Abstract
We used soybean (Glycine max) cDNA microarrays to identify candidate genes for a stable mutation at the Wp locus in soybean, which changed a purple-flowered phenotype to pink, and found that flavanone 3-hydroxylase cDNAs were overexpressed in purple flower buds relative to the pink. Restriction fragment length polymorphism analysis and RNA gel blots of purple and pink flower isolines, as well as the presence of a 5.7-kb transposon insertion in the wp mutant allele, have unequivocally shown that flavanone 3-hydroxylase gene 1 is the Wp locus. Moreover, the 5.7-kb insertion in wp represents a novel transposable element (termed Tgm-Express1) with inverted repeats closely related to those of other Tgms (transposable-like elements, G. max) but distinct in several characteristics, including the lack of subterminal inverted repeats. More significantly, Tgm-Express1 contains four truncated cellular genes from the soybean genome, resembling the Pack-MULEs (Mutator-like transposable elements) found in maize (Zea mays), rice (Oryza sativa), and Arabidopsis thaliana and the Helitrons of maize. The presence of the Tgm-Express1 element causing the wp mutation, as well as a second Tgm-Express2 element elsewhere in the soybean genome, extends the ability to acquire and transport host DNA segments to the CACTA family of elements, which includes both Tgm and the prototypical maize Spm/En.
INTRODUCTION
Soybean (Glycine max) plants display diverse coloration in their flowers, seed coats, hypocotyls, and trichome hairs (pubescence). Genetic studies have identified at least five loci affecting flower pigmentation, W1, W3, W4, Wm, and Wp, and these loci are distinct from those (I, R, and T) determining seed coat and pubescence coloration (Bernard and Weiss, 1973; Palmer and Kilen, 1987; Groose and Palmer, 1991). Cultivars with a purple flower phenotype have a W1W1 genotype, while those with white flowers have w1w1. Pink-flowered plants (W1_wpwp) were first observed in 1989 (Stephens and Nickell, 1991) and were derived from a mutable, chimeric plant having purple and pink flowers on the same plant. Interestingly, the derived pink flower lines averaged 22% higher in seed weight, 4% higher in protein, and 3% lower in oil compared with the purple-flowered lines derived from the chimeric plant (Stephens and Nickell, 1992; Stephens et al., 1993). The inheritance of the mutable phenotype and derived pink and purple lines showed a high rate of instability (Johnson et al., 1998).
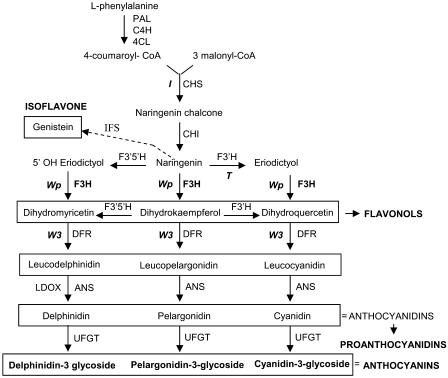
The soybean color phenotypes are likely the result of mutations affecting different enzymes of the anthocyanin and proanthocyanidin pathways. Molecular data indicated that the W3 locus encodes a dihydroflavonol reductase (Fasoula et al., 1995). The I locus corresponds to a 27-kb-long chalcone synthase gene cluster that exhibits a unique tissue-specific gene silencing mechanism in the seed coats mediated by short-interfering RNA (Todd and Vodkin, 1996; Senda et al., 2004; Tuteja et al., 2004). Recently, it has been shown that the pleitropic T locus that affects seed coat pigmentation and cell wall integrity encodes a flavonoid 3′ hydroxylase (Toda et al., 2002; Zabala and Vodkin, 2003) (Figure 1).
Figure 1.
Flavonoid Biosynthetic Pathway Indicating Cloned Loci (I, T, Wp, and W3) Influencing Flower, Hypocotyl, Pubescence, and Seed Coat Color in G. max.
Enzymes are indicated in uppercase letters. PAL, Phe ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; IFS, isoflavone synthase; F3′H, flavonoid 3′-hydroxylase; F3′, 5′H, flavonoid 3′,5′-hydroxylase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol-4-reductase; ANS, anthocyanidin synthase (also called LDOX for leucoanthocyanidin dioxygenase); UFGT, UDP-flavonoid glucosyltransferase.
An ideal use of microarray technology is the identification and isolation of cloned cDNAs representing genes with differing levels of expression, as in RNAs of two isolines varying only at a single locus. Therefore, we used soybean cDNA microarrays as preliminary screens of differential expression between the purple and pink flower isolines. We identified flavanone 3-hydroxylase (F3H) cDNAs that hybridized more strongly to RNA from young flower buds of a purple flower line (WpWp) than to RNA from a pink flower (wpwp) isoline. The differential expression of F3H was confirmed by RNA gel blotting experiments.
Fragment length polymorphisms between the purple and pink mutant isolines supported the discovery of a transposon insertion in the wp mutant allele, and the aberrant size F3H transcripts in the flower buds and seed coats of the pink-flowered line demonstrated that the Wp locus of soybean encodes an F3H gene. The DNA sequence of a 5.7-kb insertion in the mutant wp allele revealed a transposable element member of the CACTA family of transposons (Tgm, Spm, and Tam) (Vodkin et al., 1983; Rhodes and Vodkin, 1988). However, the element in the wp pink flower mutation differed from the other Tgm family members previously characterized in that it lacks the subterminal repeats and was laden with at least four genic fragments picked up from the host genome. The potential of CACTA elements to carry truncated genic fragments resembles that of the Pack-MULEs found in maize (Zea mays), rice (Oryza sativa), and Arabidopsis thaliana (Talbert and Chandler, 1988; Yu et al., 2000; Turcotte et al., 2001; Jiang et al., 2004) and the Helitron discovered more recently in maize (Lal et al., 2003; Gupta et al., 2005). It has been speculated that these types of elements have the potential to create novel genes through the rearrangement and fusion of noncontiguous genomic sequences captured by the transposons.
The discovery of the wp insertion element, named Tgm-Express1, adds a new level of transposable element complexity to the CACTA family of elements that includes the Spm/En (Suppressor-mutator/Enhancer elements), one of the original maize transposable elements first described genetically in the 1940s by Barbara McClintock and Peter Peterson (reviewed in Wessler 1988; Gierl et al., 1989). Another occurrence of the capture of genomic sequences by a CACTA-type element has been shown for the Tpn1 of the Japanese morning glory (Ipomoea tricolor) (Takahashi et al., 1999). The existence of the Tgm-Express and CACTA family elements in other plant species indicates that the ability to acquire, recombine, and replicate host genomic DNA fragments may be widespread. In addition, they represent only a few genetically described movements of cellular genes revealed as insertional inactivations of the target genes rather than by extrapolation from data mining of high-throughput genome sequencing data.
RESULTS
Identification of F3H as a Candidate for the Flower Color Gene Wp Using Soybean cDNA Microarrays as Preliminary Screens
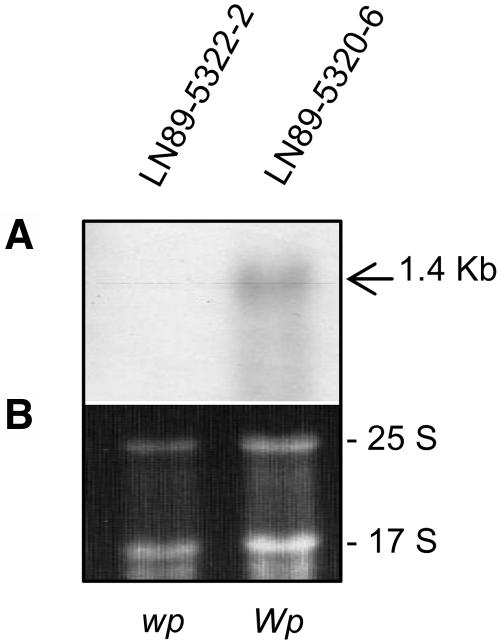
Two stable isolines, one with purple flowers (WpWp) and a second with pink flowers (wpwp), were recovered from the progeny of a mutant variegated plant that arose spontaneously in the field from a cross that was expected to produce purple- or white-flowered plants. To capture and understand the nature of such a mutational event, we attempted to identify and clone the gene at the Wp locus. To that end, we screened soybean cDNA microarrays (Vodkin et al., 2004) to obtain preliminary information on differential gene expression between the isolines. Total RNA was extracted from young flower buds of two soybean isolines varying only at the Wp locus, LN89-5320-6 (Wp) purple flower and LN-5322-2 (wp) pink flower (Table 1). After global normalization within each slide and between the dye-swap replicates, the data were displayed as log scatterplots (see Supplemental Figure 1 online for an example). The majority of the 9728 cDNAs on the array hybridized to RNAs from the purple and pink lines in a similar fashion. However, multiple cDNAs representing two F3H EST clones (Gm-c1012-683, accession number AY669324, and Gm-c1019-2646, accession number AW277481) that were each printed eight times on the arrays were found to deviate from equal expression between the two isolines. The fluorescence intensity values after background subtraction and global normalization between replicates were examined for all 16 repetitive F3H cDNAs that are present on each array (see Supplemental Table 1 and Supplemental Figure 2 online). Figure 2 confirms with an RNA gel blot using the full-length Gm-c1012-683 EST clone as probe that F3H cytoplasmic transcripts are not detectable in flower buds with the pink flower (wp/wp) genotype compared with those of the purple-flowered line (Wp/Wp).
Table 1.
Genotypes and Flower Phenotypes of Soybean Lines Used in This Study
| Lines | Genotype | Phenotype |
|---|---|---|
| (1) LN89-5320-6 | iiRtW1Wp | Stable purple flower |
| (2) LN89-5322-2 | iiRtW1wp | Stable pink flower |
| (3) LN89-5320-8-53 | iiRtW1 wpm | Multicolored, pink and purple flowers |
| (4) RM30 | i R*TW1Wp | Purple flower |
| (5) RM38 | i r*TW1Wp | Purple flower |
| (6) Williams | iiRTw1Wp | White flower |
Figure 2.
RNA Gel Blot with the Same Flower Bud RNAs Used to Hybridize to the Soybean Microarrays.
(A) RNA gel blot containing 10 micrograms of each RNA sample (cleaned by RNeasy Qiagen minicolumns) from flower buds of LN89-5322-2 (wp) and LN89-5320-6 (Wp) isolines. A 1.4-kb transcript is detected in the purple flower line (Wp) and is not visible in the pink flower (wp) isoline. The Gm-c1012-683 cDNA clone was used as probe.
(B) Ethidium bromide–stained gel prior to membrane transfer. The 25 S and 17 S rRNAs are shown to compare sample loading.
An independent experiment using even younger flower buds as the source of RNA for two additional microarrays was performed and also showed that the F3H cDNAs were differentially expressed between the purple (Wp) and pink (wp) flower buds (see Supplemental Table 2 online). As will be discussed later, we subsequently found that expression of F3H is higher in immature seed coats than in flowers; therefore, we performed a third, independent microarray screen with RNAs from the immature seed coats (see Supplemental Figures 3 and 4 and Supplemental Table 3 online). Again, the results from the arrays indicated differential expression of the F3H cDNAs in the seed coats of the Wp/Wp and wp/wp isolines. Thus, the results of the microarray screens and RNA gel blots suggested that Wp encodes F3H or affects its transcript levels.
Characterization of a G. max F3H cDNA
F3H cDNA clones have been isolated from many plant species (Britsch and Grisebach, 1986; Britsch et al., 1993; Sparvoli et al., 1994; Charrier et al., 1995; Honda et al., 2002), and the genomic sequences of only two of the genes (Medicago sativa and Arabidopsis) have been described (Charrier et al., 1995; Pelletier and Shirley, 1996). Alignment of the full-length EST clone Gm-c1012-683–derived amino acid sequence to those of 11 other F3Hs, M. sativa, Vitis vinifera, Petunia hybrida, Dianthus caryophyllus, Callistephus chinensis, Matthiola incana, Malus domestica, Prunus persica, Arabidopsis, Hordeum vulgare, and Z. mays, showed homology to all the sequences, with H. vulgare (69% identical, 81% similar) being the least and P. persica (85% identical, 93% similar) the most similar (see Supplemental Table 4 online). A multiple sequence alignment of the G. max F3H-derived amino acid sequence to a consensus F3H amino acid sequence using nine of the most closely related F3H amino acid sequences mentioned above is shown in Figure 3.
Figure 3.
G. max F3H Amino Acid Sequence Alignment.
The G. max F3H derived amino acid sequence is shown aligned to a F3H consensus amino acid sequence derived from nine other F3H amino acid sequences (see Supplemental Table 4 online). The asterisks indicate conserved His residue chelators of ferrous ions at the enzyme's active site (positions 77, 219, and 277). A conserved Asp (#) also believed to be involved in the iron binding site of the enzyme is located at position 221 (Britsch et al., 1993). A Ser residue (+) at position 289 has functional significance for 2-oxoglutarate binding (Lukacin et al., 2000). Black boxes indicate identical residues, and gray boxes indicate conserved residues between the two sequences. Amino acid numbering is shown at left.
Restriction Site Polymorphisms Associated with the Wp Locus
In order to determine if the F3H gene correlated with the Wp locus, an restriction fragment length polymorphism (RFLP) analysis was performed using five soybean lines varying at the Wp locus (Table 1). Line LN89-5320-8-53 is a mutable line, derived from a plant with a mutable allele (wpm) and a variegated flower phenotype. The stable mutant line LN89-5322-2 with the pink flower phenotype was derived from that original mutable plant and is homozygous (wpwp). Also derived from the same plant with the mutable allele was a stable isoline (LN89-5320-6) with a purple flower phenotype and homozygous at that locus (WpWp). RM30 and RM38 are not isolines of those mentioned above but are homozygous for the Wp allele and they vary at the R locus.
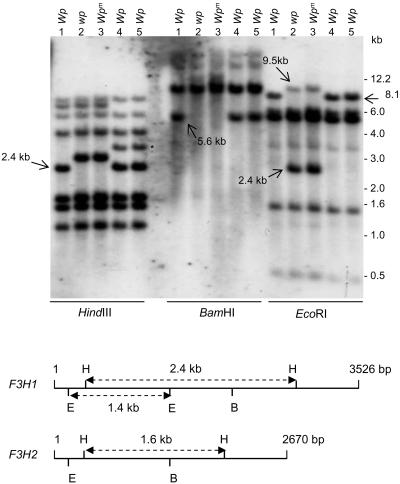
Figure 4 shows the results of hybridizing the F3H probe (PCR-amplified Gm-c1012-683) to DNA fragments resulting from digestion of genomic DNAs with three restriction enzymes: HindIII, BamHI, and EcoRI. Each enzyme digestion shows polymorphic changes that distinguish Wp from wp and wpm. With HindIII, a 2.4-kb band found in lines with the Wp allele (lanes 1, 4, and 5, Figure 4) is replaced by a higher molecular weight DNA fragment (2.78 kb) in lines with the mutant alleles wp and wpm (lines 2 and 3, Table 1, Figure 4). Likewise, an ∼5.6-kb band present in the Wp lines is absent in the mutant lines in the BamHI digests. With EcoRI, an 8.1-kb band is replaced by a 2.4-kb smaller band with a 9.5-kb fragment also detectable in the mutant lines 2 and 3. These novel polymorphic fragments observed in the mutant lines were accurately sized and corroborated by in silico restriction analysis of the transposon sequence (described later in Figure 9) inserted in the recessive wp allele. These polymorphisms associated with the DNA insertion in wp strongly support that Wp is the F3H gene.
Figure 4.
RFLP Analysis of Soybean Lines Varying at the Wp Locus.
DNA gel blot of genomic DNA from five soybean lines digested with three different restriction enzymes, HindIII, BamHI, and EcoRI. The genotype and phenotype of each line (1 to 5) are described in Table 1. The type of Wp allele of each line is indicated at the top. The sizes of marker fragments are shown at the right in kilobases. The Gm-c1012-683 cDNA clone was used as probe. Arrows point to DNA polymorphisms at the Wp locus. A polymorphism detected in the RM lines (4 and 5; asterisk) is likely due to genetic diversity, since the RM lines are not isolines with lines 1 to 3 (Table 1). The bottom panel is a map of HindIII, BamHI, and EcoRI restriction sites for F3H1 and F3H2. The size of these genomic sequences is indicated in base pairs at the right. The sizes of HindIII and EcoRI fragments are given in kilobases.
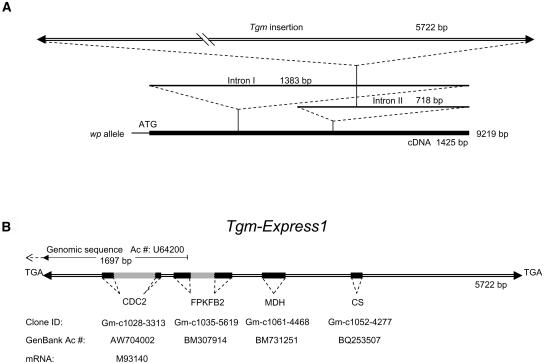
Figure 9.
Schematic Representation of the Mutant Allele wp Genomic Sequence and Details of the Tmg-Express1 Insertion in Intron II.
(A) Genomic sequence of the mutant wp allele obtained from PCR fragments amplified from the wp line 2 (Table 1). The introns are indicated and their length given in base pairs. The 5725-bp Tgm-Express1 insertion in Intron II, 231 bp into the intron, is drawn at the top as a double line, with the arrowheads representing the inverted repeats. The full length of the mutant gene, 9251 bp, is written to the right of the thicker solid line representing the cDNA if proper splicing should take place.
(B) Detailed representation of the Tgm-Express1 insertion drawn to scale and flanked by the target site duplication, TGA. Arrowheads represent the 24-bp imperfect inverted repeats. The segments of sequence with high identity scores to soybean EST sequences in GenBank are indicated by the solid blocks and intron regions by gray boxes. The annotation, clone IDs, and GenBank accession numbers are given underneath. The first portion of the transposon insertion with 97% identity to a genomic sequence entered in GenBank as G. max CDC2 (1) pseudogene is measured at top with the 1697-bp line and accession number U64200. The dashed line pointing upstream of the 1697-bp portion of the U64200 genomic fragment represents the diverging sequence with no similarity to the F3H Intron II to the left of the insertion. Instead, it matches an EST with accession number CF922959. The left border inverted repeat of U64200 identical to that in the wp insertion is represented with an arrowhead. The sequence divergence upstream of the identical left border inverted repeats reveals the existence of a second Tgm-Express2 insertion somewhere else in the genome. CDC2, cell division cycle 2; FPKFB2, fructose-6-phosphate 2-kinase/fructose-2-6-biphosphatase; MDH, malate dehydrogenase; CS, Cys synthase.
As it will be shown later, two related genes (F3H1 and F3H2) were amplified by PCR with a specific set of primers from the LN89-5320-6 line with the Wp allele. The multiple hybridizing bands observed in the DNA gel blot shown in Figure 4 are consistent with the existence of more than one F3H gene. The genomic sequence of the two genes allowed the identification of some of the restriction fragments in the DNA gel blot in Figure 4. HindIII cuts twice in both genes, generating internal fragments of different sizes in each gene, F3H1 (2452 bp) and F3H2 (1622 bp). The 1.6-kb internal HindIII fragment of the F3H2 gene is present in all lines, while the 2.4 kb of F3H1 is missing in the wp mutant lines. The higher molecular weight fragment hybridizing in the mutant lines is the result of a 5.7-kb insertion in Intron II of F3H1 that contains three additional HindIII sites. BamHI cuts only once in both genes, and the 5.6-kb fragment missing in the mutant lines is the result of the 5.7-kb insertion bringing the smaller size band to equal that of the ∼11.3-kb labeled molecular weight band. EcoRI cuts twice in F3H1, generating an internal fragment of 1458 bp, but it cuts only once in F3H2. The 1.4-kb EcoRI fragment was present in all lines and includes most of the first exon and part of the first intron that is identical in F3H1 of the Wp line and the wp allele of the mutant isoline. The 2.4- and 9.5-kb polymorphic fragments in the mutant DNAs (Figure 4, DNA gel blot) are the result of two additional EcoRI sites in the 5.7-kb transposon insertion.
G. max F3H Tissue-Specific Expression
Even though the Wp locus had been grouped with those genes that control flower color in soybean (W1, W3, W4, and Wm) that seem to be distinct from those contributing to seed color (I, R, and T), the Wp locus also affects the color of the seed coats. Seeds of plants with purple flowers, tawny pubescence, and the genotype i/i R/-T/-Wp/- are black. Seed coats of plants with pink flowers, tawny pubescence, and genotype i/iR/-T/-wp/wp are lighter and grayish. Imperfect black seed coats are characteristic of plants with purple flowers and gray pubescence with genotype i/iR/-t/tWp/-. By contrast, plants with pink flowers and gray pubescence having the genotype i/iR/-t/twp/wp have very lightly colored seed coats (Johnson et al., 1998). If the F3H cDNA we had isolated corresponds to the Wp locus, the expression of this gene in the different plant tissues and in tissues of plants with differing flower and seed coat color phenotypes should be different accordingly.
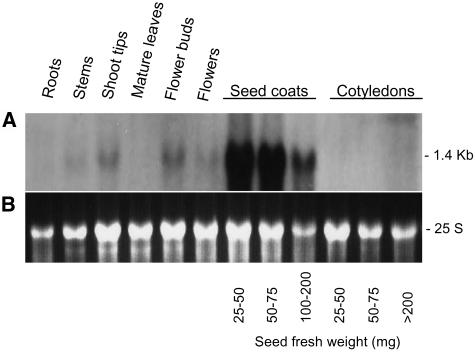
Figure 5 shows the result of an RNA gel blot containing RNAs extracted from several tissues of a Williams cultivar plant with genotype iiRTw1Wp that was hybridized to the PCR-amplified Gm-c1012-683 probe. No expression was detected in roots, mature leaves, or cotyledons. A very low abundance, 1.4-kb transcript was observed in stems and mature flowers. Shoot tips and flower buds contained more of this size transcript but little compared with the larger amount detected at the early stages of seed coat development. This pattern of transcript accumulation in shoot tips, flower buds, and seed coats agrees with the expected tissue-specific expression for the Wp allele, further supporting that F3H is Wp.
Figure 5.
G. max F3H Tissue-Specific Expression.
(A) RNA gel blot containing 10 μg of total RNA samples purified from roots, stems, shoot tips, mature leaves, flower buds, flowers, seed coats, and cotyledons of soybean plants (cv Williams) (ii, R, T, w1, and Wp). Seed coats and cotyledons from three different stages of seed development were used. Seed fresh weight in milligrams is shown at bottom. The Gm-c1012-683 cDNA clone was used as probe.
(B) Ethidium bromide–stained gel prior to membrane transfer. The 25 S rRNA is shown to compare RNA sample loading.
F3H Expression in Flower Buds and Seed Coats in Soybean Lines Varying at the Wp Locus
As described earlier, we found out that those flower buds of the soybean pink mutant line LN89-5322-2 (iiRtW1wp) had reduced amounts of transcripts of the F3H gene, while buds of the purple-flowered isoline LN89-5320-6 (iiRtW1Wp) contained a higher amount. Although the level of transcripts was not very high in flower buds, this differential was detectable in RNA gel blots (Figure 2) and sufficient to help in the identification of the F3H cDNAs in the soybean microarray experiments described above.
Once it had been determined that seed coats of the Williams cultivar contained higher levels of the 1.4-kb F3H transcript at early stages of seed development, it was relevant to determine the expression of this gene in seed coats of the soybean isolines varying at the Wp locus (Table 1, lines 1 to 3). Figure 6 shows the results of an RNA gel blot containing RNAs from seed coats at three early stages of seed development from LN89-5320-8-53 (wpmwpm), LN89-5320-6 (WpWp), and LN89-5322-2 (wpwp) isolines hybridized to the F3H probe (Gm-c1012-683). As expected, the 1.4-kb transcript was present in great abundance in the purple flower (WpWp) line, but the apparent lack of that transcript in both the mutable (wpmwpm) and pink flower mutant (wpwp) lines was very striking. The F3H tissue-specific expression and the lack of the 1.4-kb transcript in the pink mutant line together with the F3H restriction site polymorphisms associated to the pink line mutation is strong evidence that the F3H gene is the Wp locus.
Figure 6.
G. max F3H Expression during Seed Coat Development in Soybean Lines Varying at the Wp Locus.
(A) RNA gel blot containing 10 μg of total RNA from three seed coat developmental stages in three flower color isolines: LN89-5320-8-53 (wpm wpm), LN89-5320-6 (WpWp), and LN89-5322-2 (wpwp) (Table 1). Seed fresh weight of each seed coat sample in milligrams is shown at bottom. The Gm-c1012-683 cDNA clone was used as probe.
(B) Ethidium bromide–stained gel prior to membrane transfer. The 25 S rRNA is shown to compare RNA sample loading.
As it will be described below, we have isolated two related F3H genes, and based on the differences in their DNA sequences, the expected transcript size of F3H2 should be only 25 bp smaller than that of F3H1 and expressed in both Wp and wp lines. The lack of hybridization to the mutant line's RNA suggests that the amount of transcripts derived from this second family member gene is undetectably low or not existent (Figure 6).
F3H Genomic DNAs from Soybean Lines with Wp and wp Genotypes
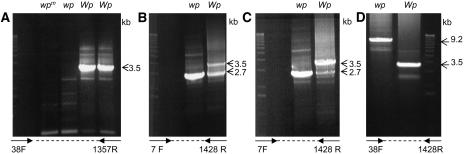
To further characterize the Wp locus, the amplification of genomic DNAs from four differing Wp lines (Table 1, lines 1 to 4) was attempted using the 38F and 1357R primer set (see Methods for reaction details). Figure 7A shows the amplification products of those reactions. A 3.5-kb fragment was amplified only from the two lines with the Wp allele. No major genomic PCR product was obtained from the lines with the wpm and wp mutant alleles. However, when the 7F and 1428R primer set was used, a smaller genomic fragment of 2.7 kb in size was amplified in the mutant line with the wp allele (Figure 7B). By contrast, two fragments, 3.5 and 2.7 kb in size, resulted from the line with the Wp allele (Figure 7B). Figure 7C shows the results of an experiment as the one described for Figure 7B except that the annealing temperature of the PCR reaction was raised 3°C. This change favored the amplification of the larger 3.5-kb fragment in the Wp line resembling the result obtained with the 38F and 1357R primers (Figure 7A). Sequence analysis of the 3.5-kb genomic fragment from the Wp line and the 2.7-kb fragment from wp have shown that the two fragments represent two related genes. The gene contained in fragment 3.5 kb was named F3H1 and the one in the 2.7 kb was named F3H2.
Figure 7.
Variation in G. max F3H Genomic Amplification between Lines Varying at the Wp Locus.
Ethidium bromide–stained gels showing the results of genomic PCR amplification using the following.
(A) The 38F and 1357R primer set. A 3.5-kb fragment was amplified in two lines with the Wp allele (LN89-5320-6 [Wp] and RM30 [Wp]) (Table 1). By contrast, no significant amplification occurred in the mutant isolines LN89-5320-8-53 (wpm) and LN89-5322-2 (wp).
(B) The 7F and 1428R primer set. A 3.5-kb fragment was amplified in the LN89-5320-6 (Wp) line, and an additional 2.7-kb fragment was amplified in both the LN89-5320-6 (Wp) and the mutant isoline, LN89-5322-2 (wp).
(C) The 7F and 1428R primer set and higher annealing temperature (58°C). This change favored the amplification of the 3.5-kb genomic fragment in the LN89-5320-6 (Wp) line.
(D) The 38F and 1428R primers and new set of PCR conditions that favor amplification of larger fragments (see Methods). A 3.5-kb fragment was amplified in the LN89-5320-6 (Wp) line, while a 9.2-kb fragment was amplified in the mutant LN89-5322-2 (wp) isoline.
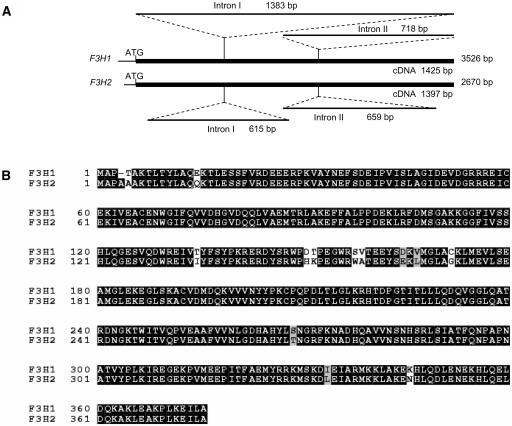
Using multiple primer sets that allowed the specific amplification of each gene, we were able to sequence both genes in their entirety. Figure 8A and Supplemental Table 5 online show that the F3H1 and F3H2 genes were 3526 and 2670 bp long, respectively (accession numbers AY669325 and AY669326). Both genes had two introns at the same location at positions 432 and 860 of the cDNA sequence (AF198451). Intron I sequences were very different, with Intron I of F3H1 being 1383 bp and that of F3H2 615 bp long. Multiple indels must have occurred in the evolution of those two intron sequences. Intron II sequences were less divergent but still contained some small indels and multiple base pair substitutions. Both introns follow the GT-AG splice site convention (Brown, 1986). The 5′ end sequence of F3H2 had a 31-bp deletion that includes the entire 22 bp of the 38F primer. This explains the inability to amplify F3H2 from the mutant wp or Wp lines with the 38F and 1357R primer set. The sequences of the three exons were more closely related than that of the introns but with a total of 38 bp substitutions. The F3H2 open reading frame with 376 amino acids is one amino acid longer than that of F3H1 (375 amino acids). In addition, it contained four conserved amino acid substitutions and eight relatively conserved amino acid changes except for the Ser-to-Trp mutation at position 158 (Figure 8B). None of those changes resulted in a stop codon that could truncate the translation product. These results imply that if F3H2 were to be expressed at low levels in the flowers, the translated protein could possibly account for the pigmentation of the pink flower mutant line.
Figure 8.
Schematic Representation of the F3H1 and F3H2 Genes and Alignment of the Two Derived Amino Acid Sequences.
(A) The top schematic represents the genomic sequence of F3H1 obtained from PCR fragments amplified from the Wp line 1 (Table 1). The introns are indicated and their length given in base pairs. The bottom schematic represents the genomic sequence of F3H2 generated from PCR fragments amplified from the wp line 2 (Table 1). The location of the introns and their length are indicated. The full length of the cDNAs is given underneath the thick solid lines representing each gene, and the entire length of each genomic sequence is given to the right of each thick solid line.
(B) Alignment of the two derived amino acid sequences from F3H1 and F3H2. Black shading indicates identical amino acids. Gray shading indicates conserved residues, and unshaded amino acids represent differences between the two gene's amino acid sequences, including the Ser-to-Trp nonconserved amino acid change at position 158. Amino acid numbering is shown at left.
The Large Insertion Element in wp Captured Four Genic Fragments
Attempts to amplify the entire wp mutant allele using the PCR conditions that successfully amplified F3H1 and 2 with the 38F primer that selectively amplified F3H1 failed. It was only when the second set of PCR conditions described in Methods was used that we were able to amplify the 9.2-kb fragment containing the wp allele (Figure 7D).
Analysis of the 9219-bp genomic sequence (accession number AY994154) revealed the existence of a 5722-bp insertion in Intron II, located 231 bp into the intron (Figure 9A), and it appears to be a member of the CACTA family of transposable elements (Tgm, Tam, and Spm) (Rhodes and Vodkin, 1988). The termini are imperfect inverted repeats flanked by a 3-bp duplication (TGA) of the target DNA. The left border sequence of the wp insertion CACTACTAAAAAAATCTGTTTTT is very similar to that of a soybean transposable element, Tgm1 (Vodkin et al., 1983; Rhodes and Vodkin, 1988), inserted in the lectin gene, CACTATTAGAAAAXTATGTTTTT, where the underlined bases are different from those in the wp insertion and the X represents a base insertion (A) in the wp element. The right border of the wp insertion is an imperfect 24-bp inverted repeat, TAAAAAACCTCTTTTGTAGTAGTG, where the underlined bases are not complementary to the corresponding ones in the left border inverted repeat. If the termini were to be displayed in a pairwise structure, the divergent bases would map to the loops of two putative stem-loop regions (see Supplemental Figure 5 online).
Despite those similarities, the wp insertion is very different from other Tgm elements. It lacks the subterminal repeats found in all other Tgm elements studied (Vodkin et al., 1983; Rhodes and Vodkin, 1985, 1988), and unlike Tgm1, which is found in a coding region, the wp insertion must have targeted the intron A-and T-rich sequences surrounding the 5.7-kb element. This is not unique to this insertion, as most of the Arabidopsis transposons mined from the DNA sequence show a distribution preference for A- and T-rich sequences (Le et al., 2000).
More importantly, we found similarities between the 5.7-kb insertion sequence and host cellular genes as shown in Figure 9B. For example, two stretches of 219 and 203 bp, separated by a 258-bp intronic sequence, had 96 to 98% identity to G. max ESTs annotated as fructose-6-phosphate 2-kinase/fructose-2-6-biphosphatase (FPKFB2) (clone ID, Gm-c1035-5619, accession number, BM307914, and Gm-c1059-4042, accession number BM521027). Further downstream and interspersed with genomic sequences showing no similarities to other sequences in GenBank, there were 314 bp 93% identical to a soybean malate dehydrogenase EST with clone ID Gm-c1061-4468 and accession number BM731251 and a shorter 101 bp also 93% identical to a soybean Cys synthase EST with clone ID Gm-c1052-4277 and accession number BQ253507. The largest region of homology of the wp insertion was to a contiguous 1697-bp region with 97% identity to a 2262-bp G. max genomic sequence previously entered into GenBank with accession number U64200 that contains similarity to cell division cycle 2 (CDC2) protein kinase cDNAs. That same region of the insertion also had 97% identity to a total of 305 bp sequence of a G. max protein kinase (p34cdc2) mRNA, accession number M93140 (Miao et al., 1993). Further inspection of the U64200 genomic sequence revealed that it contained the left border inverted repeat CACTACTAAAAAAATCTGTTTTT identical to the one found in the wp insertion (shown in Figure 9B as a solid arrowhead). In addition, we found that the upstream sequence diverged from that of wp, indicating that the U64200 sequence is not wp but is likely to represent a second related Tgm insertion somewhere else in the genome.
A transposable element containing a fragment of host genome was first isolated in maize and named MRS-A (for Mu-related sequence) (Talbert and Chandler, 1988). More recently with the availability of entire genome sequences for rice and Arabidopsis, many Mu-like elements carrying host cellular genes have been found in those two plant species (Yu et al., 2000; Turcotte et al., 2001; Jiang et al., 2004). Jiang et al. named these elements Pack-MULEs and gathered in silico information indicating that the captured genes are expressed and might be functional (Jiang et al., 2004). More recently, a novel family of transposons of maize called Helitrons has been found to be embedded with portions of cellular pseudogenes (Lal et al., 2003; Gupta et al., 2005). We have named the element found in the wp allele Tgm-Express1 and chose the word Express to designate the type of Tgm elements that transport gene fragments picked up from the host genome.
The orientation of the four genic fragments carried by the Tgm-Express element is the same as the direction of transcription of the F3H1 gene that the element interrupts. Because of the scant amount of genomic sequence available for the soybean, we could not determine the proximity of the host genes corresponding to the gene fragments embedded in the Tgm-Express element. Although extensive simple sequence repeat maps exist for soybean (Cregan et al., 1999), there is no mapping data for the CDC2, FPKFB2, malate dehydrogenase (MDH), and Cys synthase (CS) genes. A search of legume databases (http://www.comparative-legumes.org/lisg/seqlist.html) found multiple BACs containing MDH, CS, and CDC2 in Lotus japonicum and Medicago truncatula, but none of the genes were colocated in the same BAC sequence. In Arabidopsis, chromosome 3 has two CDC genes (but not CDC2), two CS loci (out of four), and MDH, but they are thousands of kilobases apart. F2KP is found on chromosome 1.
Alignment of the entire 9219-bp sequence of wp to the F3H1 genomic sequence of Wp using EMBOSS pairwise alignment algorithms from EMBL-EBI (European Bioinformatics Institute; http://www.ebi.ac.uk/emboss/align/) (see Supplemental Table 6 online) showed that except for the Tgm-Express1 insertion in wp, the two allelic sequences were identical, consistent with the DNA insertion in the F3H gene being a recent event.
F3H RT-PCR from Wp and Mutant Lines
The results of RNA gel blots (Figures 2 and 6) had shown that there was little or no expression in either flower buds or seed coats of the wp line containing the insertion when hybridizing it to the F3H cDNA probe. To verify that the insertion was affecting expression of F3H, we used a more sensitive technique, RT-PCR, with DNA-free RNAs isolated from both flower buds and seed coats and two sets of primers, 7F and 1428R or 38F and 1357R. As mentioned earlier, the 38F, 5′ end primer allows the specific amplification of F3H1, since the entire 38F primer sequence is deleted in F3H2. By contrast, 7F amplifies both F3H1 and 2 (see Methods for more details).
The results of amplifying the first-strand cDNAs synthesized from seed coat RNAs of isolines LN89-5320-8-53 (wpm), LN89-5320-6 (Wp), and LN89-5322-2 (wp) (Table 1) using the 7F and 1428R primer set showed a PCR fragment of 1.4 kb in the purple line with the Wp allele, which is in agreement with the expected size of 1422 bp (see Supplemental Figure 6A online). No PCR product of that size was detected when using the seed coat RNAs of the mutant line. Instead, a higher molecular weight PCR band of ∼2.3 kb (diffuse, not a tight band), possibly representing multiple size fragments, was obtained (see Supplemental Figure 6A online, wpm and wp). The latter were not the result of DNA amplification, since the negative control reactions lacking reverse transcriptase were devoid of amplification products. In a similar experiment in which flower bud RNAs from the pink mutant and purple lines were used instead, very similar results were obtained (data not shown).
When the PCR conditions that allowed amplification of larger fragments (9.2 kb) were used with the 7F and 1428R primer pair and first-strand cDNAs synthesized from seed coats and flower bud RNAs of the Wp and wp isolines, the larger diffused, ∼2.3-kb product would resolve into a discreet set of bands relatively close in size (see Supplemental Figure 6B online; data not shown). These results suggest that the large transposon insertion (5.7 kb) in Intron II of the wp allele may hamper intron processing, resulting in multiple sizes and wrongly processed transcripts. These aberrant, larger transcripts may be targeted for degradation, explaining the results obtained with RNA gel blots, mainly the lack or very low abundance of the fully processed 1.4-kb RNAs in the mutant lines. These RT-PCR results also show that F3H2 may not be expressed due to the lack of an amplicon of ∼1.4 kb in size in the mutant line where it can be distinguished from the fully processed F3H1 1.4-kb transcript.
DISCUSSION
The F3H Gene Is the Molecular Target of the wp Insertion
Genetic studies had determined that a rare pink flower mutation in soybean was due to a single gene locus named wp by Stephens and Nickell (1991) and that it arose spontaneously in the field from a chimeric plant segregating pink and purple flowers. We analyzed RNAs from the isogenic pink and purple lines derived from the mutable plant using soybean cDNA arrays as preliminary screens of differential gene expression. These results were then validated by RNA gel blotting experiments. Two cDNAs were identified with sequence similarity to other F3H sequences in GenBank that were overexpressed in the Wp soybean line relative to the wp mutant isoline. One of the cDNAs was full length, and its derived amino acid sequence contained all the motifs characteristic of an active F3H enzyme. RFLP analysis, hybridizations to RNA gel blots using the F3H full-length cDNA (Gm-c1012-683) as probe, RT-PCR, and genomic DNA PCR amplifications and sequencing with isolines varying at the Wp locus have proven unequivocally that the Wp locus encodes a potentially functional F3H and that a 5.7-kb transposon insertion created the wp mutant allele. The identification and isolation of this soybean gene on the basis of differential hybridization to the soybean cDNA microarrays with RNAs from variant and normal isolines is a demonstration of the potential that microarrays offer for gene discovery in instances where isogenic lines varying at a locus of interest are available, and the list of candidate genes derived from the arrays is relatively small, enabling testing by other methods, including RNA gel blotting and RT-PCR.
The identification of a full-length F3H EST (Gm-c1012-683) permitted the discovery of two family member genes, F3H1 and F3H2, through the amplification of their genomic sequences. Based on their derived amino acid sequence alignment, the amino acid differences appear to be relatively conservative except for the Ser to Trp at position 158 (Figure 8). However, no 1.4-kb transcript was hybridized to the F3H probe in RNA gel blots containing RNAs extracted from either flower buds (Figure 2) or seed coats (Figure 6) of the pink flower plants from which F3H2 was amplified and sequenced. The fact that 1.4-kb transcripts from the pink mutant line were not detected in RNA gel blots suggests that transcripts for the F3H2 gene do not accumulate or accumulate only in very low amounts. Two other pieces of evidence support no or low expression of F3H2: one, the failure to amplify its transcript's derived cDNA via RT-PCR, and the second, the lack of EST sequences in GenBank representing the F3H2 transcribed sequence. Out of >29,000 primary ESTs from flowers and immature seed coats, 52 were F3H ESTs, and none contained the F3H2 specific sequence.
Because the F3H enzyme is required in the three branches leading to the synthesis of the three groups of anthocyanins (Figure 1), the pink flower mutation in soybean (wp) cannot be a null mutation unless F3H2 or other not yet identified family members could function in at least one of the pathway's three branches. However, the RT-PCR experiments showed several bands in the mutant lines reflecting multiple, larger transcript sizes (see Supplemental Figure 6B online wpm and wp). These, most likely, are the result of the 5.7-kb insertion in Intron II of F3H1 causing improper RNA splicing. A few accurately spliced transcripts could allow translation of active peptides sufficient to allow the synthesis of some pigment resulting in the pink flower phenotype. Evidence exists of accurate removal of transcribed elements from RNA as if they were introns, permitting gene expression even when the insertion was found in an exon (Wessler, 1988).
Evidence has been mounting that the F3H gene is a key, highly regulated enzyme in controlling the flow of naringenine toward the synthesis of flavanols, anthocyanins, and proanthocyanidins in soybeans. For example, in order to substantially increase the levels of isoflavone genistein in transgenic Arabidopsis using isoflavone synthase, Chang-Jun et al. (2002) found that it was necessary to transform a line that had a knocked out F3H. Their results strongly support that F3H is critical for preferential channeling of naringenin into flavonol synthesis and away from isoflavone synthesis (Figure 1). Thus, regulation of F3H expression is of great consequence. From our work, the soybean seed F3H is highly expressed in the seed coat but is undetectable in the cotyledons (Figure 5). Likewise, the anthocyanins and proanthocyanidins accumulate in the pigmented seed coats of lines that are homozygous for the recessive i alleles and not in the cotyledons of those plants. By contrast, isoflavones, such as genistein, accumulate in the cotyledons but are not abundant in the seed coats. Interestingly, we have shown here that expression of F3H mRNA appears to be very low or absent in the cotyledons, thus supporting the hypothesis that F3H plays a role in directing the flow of substrate to the anthocyanin/proanthocyanidin branch of the pathway and away from the isoflavone branch. The plant may achieve regulation of the types of flavonoids in a particular tissue or organ by differential expression or regulation of its F3H gene(s).
Tgm-Express Elements Acquire Cellular Genes
Genomic amplification of F3H1 from normal and mutant isolines revealed the molecular nature of the wp allele to be a 5.7-kb transposon insertion in the second intron of the F3H1 gene (Figure 9A). This transposon insertion had features such as the 3-bp target site duplication and inverted repeats characteristic of the CACTA family of transposable elements (Tgm, Spm, and Tam1) but lacks the complex subterminal repeats of other Tgm elements and any remnants of a transposase (Vodkin et al., 1983; Rhodes and Vodkin, 1985, 1988). In addition, the 5.7-kb transposon insertion differed from previously reported Tgm elements in that it had accumulated at least four identifiable host gene fragments (CDC2, FPKFB2, MDH, and CS) reminiscent of the Pack-MULEs found in maize, rice, and Arabidopsis (Talbert and Chandler, 1988; Yu et al., 2000; Turcotte et al., 2001; Jiang et al., 2004), maize Helitrons (Lal et al., 2003; Gupta et al., 2005), and Tpn1 of Japanese morning glory (Takahashi et al., 1999). We have named this element Tgm-Express1 to distinguish it from other Tgms that do not carry host genes and propose the term CACTA-Express to distinguish the CACTA multigenic elements from the Pack-MULEs.
The 2262-bp G. max genomic sequence (U64200) with 1697 bp of 97% identity to the Tgm-Express1 contained an identical left border inverted repeat at the beginning of the 1697-bp stretch (arrowhead in Figure 9B). However, the 569-bp sequence upstream from the inverted repeat is completely different from that upstream of the Tgm-Express1 left border inverted repeat, revealing the existence in the G. max genome of a second Tgm-Express (Tgm-Express2) element and transposition event. This upstream region is a gene with similarity (93% identical) to an EST entered in GenBank with accession number CF922959, indicating that Tgm Express2 is inserted into a coding region. The existence of two Tgm Express elements with at least 1697 bp of almost identical sequence is an indication that these complex CACTA-Express elements have moved to multiple locations. However, Tgm-Express1 does not seem to be an autonomous element since there was no trace of transposase sequence in the entire 5.7-kb insertion; thus, its recent movement into wp must have been directed by an autonomous element elsewhere in the genome.
It is remarkable that given the scant amount of soybean genomic sequences available in GenBank that we were able to identify a second different Tgm-Express insertion. This could be pure coincidence or perhaps it predicts that in soybean this type of element may be abundant. By contrast, two computer-assisted projects in which mining of extensive Arabidopsis and rice genome sequences were performed did not find any CACTA-Express elements. In addition to generating element diversity, the ability of these transposons to capture cellular genes, replicating and transporting them to different regions of the genome in different arrangements, suggests that the CACTA-Express subfamily of elements also plays an important role in the evolution of the soybean genome and most likely in other plant species. The discovery of these two CACTA-Express elements in soybean and one in the Japanese morning glory (Takahashi et al., 1999) emphasizes that this ability of acquiring and moving host cellular genes by transposable elements is a more widespread phenomenon and that many more of these elements may exist in all plant genomes contributing to gene and genome expansion.
There are examples of Pack-MULEs where gene fragments from multiple chromosomal loci were fused to form new open reading frames, some of which could potentially be expressed as chimeric transcripts (Jiang et al., 2004). Similarly, it may be possible that a chimeric transcript could be generated from the string of gene fragments present in Tgm-Express1.
In soybean, the wp flower mutation has been correlated with increased seed protein content and seed size (Stephens and Nickell, 1992; Hegstad et al., 2000). The influence of a single locus on the anthocyanin pathway and also on seed protein accumulation and seed size has not been documented in other plant species. Now that we know the nature of the wp allele, we can further scrutinize how it may mediate those observed quantitative changes. One possibility is that the defect in expression of the F3H mRNA in the wp allele mediates changes in the flavonoid profiles of the seed coats that in turn modulate changes in the metabolism of early cotyledon development possibly through direct transfer of flavonoids from the seed coat to the cotyledons. Flavonoids and flavonols are known to have effects on a number of metabolic processes, although there is no indication in the literature of a direct involvement of flavonoids on protein synthesis and accumulation.
Another possibility is that the aberrant expression of the gene fragments carried by the Tgm-Express1 insertion may somehow mediate changes in protein content and seed size. These four gene fragments represent enzymes of an array of metabolic processes in plant cells, including cell division (CDC2 kinase), sugar metabolism (FPKFB2), Krebs cycle (MDH), and amino acid biosynthesis (CS). Partial polypeptide fragments produced from the genic fragments might signal upregulation or competitive inhibition of specific pathways. Alternatively, aberrant antisense or double-stranded transcripts may trigger the short-interfering RNA pathway leading to degradation of the corresponding homologous transcripts located elsewhere in the genome and resulting in the pleiotropic effect of the pink flower mutation on quantitative traits such as protein content and seed size.
METHODS
Plant Material and Genotypes
The Glycine max cultivars and isolines used for this study are described in Table 1 and were obtained from the USDA Soybean Germplasm Collections (Department of Crop Sciences, USDA Agricultural Research Service, University of Illinois, Urbana, IL). The genotypes and phenotypes of the lines used are shown in Table 1. All lines are homozygous, and only one of the alleles at each locus is shown for brevity in the tables and text.
The pink flower phenotype was first observed in 1989 (Stephens and Nickell, 1991) in F4-derived progeny rows from a cross that was expected to segregate only purple flowers (Stephens et al., 1993). Line LN89-5320-8-53 represents a single plant of the F6 generation that had flowers with pink and purple sectors or purple and pink flowers on the same plant. Further crosses and segregation studies showed that the original LN89-5320-8-53 plant represented a zygote with heterozygous genotype wpm/wp, where wpm was a novel unstable allele with somatic and germinal mutability (Johnson et al., 1998).
Plants were grown in the greenhouse. Seed coats dissected from seeds at varying stages of development, cotyledons of various stages of seed development, shoot tips, stems, mature leaves, flower buds, flowers, and roots were frozen in liquid nitrogen, freeze-dried (Multi-dry lyophilazer; FTS Systems), and stored at −20°C. For seed coats of developmental stages, seeds were divided into the following groups according to the fresh weight of the entire seed: 10 to 25 mg, 25 to 50 mg, 50 to 75 mg, 75 to 100 mg, and 100 to 200 mg.
RNA Extraction, Purification, and RNA Gel Blot Analysis
Total RNA was isolated from seed coats and other soybean tissues using a phenol-chloroform and lithium chloride precipitation method (McCarty, 1986; Wang et al., 1994). RNA was stored at −70°C until used.
RNA (10 μg/sample) was electrophoresed in a 1.2% agarose-3% formaldehyde gel (Sambrook et al., 1989). Size-fractionated RNAs were transferred to Optitran-supported nitrocellulose membrane (Midwest Scientific) by capillary action as described by Sambrook et al. (1989) and cross-linked in a UV Stratalinker (Stratagene). Nitrocellulose RNA gel blots were prehybridized, hybridized, washed, and exposed to Hyperfilm (Amersham) as described by Todd and Vodkin (1996).
Preparation of Microarray RNA Probes
Flower bud RNA samples used as probes to hybridize to microarrays were cleaned with RNeasy Minicolumns (Qiagen) according to the manufacturer's instructions. The eluates were concentrated to 8 μL by lyophilization in a SpeedVac (Savant Instrument). The amounts of RNA used in the two flower bud experiments differed. Samples for Experiment 1 (see Supplemental Figure 1 and Supplemental Table 1 online) contained 89 μg/sample, and those for Experiment 2 with younger flower buds had 45 μg/sample (see Supplemental Table 2 online). Seed coat RNAs (10 to 25 mg seed size) used as probes in the microarray Experiment 3 (see Supplemental Figures 3 and 4 and Supplemental Table 3 online) were not cleaned through the RNeasy column, and the concentration used was 50 μg/sample. Each RNA sample was concentrated to 8 μL by lyophilization in a SpeedVac (Savant Instrument) and reverse transcribed in the presence of Cy3 or Cy5-dUTP following the method described by Thibaud-Nissen et al. (2003).
Microarray Hybridization and Analysis
The microarrays used in this study contained 9216 spots with cDNAs from re-rack Gm-r1070 that are highly representative of RNAs from developing seeds and flowers (Vodkin et al., 2004). They also contained an additional 512 spots corresponding to 64 selected cDNAs or choice clones printed eight times each and distributed through the array (Vodkin et al., 2004). Among these 64 choice clones were 32 cDNAs corresponding to 13 different enzymes of the soybean flavonoid pathway. For some enzymes, more than one cDNA clone was chosen, and each was repeated eight times per array.
The microarray platform was entered in the Gene Expression Omnibus with accession number GPL229 (http://www.ncbi.nlm.nih.gov/geo). All cDNA clones are available from Biogenetics Services (http://www.biogeneticservices.com) or from the American Type Culture Collection (http://www.atcc.org).
The labeled cDNA probes were hybridized to the microarray cDNAs as described by Thibaud-Nissen et al. (2003). The slides were scanned in ScanArray Express 1.0 (Packard BioScience, BioChip Technologies). Fluorescence of the spots was quantitated with software provided with ScanArray Express 1.0. Local background subtraction, filtering out of spots, and correction between replicates were done as described previously (Thibaud-Nissen et al., 2003).
DNA Isolation and DNA Gel Blot Analysis
Genomic DNA was isolated from soybean freeze-dried shoot tips using the methods of Dellaporta (1993) with minor modifications (Zabala and Vodkin, 2003). Genomic DNA (12 μg) was digested with restriction endonucleases HindIII, BamHI, and EcoRI for ∼2 h at 37°C and electrophoresed in a 0.7% agarose gel (Sambrook et al., 1989).
Transfer of fractionated DNAs to supported nitrocellulose membrane was done as described for RNA gel blots.
cDNA Synthesis
cDNA copies of the F3H genes from the three isolines (LN89-5320-6, LN89-5322-2, and LN89-5320-8-53) were amplified from a first-strand cDNA pool synthesized using 1 μg of seed coat or flower bud total RNA and the Superscript first-strand synthesis system for RT-PCR (Invitrogen). The total RNAs used for these RT-PCR reactions were treated with DNAase I using Ambion's DNA-free kit and concentrated in Microcon YM-30 columns (Millipore). For each RNA sample, parallel reactions were allowed in the absence of superscript (− controls) to assess the extent of DNA contamination. The sequences of the four primers used were as follows: 5′-TACACGCACATTCTCCTCAAAG-3′ (38F), 5′-AATAAGACATAGGCAACTGAAC-3′ (1357R), 5′-GCATTGCATTCTGCTATTTAATTCC-3′ (7F), and 5′-AAAGACAGTGCCACTTATTTTCATT-3′ (1428R). The primer's numbering was based on the sequence of a cDNA with accession number AF198451. Once the DNA sequence of the Gm-c1012-683 EST clone was determined, it was used in a BLAST search. A DNA sequence identical to it with accession number AF198451 had been entered in GenBank by J.M.H. Chiu and C.S. Wang and erroneously annotated as a G. max flavonoid 3′ hydroxylase pseudogene (unpublished data). The sequence of this cDNA is 8 bp longer than that of the EST clone Gm-c1012-683. We have used this longer sequence during primer design, and the numbering of the primers was based on that sequence's length. Thus, primers 38F and 1357R start at base pairs 38 and 1357 of the AF198451 sequence, respectively.
The complete sequence of EST clone Gm-c1012-683 was determined and entered in GenBank with accession number AY669324. A partial 5′ end sequence for this clone had been entered in GenBank with accession number AI900038. This accession number was used in the microarray annotation (see Supplemental Tables 1 to 3 online). This is the reason why two different accession numbers are used in referring to the Gm-c1012-683 EST clone at different locations in the article.
Probes for DNA and RNA Gel Blots
Cloned DNAs used as probes were digested from their vectors or PCR amplified, electrophoresed, and purified from agarose using the QIAquick gel extraction kit (Qiagen). DNA concentration of the final eluate was determined with NanoDrop (NanoDrop Technologies). Purified DNA fragments (25 to 250 ng) were labeled with [α-32P]dATP by random primer reaction (Feinberg and Vogelstein, 1983).
Of the two EST clones representing F3H, Gm-c1012-683 was a full-length cDNA including the ATG and 57 upstream base pairs. The Gm-c1019-2646 clone starts a base pair after the ATG and therefore is 60 bp shorter than the other one, but both represent the same gene. The full-length Gm-c1012-683 EST clone was chosen to be used as probe in the experiments to determine the true identity of the clone and its correspondence to the Wp locus.
Primer Synthesis, PCR Reaction Conditions, and DNA Sequencing
Oligonucleotide primers were synthesized on an Applied Biosystems model 394A DNA synthesizer at the Keck Center, a unit of the University of Illinois Biotechnology Center. Multiple primer pairs were synthesized to complete the F3HcDNA sequence of two ESTs (GenBank accession numbers AI900038 and AW277481) as well as to amplify and sequence the F3H genomic DNAs from two of the isolines (LN89-5320-6 and LN89-5322-2).
Soybean genomic DNA fragments encoding the F3H1 or F3H2 gene were obtained via PCR from the LN89-5320-6, LN89-5322-2, and LN89-5320-8-53 lines. Most PCR reactions were performed by an initial denaturation step at 96°C for 2 min followed by 39 cycles of denaturing at 96°C for 20 s, annealing at 55°C for 1 min, and polymerization at 72°C for 2 min, to end with a 7-min extension at 72°C. In an experiment design to favor amplification of the larger F3H1 gene (3.5 kb) from the LN89-5320-6 line with primers 7F and 1428R, the annealing temperature of the PCR reaction was raised to 58°C. To amplify the wp mutant allele with a 5.7-kb insertion and a total length of 9.2 kb, the following PCR conditions were used: denaturation at 94°C for 1 min followed by 31 cycles of denaturing at 94°C for 30 s, annealing at 68°C for 10 min, to end with a 10-min extension at 72°C. High-fidelity and high-efficiency ExTaq (Takara Bio) polymerase was used for all above PCR reactions.
Genomic DNA fragments resulting from PCR amplification were fractionated in a 0.7% agarose gel, purified with a QIAquick gel extraction kit (Qiagen), and sequenced in ABI 3730 × l (Applied Biosystems) at the Keck Center. Two sequence alignment programs were used: MultAlin version 5.4.1 alignment program (Corpet, 1988; http://prodes.toulouse.inra.fr/multalin/multalin.html) and EMBOSS pairwise alignment algorithms from EMBL-EBI (http://www.ebi.ac.uk/emboss/align/). The BOXSHADE server of the BCM Search Launcher (http://www.ch.embnet.org/software/BOX_form.html) was used to highlight identical and conserved amino acids.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY669324 (F3H EST clone Gm-c1012-683), AY669325 (F3H1 genomic sequence), AY669326 (F3H2 genomic sequence), and AY994154 (wp mutant allele genomic sequence).
Supplementary Material
Acknowledgments
We thank Anupama Khanna, Robin Shealy, Steve Clough, and Renna Philip for their work during the development and printing of the soybean cDNA arrays used in the experiment leading to the identification of F3H. Special mention to Robin Shealy who created the normalization and other software programs to aid in the analysis of the ScanArray Express output data. Special thanks to Laura Guest and Virginia Lukas who directed the DNA sequencing and synthesis of the oligonucleotide primers. We gratefully acknowledge support from grants of the Illinois Missouri Biotechnology Alliance, the Illinois Soybean Program Operating Board, and National Science Foundation Grant DBI9872565.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lila O. Vodkin (l-vodkin@uiuc.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033506.
References
- Bernard, R.L., and Weiss, M.G. (1973). Quantitative genetics. In Soybeans: Improvement, Production and Uses, B.E. Caldwell, ed (Madison, WI: American Society of Agronomy), pp. 117–154.
- Britsch, L., Dedio, J., Saedler, H., and Forkmann, G. (1993). Molecular characterization of flavanone 3β-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues. Eur. J. Biochem. 217, 745–754. [DOI] [PubMed] [Google Scholar]
- Britsch, L., and Grisebach, H. (1986). Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur. J. Biochem. 156, 569–577. [DOI] [PubMed] [Google Scholar]
- Brown, J. (1986). A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res. 14, 9549–9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Jun, L., Blount, J.W., Steele, C.L., and Dixon, R.A. (2002). Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 14578–14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier, B., Coronado, C., Kondorosi, A., and Ratet, P. (1995). Molecular characterization and expression of alfalfa (Medicago sativa L.) flavanone-3-hydroxylase and dihydroflavonol-4-reductase encoding genes. Plant Mol. Biol. 29, 773–786. [DOI] [PubMed] [Google Scholar]
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan, P.B., Jarvik, T., Bush, A.L., Shoemaker, R.C., Lark, K., Kahler, G., Kaya, A.L., Van, N., Toai, T.T., Lohnes, D.G., Chung, J., and Specht, J.E. (1999). An integrated genetic linkage map of the soybean genome. Crop Sci. 39, 1464–1490. [Google Scholar]
- Dellaporta, S.L. (1993). Plant DNA miniprep version 2.1–2.3. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 522–525.
- Fasoula, D.A., Stephens, P.A., Nickell, C.D., and Vodkin, L.O. (1995). Cosegregation of purple-throat flower color with DNA polymorphism in soybean. Crop Sci. 35, 1028–1031. [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Gierl, A., Saedler, H., and Peterson, P.A. (1989). Maize transposable elements. Annu. Rev. Genet. 23, 71–85. [DOI] [PubMed] [Google Scholar]
- Groose, R.W., and Palmer, R.G. (1991). Gene action governing anthocyanin pigmentation in soybean. J. Hered. 82, 498–500. [Google Scholar]
- Gupta, S., Gallavotti, A., Stryker, G.A., Schmidt, R.J., and Lal, S.K. (2005). A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol. Biol. 57, 115–127. [DOI] [PubMed] [Google Scholar]
- Hegstad, J.M., Tarter, J.A., Vodkin, L.O., and Nickell, C.D. (2000). Positioning the wp flower color locus on the soybean genome map. Crop Sci. 40, 534–537. [Google Scholar]
- Honda, C., Kotoda, N., Wada, M., Kondo, S., Kobayashi, S., Soejima, J., Zhang, Z., Tsuda, T., and Moriguchi, T. (2002). Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biochem. 40, 955–962. [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S.R., and Wessler, S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431, 569–573. [DOI] [PubMed] [Google Scholar]
- Johnson, E.O.C., Stephens, P.A., Fasoula, D.A., Nickell, C.D., and Vodkin, L.O. (1998). Instability of a novel multicolored flower trait in inbred and outcrossed soybean lines. J. Hered. 89, 508–515. [Google Scholar]
- Lal, S.K., Giroux, M.J., Brendel, V., Vallejos, E., and Hannah, L.C. (2003). The maize genome contains a Helitron insertion. Plant Cell 15, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacin, R., Groning, I., Pieper, U., and Matern, U. (2000). Site-directed mutagenesis of the active site serine290 in flavanone 3b-hydroxylase from Petunia hybrida. Eur. J. Biochem. 267, 853–860. [DOI] [PubMed] [Google Scholar]
- McCarty, D. (1986). A simple method for extraction of RNA from maize tissue. Maize Genet. Coop. Newsl. 60, 61. [Google Scholar]
- Miao, G.H., Hong, Z., and Verma, D.P. (1993). Two functional soybean genes encoding p34cdc2 protein kinases are regulated by different plant developmental pathways. Proc. Natl. Acad. Sci. USA 90, 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, R.G., and Kilen, T.C. (1987). Quantitative genetics and cytogenetics. In Soybeans: Improvement, Production and Uses, 2nd ed., J.R. Wilcox, ed (Madison, WI: American Society of Agronomy), pp. 135–209.
- Pelletier, M.K., and Shirley, B.W. (1996). Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiol. 111, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, P.R., and Vodkin, L.O. (1985). Highly structured sequence homology between an insertion element and the gene in which it resides. Proc. Natl. Acad. Sci. USA 82, 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, P.R., and Vodkin, L.O. (1988). Organiztion of the Tgm family of transposable elements in soybean. Genetics 120, 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Senda, M., Masuta, C., Ohnishi, S., Goto, K., Kasai, A., Sano, T., Hong, J.-S., and MacFarlane, S. (2004). Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli, F., Martin, C., Scienza, A., Gavazzi, G., and Tonelli, C. (1994). Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol. Biol. 24, 743–755. [DOI] [PubMed] [Google Scholar]
- Stephens, P.A., and Nickell, C.D. (1991). A pink flower-color mutant in soybean. Soybean Genet. Newsl. 18, 226–228. [Google Scholar]
- Stephens, P.A., and Nickell, C.D. (1992). Inheritance of pink flower color in soybean. Crop Sci. 32, 1131–1132. [Google Scholar]
- Stephens, P.A., Nickell, C.D., and Vodkin, L.O. (1993). Pink flower color associated with increased protein and seed size in soybean. Crop Sci. 33, 1135–1137. [Google Scholar]
- Takahashi, S., Inagaki, Y., Satoh, H., Hoshino, A., and Iida, S. (1999). Capture of a genomic HMG domain sequence by the En/Spm-related transposable element Tnp1 in the Japanese morning glory. Mol. Gen. Genet. 261, 447–451. [DOI] [PubMed] [Google Scholar]
- Talbert, L.E., and Chandler, V.L. (1988). Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol. Biol. Evol. 5, 519–529. [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen, F., Shealy, R.T., Khanna, A., and Vodkin, L.O. (2003). Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol. 132, 118–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, K., Yang, D., Yamanaka, N., Watanabe, S., Harada, K., and Takahashi, R. (2002). A single-base deletion in soybean flavonoid 3′-hydroxylase gene is associated with gray pubescence color. Plant Mol. Biol. 50, 187–196. [DOI] [PubMed] [Google Scholar]
- Turcotte, K., Srinivasan, S., and Bureau, T. (2001). Survey of transposable elements from rice genomic sequences. Plant J. 25, 169–179. [DOI] [PubMed] [Google Scholar]
- Tuteja, J., Clough, S.J., Chan, W.-C., and Vodkin, L.O. (2004). Tissue specific gene silencing mediated by a naturally occurring chalcone synthase cluster in soybean. Plant Cell 16, 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, J.J., and Vodkin, L. (1996). Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin, L.O., et al. (2004). Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genomics 5, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin, L.O., Rhodes, P.R., and Goldberg, R.B. (1983). A lectin gene insertion has the structural features of a transposable element. Cell 34, 1023–1031. [DOI] [PubMed] [Google Scholar]
- Wang, C., Todd, J., and Vodkin, L.O. (1994). Chalcone synthase mRNA and activity are reduced in yellow soybean seed coats with dominant I alleles. Plant Physiol. 105, 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S.R. (1988). Phenotypic diversity mediated by the maize transposable elements Ac and Spm. Science 242, 399–405. [DOI] [PubMed] [Google Scholar]
- Yu, Z., Wright, S.I., and Bureau, T.E. (2000). Mutator-like elements in Arabidopsis thaliana: Structure, diversity, and evolution. Genetics 156, 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala, G., and Vodkin, L.O. (2003). Cloning of the pleitropic T locus in soybean and two recessive alleles that differentially affect structure and expression of the encoded flavonoid 3′ hydroxylase. For. Genet. 163, 295–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.