monitor the steady-state subcellular localization of hTERT. Addition of GFP to the N-terminus of hTERT did not alter its catalytic function, because the GFP-hTERT fusion reconstituted human telomerase activity when expressed in telomerase-negative human fibroblasts (data not shown). The GFP-hTERT fusion protein showed a diffuse nucleoplasmic distribution both in untransfected (data not shown) and in Myc-SMN–transfected HeLa cells (Figure 4B, b), consistent with previous reports of TERT localization (Harrington et al., 1997b; Martin-Rivera et al., 1998). GFP-tagged hTERT did not specifically colocalize with Myc-SMN in gems and CBs (Figure 4B, a–d) but did frequently localize to the nucleolus of transfected cells (Figure 4B and data not shown). GFP-hTERT expressed in Myc-SMNΔN27–transfected cells accumulated prominently in the cytoplasm in structures that colocalized with the Myc-SMNΔN27 protein (Figure 4B, e–l), in striking contrast to the restricted and diffuse nucleoplasmic localization of hTERT in Myc-SMN–transfected and untransfected cells. The colocalization of hTERT and Myc-SMNΔN27 was also observed in the nucleus, where GFP-hTERT accumulated in gems and CBs (Figure 4B, f–h). The observation that hTERT subcellular localization is affected by expression of SMNΔN27 suggests a functional relationship between the SMN complex and human telomerase.
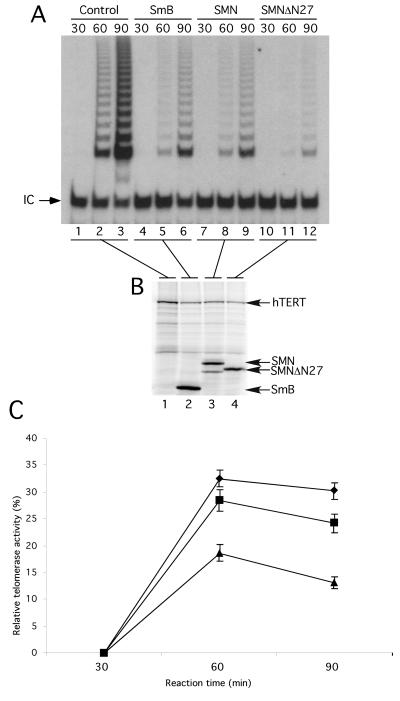
SMNΔN27 Affects Human Telomerase Reconstitution In Vitro
Coexpression of hTERT and hTR reconstitutes human telomerase activity in RRLs (Weinrich et al., 1997; Beattie et al., 1998). Studies of human and Tetrahymena telomerase suggest that proteins present in reticulocyte extracts may be involved in reconstitution of telomerase assembly and/or activity (Holt et al., 1999; Licht and Collins, 1999). Western blot analysis of crude RRL extracts using a monoclonal SMN antibody revealed the presence of a single 38-kDa protein (data not shown), suggesting the presence of rabbit SMN in reticulocyte lysates. This latter observation and the profound effects of SMNΔN27 on hTERT subcellular localization led us to examine whether expression of SMNΔN27 in RRL would affect the reconstitution of human telomerase activity.
In vitro–transcribed hTR was added to RRL programmed to express hTERT alone or to coexpress hTERT with SmB, wild-type human SMN, or the SMNΔN27 mutant. Protein synthesis was allowed to proceed for different lengths of time, followed by analysis of telomerase activity by TRAP. Human telomerase activity was undetectable after 30 min, whether hTERT was expressed alone or with another protein (Figure 5A, lanes 1, 4, 7, and 10). At 60 min, robust telomerase activity was observed in the control RRL reaction, in which hTERT was expressed alone (lane 2). Telomerase activity was also detected when SmB or SMN was coexpressed with hTERT (lanes 5 and 8). Lower levels of telomerase activity were observed in RRL reactions coexpressing hTERT and SmB/SMN than in the control reaction (compare lanes 5 and 8 with lane 2) because of the lower levels of hTERT protein synthesized when additional DNA is present in the RRL reaction (Figure 5B and data not shown). Human telomerase activity reconstituted after 60 min was barely detectable from the reaction that coexpressed hTERT and SMNΔN27 (lane 11). Similarly, after 90 min, the amount of telomerase activity reconstituted in the SMNΔN27-programmed RRL was considerably lower than in SmB- and SMN-containing RRLs (compare lane 12 with lanes 6 and 9). As can be seen in Figure 5B, these differences were not attributed to drastically different levels of hTERT protein (lanes 2–4). Telomerase activity was similar to the results seen in Figure 5A when luciferase was expressed with hTERT rather than SmB (data not shown). The decreased efficiency of human telomerase reconstitution when SMNΔN27 is expressed in RRL was reproduced in three independent experiments. Telomerase activity levels were quantified from the three independent experiments and averaged, and a percentage was calculated relative to the levels of activity reconstituted in the control RRL reaction, in which hTERT was expressed alone (Figure 5C). These results indicate that a previously characterized dominant-negative SMN mutant, SMNΔN27, significantly decreases the efficiency of human telomerase reconstitution in vitro.
Figure 5.
Expression of SMNΔN27 decreases human telomerase activity reconstitution in RRLs. (A) hTERT was synthesized in RRL in the presence of hTR, [35S]methionine, and equal amounts of DNA constructs expressing Myc-SmB (lanes 4–6), Myc-SMN (lanes 7–9), Myc-SMNΔN27 (lanes 10–12), or no additional DNA (lanes 1–3). At 30, 60, and 90 min after the start of the RRL reactions, telomerase activity was assayed by the TRAP assay. Each TRAP reaction included an internal control (IC) to normalize for variation in PCR efficiency. (B) At 90 min after the start of the RRL reactions, equal amounts were analyzed by SDS-PAGE and autoradiography. (C) The telomerase activity was calculated as the ratio between the intensity of the telomerase ladder products and the intensity of the internal PCR control. A ratio of this telomerase activity value to the amount of in vitro–translated hTERT measured by the intensity of the S35-labeled hTERT was calculated to generate the relative telomerase activity. The activities from three independent experiments performed in the presence of SmB (diamonds), wild-type SMN (squares), and SMNΔN27 (triangles) were averaged and compared relative to RRL reactions in which no additional DNA construct was included.
DISCUSSION
Recent results suggest that spliceosomes and transcriptosomes are preassembled in a substrate-independent manner (Gall et al., 1999; Stevens et al., 2002). Similarly, telomerase is most likely preassembled into a functional RNP before recruitment to its site of action, the telomere. The assembly and maturation of many RNP particles is believed to occur in nuclear structures such as the CBs and nucleoli (Matera, 1999; Olson et al., 2000). Vertebrate telomerase RNA is a member of the H/ACA box family of sno RNA (Chen et al., 2000), and a fraction of hTR localizes to the nucleolus (Mitchell et al., 1999a; Narayanan et al., 1999), suggesting that vertebrate telomerase assembly and/or maturation transits through the nucleolus. Furthermore, Lukowiak et al. (2001) recently reported that in vitro–transcribed human and Xenopus telomerase RNAs microinjected into Xenopus oocyte nuclei localize not only to nucleoli, but also to CBs.
Our results support a model in which the human telomerase RNP is assembled and/or matured into a functional enzyme by transit through the nucleoli and/or the CBs. The physical association between endogenous hTERT and SMN, a protein involved in RNP assembly that localizes in both nucleoli and CBs, strongly suggests that SMN plays a role in human telomerase biogenesis. Two experimental observations are in support of the association between SMN and telomerase does not merely reflect the fact they both colocalize to similar nuclear structures: nucleoli and/or CBs. First, recombinant telomerase can specifically bind in vitro–translated SMN (Figure 1A). Second, hTERT protein and telomerase activity is undetectable in immunoprecipitates performed using antibodies specific to the box C/D snoRNP nucleolar protein fibrillarin (data not shown). The effect of SMNΔN27 on hTERT subcellular localization and telomerase reconstitution in vitro further supports a functional role for SMN in telomerase assembly. The mechanism by which SMNΔN27 disturbs the cellular organization of snRNPs (Pellizzoni et al., 1998) and snoRNPs (Pellizzoni et al., 2001a) has not yet been defined. GFP-hTERT did not specifically colocalize with wild-type SMN in gems and CBs, yet it accumulated and colocalized with SMNΔN27 (Figure 4). These results suggest dynamic and transient interactions between the SMN complex and components of the telomerase RNP. Similar results are observed in immunofluorescence analyses of several components of the RNA polymerase II transcription machinery upon overexpression of SMN and SMNΔN27 (Pellizzoni et al., 2001b). The effect of SMNΔN27 on hTERT cellular localization is consistent with the proposed view that this SMN mutant sequesters associated proteins in cellular bodies by blocking or retarding their release and/or their transport between different nuclear bodies (Pellizzoni et al., 1998, 2001a, 2001b; Terns and Terns, 2001). SMNΔN27 expression also resulted in the accumulation and the detection of GFP-hTERT in the cytoplasm of cotransfected cells (Figure 4B). The accumulation of GFP-hTERT in the cytoplasm was never observed in cells either cotransfected with wild-type SMN (Figure 4B) or transfected with the GFP-hTERT construct alone (data not shown). Sm proteins also colocalize with the SMNΔN27-induced cytoplasmic accumulations (Figure 4A) (Pellizzoni et al., 1998), possibly as a result of a perturbed interaction between endogenous SMN and Sm proteins in the cytoplasm (Fischer et al., 1997; Liu et al., 1997). Yet, SMNΔN27 does not elicit the cytoplasmic accumulation of other SMN-associated proteins such as p80coilin (Pellizzoni et al., 1998), components of the RNA pol II complex (Pellizzoni et al., 2001b), and snoRNP proteins (Pellizzoni et al., 2001a). Our results demonstrating the accumulation of GFP-hTERT in cytoplasmic SMNΔN27-containing aggregates suggest that the interaction between the SMN complex and hTERT could be initiated in the cytosol. The SMN-hTERT complex could then relocalize to the nucleus and encounter a fully processed hTR-snoRNP protein complex in subnuclear domains such as the nucleolus (Mitchell et al., 1999a; Narayanan et al., 1999) and/or the CBs (Lukowiak et al., 2001).
How might SMN be involved in human telomerase biogenesis? The best-characterized function of SMN is its role in snRNP assembly. Experiments in Xenopus oocytes and those using a cell-free system for in vitro reconstitution of UsnRNP assembly suggest that the SMN complex is involved in facilitating the association of distinct snRNAs with Sm proteins (Fischer et al., 1997; Meister et al., 2001). hTERT, hTR, and human telomerase activity are not detected in anti-Sm immunoprecipitates (Figures 1 and 3), suggesting that the Sm protein complex is not involved in human telomerase biogenesis. The lack of association between hTR and Sm proteins was previously noted (Le et al., 2000; Lukowiak et al., 2001). Thus, on the basis of our results and recent studies that report interactions between SMN and snoRNP proteins (Jones et al., 2001; Pellizzoni et al., 2001a), we propose that SMN may function in human telomerase assembly through the association of SMN with snoRNP proteins such as hGAR1. The human GAR1 snoRNP protein is an attractive candidate, because it associates with hTR (Pogacic et al., 2001). Using antibodies specific for H/ACA snoRNP proteins, we were unable to immunodeplete the SMN-associated telomerase activity (data not shown). However, hGAR1 and dyskerin, two hTR-associated H/ACA snoRNP proteins (Mitchell et al., 1999b; Pogacic et al., 2001), and SMN are present in partially purified human telomerase fractions generated using anion and size exclusion chromatography followed by differential-density ultracentrifugation through a cesium sulfate gradient (data not shown). These observations, coupled with the nucleolar localization of hTR (Mitchell et al., 1999a; Narayanan et al., 1999; Lukowiak et al., 2001), hTERT (Figure 4 and data not shown), SMN (Pellizzoni et al., 2001a; Young et al., 2001), and hTR-associated snoRNP proteins (Pogacic et al., 2001), suggest that SMN is involved in human telomerase biogenesis as an H/ACA snoRNP.
Our results also establish major differences between S. cerevisiae and vertebrate telomerase RNPs. The budding yeast telomerase RNA associates with Sm proteins and gains a 5′-TMG cap structure (Seto et al., 1999); both events are characteristic of snRNP assembly. However, hTR was not recovered from anti-Sm and anti-TMG immunoprecipitates (Figure 3), suggesting that human telomerase is not processed as an snRNP. This conclusion is supported by recent experiments in which in vitro–synthesized hTR was microinjected into Xenopus oocyte nuclei (Lukowiak et al., 2001). SnoRNAs are generated from two different genomic contexts: pre-mRNA introns or their own independent transcription unit (Weinstein and Steitz, 1999). The U3, U8, and U13 box C/D snoRNAs are transcribed from their own promoters and receive a TMG cap, whereas most intron-generated snoRNAs do not undergo 5′ hypermethylation (Yu et al., 1999; Speckmann et al., 2000). The lack of a TMG cap at the 5′ end of hTR is thus surprising, because it is expressed from its own promoter as an RNA polymerase II transcript (Feng et al., 1995; Hinkley et al., 1998). However, to the best of our knowledge, none of the metazoan H/ACA box snoRNAs have been shown to receive a TMG cap. Further studies will be necessary to better understand the processing and maturation events required for functional hTR formation.
Previous data support the view that telomerase reconstitution in RRLs is facilitated by the action of proteins present in the extracts (Holt et al., 1999; Licht and Collins, 1999). The negative effect of SMNΔN27 on in vitro telomerase reconstitution in RRL (Figure 5) is consistent with this view. However, when SMNΔN27-containing extracts from RRL or 293 cells were added to previously reconstituted human telomerase, the activity of telomerase was not affected (data not shown). Thus, the mutant form of SMN may affect telomerase not by inhibiting its catalytic activity but rather by affecting assembly in vitro. The incomplete inhibition of human telomerase reconstitution by SMNΔN27 in vitro could reflect a partial decrease in the efficiency of reconstitution or the inability to obtain concentrations of SMNΔN27 sufficient to completely sequester the endogenous RRL proteins involved in telomerase assembly.
The detailed characterization of the cellular components and progressive steps involved in human telomerase assembly will be critical for the rational design of new telomerase inhibitors. The identification of SMN as a telomerase-associated protein suggests that it will be an important player in the functional assembly and activation of human telomerase. Future studies will focus on understanding the specific role performed by SMN and its associated proteins in human telomerase biogenesis.
ACKNOWLEDGMENTS
We thank Drs. Robin Reed, Kathy Collins, and Witold Filipowicz for the Y12 serum, anti-dyskerin, and anti-hGAR1, respectively. We also thank Dr. Lea Harrington for the FLAG-hTERT construct as well as for the anti-TEP1 and anti-TEP2 antibodies. We are grateful to Sophie Dupuis for excellent technical assistance and encouragement. We thank the members of the Autexier laboratory for critical reading of the manuscript. F.B. is the recipient of a studentship from the Canadian Institutes of Health Research. F.-M.B. and J.C. are supported by an NCIC studentship and postdoctoral fellowship, respectively. S.R. is a Chercheur-Boursier of the Fonds de Recherches en Santé du Québec and is supported by a grant from the National Cancer Institute of Canada with funds from the Cancer Research Society. C.A. is the recipient of a CIHR scholarship and a Boehringer Ingelheim (Canada) Young Investigator Award. This work was supported by a grant from the CIHR (MOP-14026) to C.A.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–04–0216. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–04–0216.
REFERENCES
- Bachand F, Autexier C. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J Biol Chem, 1999;274:38027–38031. doi: 10.1074/jbc.274.53.38027. [DOI] [PubMed] [Google Scholar]
- Bachand F, Autexier C. Functional regions of human telomerase reverse transcriptase, and human telomerase RNA required for telomerase activity, and RNA-protein interactions. Mol Cell Biol, 2001;21:1888–1897. doi: 10.1128/MCB.21.5.1888-1897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol, 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B', and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA, 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-L, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell, 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- Chen T, Richard S. Structure-function analysis of Qk1: a lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol, 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm K, et al. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J, 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, et al. The human telomerase RNA component. Science, 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell, 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Friesen W, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell, 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Gall J, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell, 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein. Science, 1997a;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- Harrington L, Robinson M. Telomere dysfunction: multiple paths to the same end. Oncogene, 2002;21:592–597. doi: 10.1038/sj.onc.1205084. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DSK, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev, 1997b;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M, Szymczyk P, Shpargel K, Matera A. Coilin forms the bridge between Cajal bodies, and SMN, the spinal muscular atrophy protein. Genes Dev, 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss N, Knight S, Vulliamy T, Klauck S, Wiemann S, Mason P, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet, 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Hinkley C, Blasco M, Funk W, Feng J, Villeponteau B, Greider C, Herr W. The mouse telomerase RNA 5′-end lies just upstream of the telomerase template sequence. Nucleic Acids Res, 1998;26:532–536. doi: 10.1093/nar/26.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SE, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev, 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K, Gorzynski K, Hales C, Fischer U, Badbanchi F, Terns R, Terns M. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J Biol Chem, 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol, 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis J, de la Fortelle E, Raker V, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Le S, Sternglanz R, Greider CW. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol Biol Cell, 2000;11:999–1010. doi: 10.1091/mbc.11.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M, Boyle J, Hardin J, Steitz J. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science, 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- Lerner M, Steitz J. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA, 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev, 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J, 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell, 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Lukowiak A, Narayanan A, Li Z, Terns R, Terns M. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA, 2001;7:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat Res, 2000;451:227–240. doi: 10.1016/s0027-5107(00)00052-x. [DOI] [PubMed] [Google Scholar]
- Martin-Rivera L, Blasco M. Identification of functional domains, and dominant negative mutations in vertebrate telomerase RNA using an in vivo reconstitution system. J Biol Chem. 2001;276:5856–5865. doi: 10.1074/jbc.M008419200. [DOI] [PubMed] [Google Scholar]
- Martin-Rivera L, Herrera E, Albar JP, Blasco MA. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc Natl Acad Sci USA, 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. Nuclear bodies: multifaced subdomains of the interchromatin space. Trends Cell Biol, 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol, 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Culetto E, Walker D, Baylis H, Sattelle D, Davies K. The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum Mol Genet, 1999;8:2133–2143. doi: 10.1093/hmg/8.12.2133. [DOI] [PubMed] [Google Scholar]
- Mitchell J, Cheng J, Collins K. A Box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol, 1999a;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature, 1999b;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Moriarty T, Huard S, Dupuis S, Autexier C. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol Cell Biol, 2002;22:1253–1265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura C, Cascio D, Sawaya M, Eisenberg D. The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc Natl Acad Sci USA, 2001;98:5532–5537. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Lukowiak A, Jady B, Dragon F, Kiss T, Terns R, Terns M. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J, 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev, 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- Olson M, Kundr M, Szebene A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol, 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Oulton R, Harrington L. Telomeres, telomerase, and cancer: life on the edge of genomic stability. Curr Opin Oncol, 2000;12:74–81. doi: 10.1097/00001622-200001000-00013. [DOI] [PubMed] [Google Scholar]
- Paushkin S, Charroux B, Abel L, Perkinson R, Pellizzoni L, Dreyfuss G. The survival motor neuron protein of Schizosaccharomyces pombe: conservation of survival motor neuron interaction domains in divergent organisms. J Biol Chem, 2000;275:23841–23846. doi: 10.1074/jbc.M001441200. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin, and GAR1. Curr Biol, 2001a;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci USA, 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol, 2001b;152:75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell, 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs, and telomerase share evolutionarily conserved proteins NHP2, and NOP10. Mol Cell Biol, 2001;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B, Gotz R, Gunnersen J, Ure J, Toyka K, Smith A, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci USA, 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature, 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci USA, 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann W, Terns R, Terns M. The box C/D motif directs snoRNA 5′-cap hypermethylation. Nucleic Acids Res, 2000;28:4467–4473. doi: 10.1093/nar/28.22.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S, Ryan D, Ge H, Moore R, Young M, Lee T, Abelson J. Composition, and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell, 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Terns M, Terns R. Macromolecular complexes: SMN—the master assembler. Curr Biol, 2001;11:R862–864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet, 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Weinstein L, Steitz J. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol, 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- Will C, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure, and function. Curr Opin Cell Biol, 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Young P, Le T, Dunckley M, Nguyen T, Burghes A, Morris G. Nuclear gems, and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res, 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- Yu Y-T, Scharl E, Smith C, Steitz J. The growing world of small nuclear ribonucleoproteins. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 487–524. [Google Scholar]