Abstract
Photosystem II (PSII), the enzyme responsible for photosynthetic oxygen evolution, is a rapidly turned over membrane protein complex. However, the factors that regulate biogenesis of PSII are poorly defined. Previous proteomic analysis of the PSII preparations from the cyanobacterium Synechocystis sp PCC 6803 detected a novel protein, Psb29 (Sll1414), homologs of which are found in all cyanobacteria and vascular plants with sequenced genomes. Deletion of psb29 in Synechocystis 6803 results in slower growth rates under high light intensities, increased light sensitivity, and lower PSII efficiency, without affecting the PSII core electron transfer activities. A T-DNA insertion line in the PSB29 gene in Arabidopsis thaliana displays a phenotype similar to that of the Synechocystis mutant. This plant mutant grows slowly and exhibits variegated leaves, and its PSII activity is light sensitive. Low temperature fluorescence emission spectroscopy of both cyanobacterial and plant mutants shows an increase in the proportion of uncoupled proximal antennae in PSII as a function of increasing growth light intensities. The similar phenotypes observed in both plant and cyanobacterial mutants demonstrate that the function of Psb29 has been conserved throughout the evolution of oxygenic photosynthetic organisms and suggest a role for the Psb29 protein in the biogenesis of PSII.
INTRODUCTION
All oxygenic photosynthetic organisms, from cyanobacteria to vascular plants, rely on the activity of the photosystem II (PSII) pigment protein complex to transfer electrons from water to plastoquinones, a process that is driven by solar energy (Rutherford, 1989). Prior to the evolution of PSII, bacterial photosynthesis utilized reduced sulfide compounds as electron donors in photosynthetic processes. The evolution of oxygen evolving cyanobacteria ∼3 billion years ago transformed the atmosphere and chemical composition of the planet by causing O2 levels to increase (Blankenship and Hartman, 1998). Later, endosymbiosis of a cyanobacterium into a eukaryotic cell led to the evolution of eukaryotic algae and vascular plants (Yoon et al., 2002).
A number of recent structural studies have provided a detailed picture of the ∼640-kD PSII dimer complex (Rutherford and Boussac, 2004). Electron transport in PSII is initiated by an assembly of chlorophylls located at the center of the PSII monomer, termed P680. Electrons are transferred from P680 through a succession of cofactors to the QB plastoquinone binding site. P680+ is rereduced through the action of the water-splitting apparatus on the luminal side of PSII, which contains four manganese ions and one calcium ion as catalytic cofactors (Rutherford and Boussac, 2004). These cofactors, together with the proteins that ligate them and participate in electron transfer reactions, define the core of the PSII reaction center. In addition, PSII contains a proximal chlorophyll-containing antenna system composed of the CP43 and CP47 proteins. The proximal antenna system participates in light harvesting and in excitation energy transfer from distal antenna components to the reaction center core (Ferreira et al., 2004). The distal antenna complexes, phycobilisomes in cyanobacteria, and light-harvesting complex II (LHCII) in plants extend the spectral range and efficiency of PSII (Grossman et al., 1995).
Structural studies have provided a detailed but static description of PSII. In parallel, the study of the dynamic features of the photosynthetic apparatus has revealed a number of mechanisms that participate in the modulation of PSII activity. Among these are processes involved in the fine-tuning of light harvesting through LHCII phosphorylation and carotenoid deepoxidation (Keren and Ohad, 1998), long-term adaptations to changes in light intensity and quality by synthesis and degradation of specific antenna components (Grossman et al., 1995), and mechanisms that protect against light damage by rapid turnover of the PSII reaction center core protein D1 (Keren and Ohad, 1998). The extent to which PSII regulatory processes take place suggests the involvement of a large number of modifying enzymes and regulatory pathways. The identities of only a few of these proteins have been revealed in a number of recent studies. Among these are the Stt7 kinase that regulates the antenna system (Depege et al., 2003), the PsbS protein that is involved in the quenching of excitation energy (Li et al., 2002), Alb3.1p that is involved in the assembly of PSII (Ossenbuhl et al., 2004), and the DegP2 and FtsH proteases that are responsible for the degradation of the D1 protein as a part of a repair cycle (Adam and Ostersetzer, 2001).
In order to extend our knowledge of the protein subunits involved in the biogenesis and regulation of PSII, we have embarked on a proteomic study of an isolated His-tagged PSII preparation from the cyanobacterium Synechocystis sp PCC 6803 (Bricker et al., 1998). The isolated PSII preparation contained all of the confirmed PSII subunits as well as a number of novel proteins (Kashino et al., 2002). Among these were two proteins homologous to the PSII oxygen-evolving enhancer proteins, PsbP and PsbQ, which were previously considered to be present only in chloroplasts. Analysis of disruption mutants in both proteins has verified their functions in enhancing oxygen evolution in cyanobacteria, much like their vascular plant counterparts (Thornton et al., 2004).
Among the proteins found in isolated PSII complexes are a number of proteins that did not show significant homology to any other protein with known function (Kashino et al., 2002). Following the nomenclature for PSII proteins, these proteins were referred to as Psb27 through Psb30 (Kashino et al., 2002). These novel proteins were found in all of the PSII preparations analyzed in that study, but their concentrations were substoichiometric. Therefore, while the function of these proteins might be related to PSII activity, it remains possible that they were contaminants present in the isolated PSII preparation. In this work, we have examined one of these proteins, Psb29, using disruption mutants in psb29 genes in Synechocystis 6803 and in the vascular plant Arabidopsis thaliana. The results presented in this article demonstrate that disruption of these genes in both model organisms results in impairment of PSII function under high light intensities, confirming a conserved role for Psb29 proteins in PSII biogenesis.
RESULTS
Bioinformatic Analysis of Psb29 Proteins Reveals Well-Conserved Homologs in Many Photosynthetic Organisms
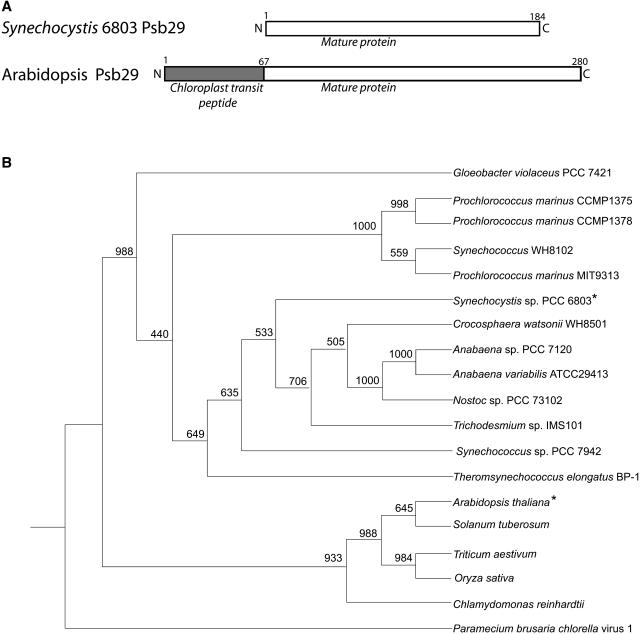
The sll1414 gene in Synechocystis 6803 encodes the Psb29 protein that shows a high degree of similarity to the Arabidopsis At2g20890 protein (see supplemental data for sequence alignments). The Psb29 protein in Arabidopsis contains a predicted chloroplast transit peptide at its N terminus (Figure 1A). Thylakoid lumen targeting sequences were not detected in the Psb29 protein from either organism. Both proteins are predicted to be soluble with no extensive hydrophobic stretches. The cyanobacterial Psb29, identified as a component of the purified PSII preparation, could be removed from PSII by treatment with high salt concentrations (Kashino et al., 2002). These data suggest that Psb29 is a peripheral subunit bound to the stromal side of PSII. Consistent with our analysis, two recent publications identified the Arabidopsis At2g20890 protein as a thylakoid-associated protein in plastids (Peltier et al., 2004; Wang et al., 2004).
Figure 1.
Bioinformatic Analysis of Psb29 Family Proteins.
(A) A schematic diagram presenting domains in the structure of Psb29 proteins in Synechocystis and Arabidopsis. Chloroplast target sequence prediction was performed using the TargetP program.
(B) Extended majority rule consensus tree. Bootstrap values are indicated at branching points. The two proteins studied in this article are marked with asterisks. The sequence alignment data on which the phylogenetic tree is based can be found in the supplemental data online.
Searching genome databases for homologs of the Synechocystis 6803 Psb29 protein yielded no significant matches to any protein with a well-characterized function (E value cutoff of 10−6). Nevertheless, Psb29 homologs are found in all cyanobacteria with a fully sequenced genome. These include the Gloeobacter violaceus PCC 7421 species, which does not have a differentiated thylakoid membrane system and is considered to be a member of an early branching lineage (Nakamura et al., 2003). Psb29 proteins have been conserved throughout evolution and are present in all vascular plant species for which genome data are available, as well as in the green alga Chlamydomonas reinhardtii. Based on the alignment of Psb29 sequences (see supplemental data online), a phylogenetic tree was constructed (Figure 1B). A sequence found in the Paramecium bursaria chlorella virus-1 was used as an outgroup. While the sequence of this viral protein shows similarities to other Psb29 proteins, it lacks the putative targeting sequence that would allow it to function in the Chlorella chloroplast. As expected, cyanobacterial sequences and plant sequences branched into separate clades.
Disruption Mutants in the psb29 Genes in Synechocystis 6803 and Arabidopsis
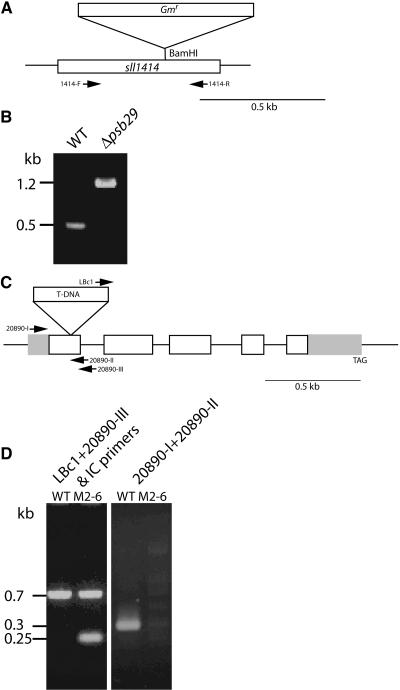
To study the function of the Psb29 proteins, we examined targeted disruption mutants in Synechocystis 6803 and Arabidopsis (Figure 2). In Synechocystis 6803, an antibiotic resistance cassette was introduced into the BamHI site in the sll1414 (psb29) gene via double homologous recombination (Figure 2A). PCR was used to validate the insertion site and the segregation of the mutant (Figure 2B). The wild-type band was not detected in the Δpsb29 strain. The 1.2-kb band observed in the mutant sample corresponds to the added size of the insert.
Figure 2.
Construction of Mutant Strains.
(A) Construction of the cyanobacterial Δpsb29 mutant. The diagram shows the site of the insertion and the positions of the PCR primers.
(B) PCR analysis of the segregation of the Δpsb29 mutant, using the same set of primers.
(C) Illustration of the genomic region of the At2g20890 gene. Gray boxes represent the 5′ and 3′ untranslated regions, and white boxes represent the coding sequence. The SALK_094925 line contains a T-DNA insert (not drawn to scale) in the first exon of the gene.
(D) PCR analysis of the segregation of the Arabidopsis M2-6 mutant. Primer sets used for the analysis are in slanted script above the gel.
In Arabidopsis, a T-DNA insertion line was identified in the At2g20890 gene (Salk_094925, Figure 2C). We tested for a homozygous line using the LBc1 and 20890-III primers. As an internal PCR control, the single copy At1g63020 gene encoding the RNA polymerase IIA large subunit was amplified, giving rise to the 0.7-kb band observed in both lanes (Figure 2D). A 0.25-kb band originating from the LBc1 and 20890-III primers was observed only in the M2-6 mutant lane. PCR amplification with the 20890-I and -II primers gave rise to a 0.3-kb fragment that was observed only in the wild-type sample. Insertion of the T-DNA fragment in the M2-6 mutant increased the distance between primers I and II by ∼5 kb and prevented amplification by PCR under our experimental conditions.
Δpsb29 Cells Display Slower Growth Rates and Lower PSII Activity under High Light Intensities
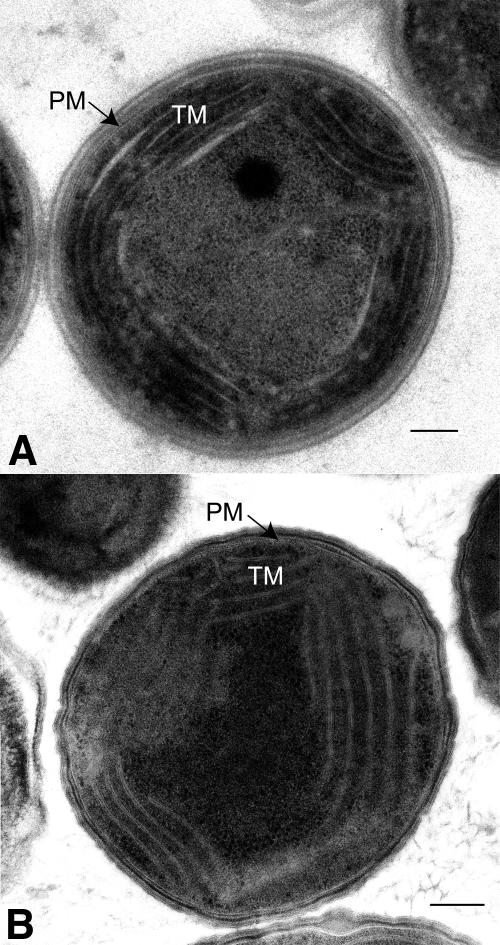
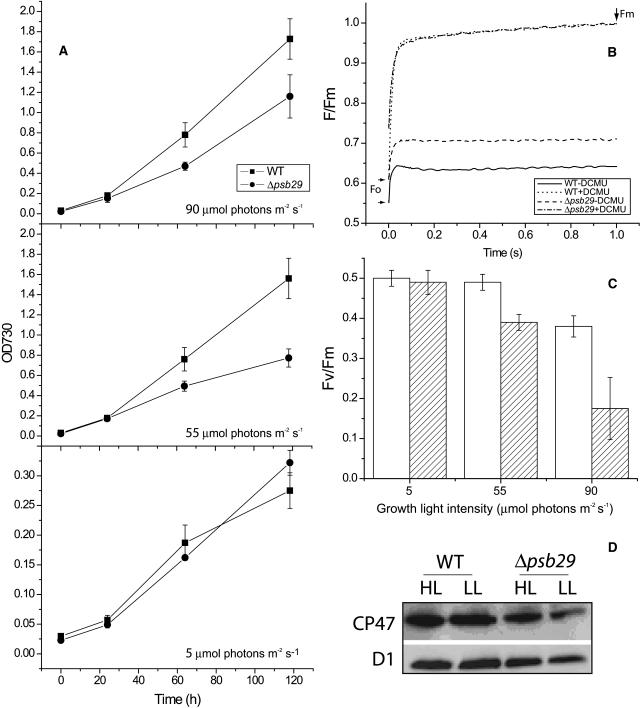
The architecture of Synechocystis wild-type and Δpsb29 cells was studied using transmission electron microscopy (Figure 3). Thylakoid membrane ultrastructure in wild-type and mutant cells appears similar. The growth rate of Δpsb29 cells, however, did differ from that of wild-type cells (Figure 4A). Under normal growth light intensities (55 μmol photons m−2 s−1), mutant cultures grew slower than wild-type cultures. Similar results were observed for cultures grown under high light intensities (90 μmol photons m−2 s−1). Only when grown under 5 μmol photons m−2 s−1 did wild-type and mutant cultures exhibit similar, albeit slow, growth rates. As a test for PSII activity under these growth conditions, we measured the fluorescence induction rates of cultures after 5 d of growth (Figure 4B). Analysis of the induction traces took into account two parameters: Fm, the maximal value of PSII fluorescence when electron transfer is blocked by the inhibitor DCMU, and F0, the contribution of photochemically inactive chlorophylls. The ratio of (Fm − F0)/Fm = Fv/Fm is a measure of PSII activity (Maxwell and Johnson, 2000). As evident from Figure 4B, Fv/Fm values of Δpsb29 cells grown under normal growth light conditions were lower due to higher F0 values. The time course of fluorescence rise in the presence of DCMU was similar in both cases. The differences between Fv/Fm values of wild-type and Δpsb29 cells was even more pronounced in cultures grown under high light intensities but could be rescued by growth under low light intensities (Figure 4C).While the photosynthetic competence of mutant cells is reduced in high light intensities, the relative content of the reaction center D1 and the proximal antenna CP47 proteins in mutant cultures are not significantly different than wild-type cultures (Figure 4D).
Figure 3.
Membrane Architecture of Synechocystis 6803 Wild-Type and Δpsb29 Mutant Cells.
Transmission electron micrographs of wild-type (A) and mutant (B) cells grown on solid media under 55 μmol photons m−2 s−1. PM, plasma membrane; TM, thylakoid membrane. Bars = 200 nm.
Figure 4.
Growth Rates and Photosynthetic Competence of Wild-Type and Δpsb29 Mutant Cultures of Synechocystis 6803.
(A) Growth of wild-type and mutant cultures were monitored by measuring absorption at 730 nm under three light intensities, indicated in bottom right corner of each set of traces. Error bars represent sd based on mean values of three replicate cultures.
(B) Fluorescence induction profile of wild-type and mutant cultures grown under normal growth light conditions. The time points for F0 and Fm are marked by arrows.
(C) Fv/Fm values as a function of growth light intensity. Open bars, wild type; hatched bars, mutant. Values were measured after 5 d of growth (n = 3).
(D) Immunoblot analysis of the relative content of D1 and CP47 proteins in total membrane extracts from wild-type and Δpsb29 cultures grown under low light (LL) or high light (HL) conditions for 3 d. Samples were loaded on an equal chlorophyll basis.
Electron Transfer Activities in PSII Reaction Center Cores Are Not Impaired in Δpsb29 Cells
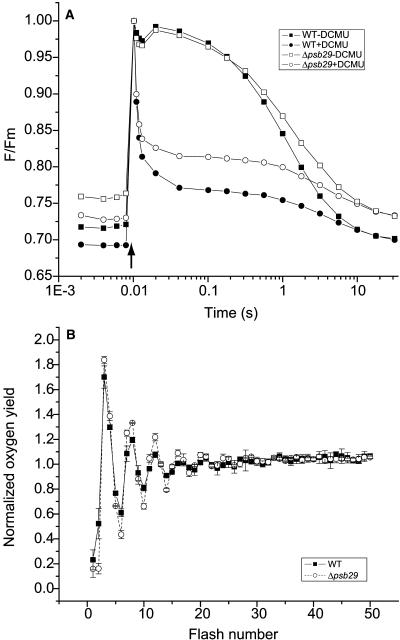
Reduced PSII activity can result from impaired electron transfer reactions in the PSII reaction center core or from the contribution of uncoupled antenna chlorophylls. In order to explore the first possibility, we tested the fluorescence decay kinetics of wild-type and Δpsb29 cells (Figure 5A). While F0 values measured in the mutant cultures were higher than in wild-type cultures (in accordance with the continuous light kinetic measurements in Figure 4B), the decay rates were similar in both strains. In the absence of DCMU, fluorescence decays through a mixture of forward electron transfer to plastoquinones and backward electron transfer through charge recombination. In the presence of DCMU, forward electron transfer reactions are blocked, and fluorescence decays solely through back-reactions (van Best and Duysens, 1975). In both cases, the rates of fluorescence decay are similar for wild-type and mutant cells. Correlation coefficients between pairs of wild-type and mutant decay curves were >0.93 for −DCMU curves and >0.98 for +DCMU curves (n = 2).
Figure 5.
PSII Activity of Synechocystis 6803 Wild-Type and Δpsb29 Cells Grown under Normal Growth Light.
(A) Fluorescence decay rates in the presence or absence of 2 μM DCMU following a single high intensity light flash (indicated by an arrow). Half times for fluorescence decay were 0.012 ± 0.0002 s and 0.012 ± 0.001 s in the absence of DCMU and 1.11 ± 0.13 s and 1.13 ± 0.08 s in the presence of DCMU for wild-type and Δpsb29 cultures, respectively (n = 2).
(B) Oxygen flash yield pattern measured on a Joliot-type electrode. The oxygen flash yield signals were normalized to the average of the signals. Error bars represent sd based on three replicate measurements.
In addition, we measured the oxygen evolution capacity of wild-type and mutant cultures using both continuous light and periodic flashes for excitation. Vmax values for steady state oxygen evolution from wild-type and Δpsb29 cultures were 364 ± 45 and 365 ± 24 μmol O2 mg chlorophyll−1 h−1, respectively (n = 3). Km values were slightly higher in Δpsb29 than in wild-type cultures (wild type, 237 ± 47; Δpsb29, 305 ± 28 μmol photons m−2 s−1). In flash-induced oxygen evolution measurements (Joliot, 1972), both wild-type and mutant cultures display virtually identical period 4 oscillation patterns (Figure 5B). Taken together, these three assays indicate that PSII electron transfer activities are not perturbed in the Δpsb29 cells.
Δpsb29 Cells Exhibit a Light-Dependent Increase in Uncoupled Proximal Antenna Fluorescence
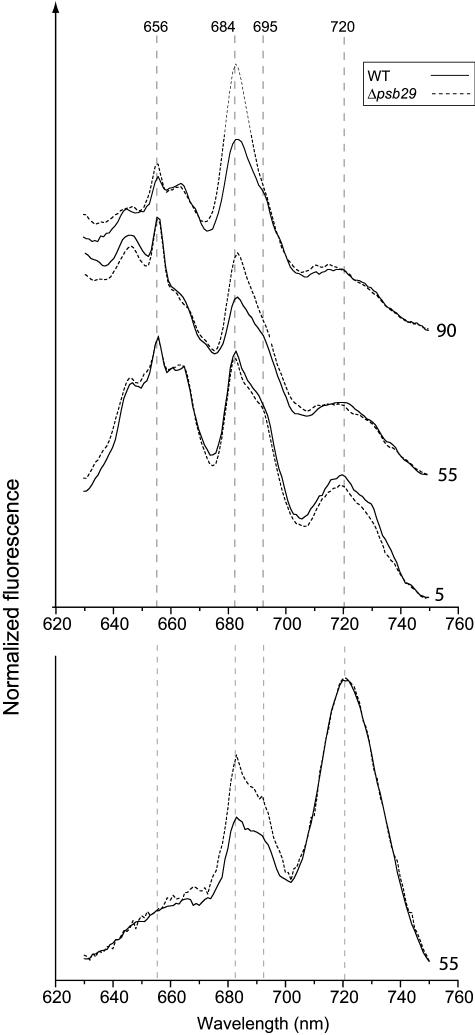
The excitonic connection between antenna pigments and reaction center cores was examined in a series of 77K fluorescence emission spectral measurements (Figure 6). These measurements probe coupling of antenna complexes to photosystem cores. The peak of phycobilisome light-harvesting antenna absorption is at 620 nm. We selected to set the excitation wavelength to 600 ± 1 nm to minimize the contribution from chlorophylls, the absorption peak of which is in the 670 to 680 nm range. The fluorescence emission spectra presented in Figure 6 contains a number of features. The three peaks centered around 656 nm originate from phycobilisomes. The peak at 684 nm contains contributions from the phycobilisome linker pigment and from the PSII proximal antenna system. The two antenna systems cannot be spectrally resolved in the 77K fluorescence spectra. The peaks at 695 and 720 nm originate from the PSII-P680 and PSI-P700 reaction center core chlorophylls, respectively (Hall and Rao, 1995). When grown under low light intensities (5 μmol photons m−2 s−1), the fluorescence spectrum of Δpsb29 cells was virtually identical to that of the wild type. Growth under normal light conditions (55 μmol photons m−2 s−1) induced a number of changes in the contribution of the different fluorescing species to the spectrum. However, only for PSII antenna fluorescence at 684 nm could a differential increase in the mutant as compared with the wild type be observed (Figure 6, top panel). The disproportional increase in 684-nm fluorescence in the mutant was more pronounced under high growth light intensities (90 μmol photons m−2 s−1). A similar differential increase in the fluorescence intensity at 684 nm was observed with excitation of chlorophylls at 420 nm instead of phycobilisomes at 600 nm (Figure 6, bottom panel). The gradual increase in 684-nm fluorescence as a function of light intensity was inversely proportional to the decrease in Fv/Fm values.
Figure 6.
77K Fluorescence Spectra of Synechocystis 6803 Wild-Type and Δpsb29 Cultures as Functions of Growth Light Intensity.
Excitation wavelength was set at 600 ± 1 nm (top) or 420 ± 10 (bottom). Vertical broken lines mark the emission maxima from phycobilisomes at 656 nm, the phycobilisome linker and PSII proximal antenna at 684 nm, PSII reaction center at 695 nm, and PSI reaction center at 720 nm. The curves are baseline shifted for clarity. The numbers on the right side of the curves indicate growth light intensity in μmol photons m−2 s−1. Curves were normalized to the fluorescence intensity at 750 nm.
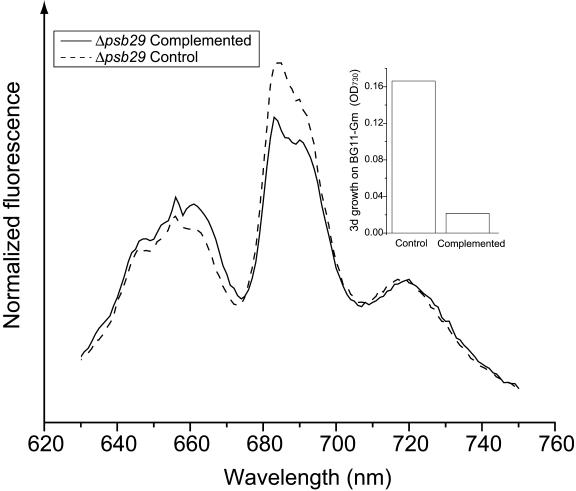
To ensure that this phenotype is a direct result of the insertion into the psb29 open reading frame, we complemented Δpsb29 cells with wild-type psb29 DNA and exposed them to high light intensities (Figure 7). Complemented cultures exhibited a lower 684-nm peak than control noncomplemented cultures. The loss of the 77K fluorescence phenotype coincided with the loss of the ability to grow on gentamycin (Gm)-containing media (Figure 7, inset). These results demonstrate a selective pressure against the gentamycin resistance (Gmr) insertion mutation in the psb29 gene under high light conditions.
Figure 7.
Complementation of Δpsb29 Eliminates the 77K Fluorescence Phenotype.
Complemented and noncomplemented control Δpsb29 cells were grown under high light intensities. After 8 d of high light growth, the 77K fluorescence spectra of complemented and control cells were measured. Aliquots from the cultures were subcultured into media containing Gm. Growth on BG11-Gm was measured as OD730 after an additional 3 d of growth (inset). The experiment was repeated twice with similar results.
PSII Activity Is Light Sensitive in Both Cyanobacteria and Plant Psb29 Mutants
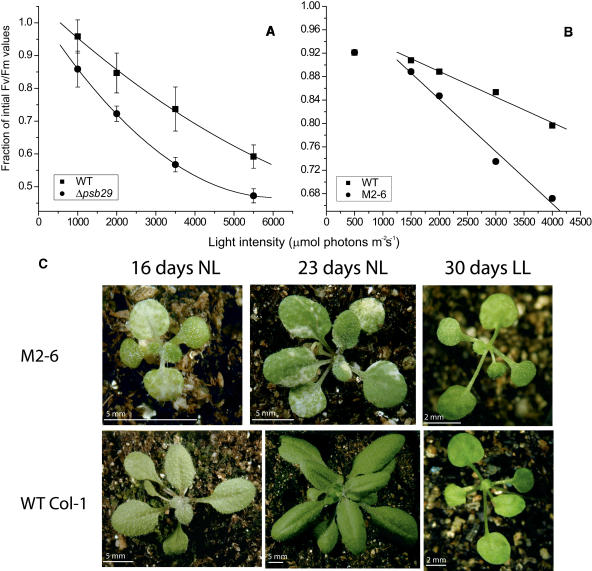
The reduced PSII activity observed in mutant cells (Figure 4) and the selective pressure against the Gmr insertion into the psb29 gene (Figure 7) at high light intensities indicate an increased sensitivity of PSII to light stress in the mutant. Reduced PSII activity under high light conditions could be the cause of the overall slower growth rate. To verify this, we challenged cultures grown under normal growth light intensities with high light intensities (Figure 8A). Over a range of high light intensities, mutant cultures lost a larger fraction of the initial Fv/Fm value than wild-type cultures.
Figure 8.
Light Sensitivity of Synechocystis and Arabidopsis Wild-Type and Mutant Strains.
(A) Light sensitivity of Synechocystis 6803 wild-type and Δpsb29 cultures measured as the ratio of Fv/Fm values after treatment with high light intensities for 20 min followed by 7 min dark adaptation to Fv/Fm values before the treatment. Fm values were determined by a single high intensity light flash. Starting Fv/Fm values were in the range of 0.35 ± 0.003 and 0.29 ± 0.03 for wild-type and Δpsb29 cells, respectively. Error bars represent sd based on mean values of two replicate cultures.
(B) Light sensitivity of Arabidopsis wild-type and M2-6 plants calculated as the residual Fv/Fm value after exposure to high light intensities for 15 min, followed by 7 min dark adaptation. Starting Fv/Fm values were in the range of 0.8 ± 0.01 and 0.79 ± 0.02 for wild-type and M2-6 plants, respectively. The experiment was performed under five different light intensities using different wild-type and M2-6 plants for each light treatment. The data between 1500 to 4000 μmol photons m−2 s−1 were fitted by linear regression, resulting in slope coefficients of −4.3E-5 ± 3.7E-6 for the wild type and −8.9E-5 ± 7.4E-6 for M2-6 plants. These values are significantly different at P < 0.01 based on a two-sample, two-tailed Student's t test.
(C) Growth of wild-type Columbia-I and M2-6 mutant plants under normal (NL; 50 μmol photons m−2 s−1) or low (LL; 10 μmol photons m−2 s−1) growth light conditions.
A parallel study was conducted on Arabidopsis plants. As in the cyanobacterial Δpsb29 mutant, PSII activity in the Arabidopsis M2-6 mutant plants exhibited higher light sensitivity. M2-6 plants lost a larger fraction of Fv/Fm values with increasing light intensities as compared with Columbia-1 wild-type plants (Figure 8B). Furthermore, M2-6 plants grew much slower than wild-type plants (Figure 8C). The mutant plants were variegated, containing large areas lacking chlorophyll. Recently, Wang and coworkers have analyzed the same insertion line and have reported a similar phenotype for mutant plants under normal growth light conditions (Wang et al., 2004). Interestingly, we were able to rescue the variegated phenotype under low growth light intensities. Under such conditions both wild-type and mutant plants grew at a comparable, albeit slow, rate and exhibit green nonvariegated leaves (Figure 8C).
PSII Antenna Quenching and Electron Transfer Reactions Downstream of PSII Are Not Effected in M2-6 Plants
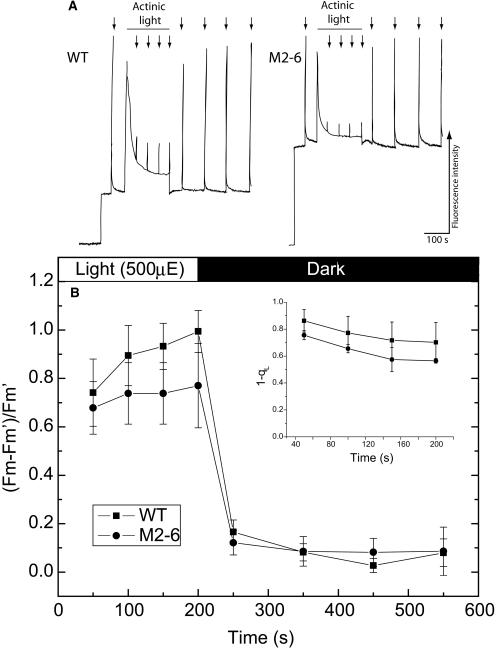
While M2-6 plants exhibit a dramatic growth phenotype, Fv/Fm values originating from the green part of the leaves grown under normal growth light intensities were not significantly different from the values measured in wild-type leaves (wild type 0.8 ± 0.01; M2-6, 0.79 ± 0.01; n = 3). Increasing the growth light intensity from 50 to 220 μmol photons m−2 s−1 resulted in an increase in the F0 values and, hence, a decrease in the Fv/Fm values (Figure 9A).
Figure 9.
Fluorescence Quenching in Wild-Type and M2-6 Plants Grown under High Light Conditions.
(A) Pulse-amplitude modulated fluorescence traces from wild-type and M2-6 mutants transiently exposed to 500 μmol photons m−2 s−1 actinic light. F0 values were measured before the actinic light treatment. The gray bar indicates the period of exposure to actinic light, and the arrows indicate supersaturating light pulses for Fm and Fm' measurements. Wild-type Fv/Fm values were 0.74 ± 0.02, and M2-6 values were 0.56 ± 0.03 (n = 3).
(B) Analysis of quenching parameters. The NPQ parameter = (Fm − Fm')/Fm'. The photochemical quenching parameter qL (inset) was calculated as [(1/Fs) − (1/Fm')/(1/Fo' − 1/Fm')]. Error bars represent sd based means of measurements on three plants.
The photosynthetic apparatus can adapt to varying light intensities by modulating the fraction of excitation energy that is utilized for photosynthesis (photochemical quenching) or lost as heat (nonphotochemical quenching). A number of PSII mutants with reduced variable fluorescence values have been found to be defective in nonphotochemical quenching (NPQ) (Shikanai et al., 1999). These processes reduce the effective antenna size in response to high light intensities. We measured the time course of the Fm' fluorescence quenching response of wild-type and M2-6 plants following exposure to high light (Figure 9A). Calculating the extent of NPQ from a series of such experiments did not reveal any significant difference between wild-type and M2-6 plants (Figure 9B). Furthermore, once the plants were returned to darkness, no difference in the rate of NPQ relaxation could be observed. Calculations of the photochemical quenching parameter, qL (Kramer et al., 2004), indicated that the rates of photosynthetic reactions downstream of PSII were not significantly different in the mutant as compared with the wild-type plants (Figure 9B, inset). These results indicate that both photochemical and NPQ processes take place to the same extent in both wild-type and mutant plants.
The Increase in F0 Fluorescence Values Is Correlated to Antenna Fluorescence in M2-6 Plants
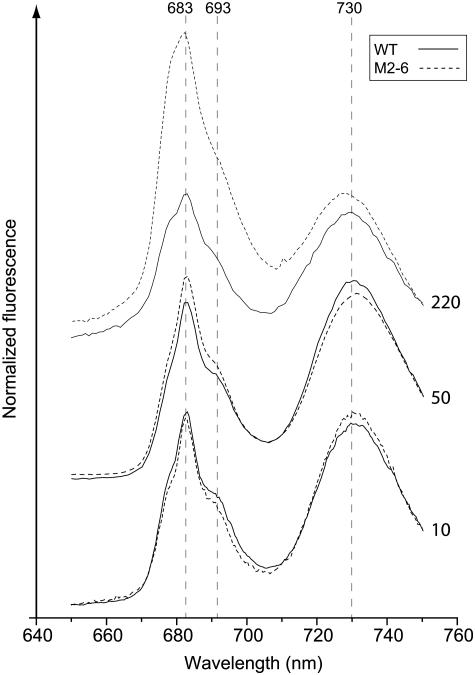
Vascular plants, unlike cyanobacteria, utilize the LHCII chlorophyll a/b pigment protein antenna instead of phycobilisomes. Therefore, in order to measure the chlorophyll fluorescence spectra at 77K, we excited isolated membranes at the Soret absorption band of chlorophyll at 420 ± 10 nm (Figure 10). The 683-nm peak is a result of contributions from distal (light harvesting II) and proximal antenna components. As in cyanobacteria, the contribution of the two antenna systems cannot be resolved at 77K (Hall and Rao, 1995). Fluorescence peaks from PSII-P680 and PSI-P700 are observed at 693 and 730 nm, respectively. In plants grown under low light intensities, the fluorescence traces were virtually identical. The 683-nm fluorescence peak of M2-6 plants grown under growth light intensities was only slightly higher than that observed in wild-type plants. The difference is much more pronounced in plants grown under high light intensities. As in the case of the cyanobacterial mutant, these differences correlated well with PSII activity as reported by Fv/Fm values. No differential effect in photosystem I fluorescence at 730 nm was observed.
Figure 10.
77K Fluorescence Spectra of Wild-Type and M2-6 Plants Grown under High and Normal Growth Light Intensities.
Excitation wavelength was set at 420 ± 10 nm and energized both chlorophyll a and b. Vertical broken lines mark the emission maxima from the PSII antenna terminal chlorophyll at 683 nm, PSII reaction center at 693 nm, and PSI at 730 nm. The curves are baseline shifted for clarity. The numbers on the right-hand side of the curves indicate growth light intensity in μmol photons m−2 s−1. Curves are normalized to the fluorescence intensity at 750 nm.
DISCUSSION
The data presented in this study indicate that Psb29 proteins, originally described as minor Synechocystis 6803 PSII copurifying proteins, are indeed relevant to the function of PSII. The absence of these proteins results in slow, light sensitive growth (Figures 4A and 8C). Furthermore, the light sensitivity is correlated specifically to PSII activity as measured by both room temperature and 77K fluorescence (Figures 6, 8, and 10). It is important to note that the phenotypes observed both in Synechocystis 6803 and in Arabidopsis mutants are very similar, indicating that the function of Psb29 proteins is conserved in cyanobacteria and plants.
The source of defects in PSII activity can be in the reaction center core that catalyzes electron transfer reactions or in the light-harvesting antenna systems. Measurements of oxygen evolution and room temperature fluorescence did not indicate a problem in the reaction center core electron transfer capabilities (Figures 5 and 9). In addition, PSII activity (Fv/Fm values) in mutant plants and cyanobacteria grown under low light intensities demonstrate that PSII can function normally in these mutants. The best corollary to Fv/Fm values measured under different growth light conditions can be found in 77K fluorescence spectra (Figures 6 and 10). The intensity of the peak at 683 to 684 nm is inversely correlated to the Fv/Fm values in both cyanobacteria and plants. This peak contains contributions from both proximal and distal antenna systems. Considering the similar phenotypes observed in both mutants, it would be difficult to explain how homologous proteins affect two very different antenna systems, phycobilisomes in cyanobacteria and LHCII in plants. Therefore, it is more probable that the increase in the 683 to 684 nm fluorescence is related to the conserved CP43 and CP47 proximal chlorophyll antenna system. This conclusion is supported by the observation that increased 683-nm fluorescence in the cyanobacterial mutant could be detected with excitation of phycobilisomes at 600 nm or with excitation of chlorophyll at 420 nm (Figure 6). Increase in uncoupled proximal antenna proteins with increasing light intensities can explain the increase in 683-nm fluorescence, the diminishing Fv/Fm values, and the lower Km values for oxygen evolution.
Wang and coworkers have recently reported on a study of the same Arabidopsis T-DNA insertion line studied by us (Wang et al., 2004). They have observed the variegated phenotype and demonstrated the chloroplast localization of the Psb29 protein. Based on their results, they have suggested that the At2g20890 gene product be called Thf1, for its putative function in thylakoid formation. However, thylakoid membranes are present and functional in the green parts of the Arabidopsis mutant as well as in the cyanobacterial mutant (Figure 3). Furthermore, thylakoid structure appears to be normal both in the green parts of Arabidopsis (Wang et al., 2004) and in Synechocystis 6803 Psb29 mutants (Figure 3). Thylakoid membrane formation occurs through very different mechanisms in cyanobacteria and vascular plants (Westphal et al., 2003), making it hard to explain the similar phenotypes observed in both mutants. Moreover, a lesion in the membrane formation process would result in a decrease in the activity of all thylakoid membrane protein complexes. The data presented here demonstrate that the problem is restricted to PSII function. No effect on downstream components of the photosynthetic apparatus could be observed.
Therefore, we propose that the occurrence of variegated leaves is not due to problems in thylakoid membrane formation but rather to light-induced damage resulting in the disintegration of thylakoid membranes. Excitation of uncoupled antenna chlorophylls can generate active oxygen species that cause membrane peroxidation (Hundal et al., 1995). In rapidly dividing young leaf cells, the concentration of chloroplasts is relatively low (Mullet, 1988). Therefore, the effective light intensity that chloroplasts in these cells are exposed to is higher than that which the chloroplasts in mature cells are exposed to, due to self shading. Under high light conditions, light-induced oxidative stress can lead to the loss of chloroplasts in some cells, generating the variegated phenotype observed in the mutant. Lowering the growth light intensity reduces the rate of photodamage and rescues the variegated phenotype in the mutant (Figure 8B).
A number of leaf variegated mutant classes are described in the literature. Among them are the immutants (im) and yellow variegated (var) mutants, in which genes encoding for chloroplast proteins are disrupted (Sakamoto, 2003). The M2-6 variegated phenotype is similar to that observed in var plants, rather than that observed in im plants, in which the white sectors dominate the leaf (Sakamoto, 2003). Interestingly, both var mutants identified in Arabidopsis plants contain disruptions in ftsH genes (Sakamoto et al., 2003). FtsH proteases are involved in the degradation of the D1 protein following photodamage to PSII.
Sequence analysis of Psb29 proteins failed to reveal their function. Since the protein is found in substoichiometric amounts in PSII preparations (Kashino et al., 2002), its function cannot be structural. Hence, the most probable role for these proteins is in biosynthetic pathways that control either the synthesis or the degradation of PSII components. Since the relative amounts of the D1 and CP47 proteins in total membrane preparations is not significantly altered in mutant cultures (Figure 4C), a role in control over translation or transcription can be ruled out. However, while the total amount of PSII proteins is unchanged, the assembly state of PSII complexes in the membranes can be. PSII assembly during biogenesis and disintegration following photodamage occur through a number of intermediary steps. Partially assembled intermediary complexes were observed in cyanobacteria (Zak et al., 2001; Keren et al., 2005), green algae (Adir et al., 1990), and higher plants (van Wijk et al., 1997; Rokka et al., 2005). Accumulation of uncoupled proximal antenna can be caused either by improper assembly of PSII supercomplexes or by inefficient disassembly of photodamaged PSII that results in a reduced fraction of active reaction center cores into which the excitation energy is transferred. Future studies will help elucidate the exact function of this protein in the biogenesis of PSII.
METHODS
Bioinformatics Analysis
WUBLAST (http://blast.wustl.edu) was used to search the National Center for Biotechnology Information (NCBI) nonredundant protein database for homologous sequences using an E value cutoff of 10−10. An additional sequence was found through searching the Chlamydomonas reinhardtii genome (http://www.chlamy.org/). An initial multiple sequence alignment was generated using ClustalW (Thompson et al., 1994), then edited by hand to identify three conserved regions, totaling 165 residues in length. These three conserved regions were concatenated together to give the final alignment used for tree building (see supplemental data online). The PHYLIP package was used to construct and bootstrap the phylogenetic tree (PHYLIP [Phylogeny Inference Package] version 3.6, distributed by J. Felsenstein, University of Washington, Seattle, WA). The SEQBOOT program was used to generate 1000 bootstrap alignments. Distances between sequences were calculated using the PROTDIST program. The Fitch-Margoliash method implemented in the FITCH program was used to generate the bootstrap trees, for each of which the sequence order was jumbled 100 times, with the global rearrangement option enabled. The CONSENSE program was then used to generate the final extended majority rule consensus tree (Figure 1B). The Chlorella virus sequence was chosen as the outgroup, as it is the most distant member of the sequence family, is the earliest branching leaf of a UPGMA generated tree, and may be under different evolutionary pressures than the nonviral sequences. Subcellular localization was predicted using the TargetP program (Emanuelsson et al., 2000). Hydropathy analysis was performed using the TopPred program (Claros and von Heijne, 1994).
Construction of Disruption Mutants
A segment of the sll1414 gene was amplified by PCR using the sll1414-R and sll1414-F primers (Figure 2A). The PCR product was cloned into a pGEM-T vector (Promega). A Gmr cassette (Schweizer, 1993) was cloned into the BamHI site in the sll1414 gene (Figure 2A). This vector was used to transform Synechocystis 6803 cells and produce the Δpsb29 strain. Segregation was verified by PCR using the cloning primers (Figure 2B). Transcript levels from the adjacent sll1415 gene were not affected by the insertion of the Gmr cassette (data not shown).
For the complementation assay, 400-μL aliquots of Δpsb29 cells were incubated for 1 h with 1.4 μg PCR product containing the complete coding sequence for the sll1414 open reading frame. Synechocystis 6803 is naturally transformable and takes up DNA from the media. Aliquots were diluted into 6 mL of BG11 media and transferred to a six-well plate for growth under high light intensities.
In Arabidopsis thaliana, a T-DNA insertion (Alonso et al., 2003) was identified in the At2g20890 gene in a line from the Salk Institute collection (SALK_094925; Figure 2C). SALK_094925 plants were screened by PCR to identify homozygous lines. One plant, M2-6, was found to be homozygous (Figure 2D). The same Salk T-DNA insertion line has been analyzed by Wang and coworkers with identical results (Wang et al., 2004). Primer sequences are indicated in Table 1.
Table 1.
Sequences of Primers
| Primer Name | Sequence |
|---|---|
| 1414-F | 5′-TCATGCAGGGCTATCAACCC-3′ |
| 1414-R | 5′-TTCTTCCGTTCTGCTTCGGC-3′ |
| 2890-I | 5′-CCATCATCATCCTTCCAATGG-3′ |
| 2890-II | 5′-GCAATGTGAAGTTCAAGAAGA-3′ |
| 2890-III | 5′-GCTGGACAATGAGCTCCTGCA-3′ |
| LBc1 | 5′-CGTCCGCAATGTGTTATTAAG-3′ |
| IC-F | 5′-CCGCATTCCTGAAAGATATCA-3′ |
| IC-R | 5′-CCGGAATTTGCATGGTTTACATAC-3′ |
Electron Microscopy
Synechocystis 6803 cells grown on solid BG11 media were fixed for transmission electron microscopy by high-pressure freezing (RMC Bockler), freeze-substituted in 2% osmium/acetone, and embedded in Epon/Araldite. Thin sections were stained with uranyl acetate and lead citrate and viewed using a LEO 912 transmission electron microscope (Zeiss) equipped with a CCD camera.
Culture Conditions
Synechocystis 6803 cultures were grown in BG11 mineral medium (Thornton et al., 2004) at 30°C with constant shaking. Illumination was at 55 μmol photons m−2 s−1 unless otherwise noted. Growth curve experiments were performed in six-well plates containing 3 mL of culture.
Arabidopsis plants were grown in a climate-controlled growth chamber at 20°C and 70% relative humidity. For growth light conditions, illumination was set at 50 μmol photons m−2 s−1 under a 16-h-day/8-h-night regime. Light intensity was lowered to 10 μmol photons m−2 s−1 by neutral density screens. For high light conditions, 220 μmol photons m−2 s−1 and continuous illumination were used. Immunoblots of total membrane preparations were isolated according to the method described by Thornton et al. (2004).
Measurements of Photosynthetic Activities
Chlorophyll concentrations and cell densities were calculated as previously described (Keren et al., 2002). Growth curve parameters were measured using a μQuant plate reader (Bio-Tek instruments). The 77K fluorescence spectra were obtained using a FluoroMax-2 fluorometer (Jobin Ivon). Fluorescence induction kinetics at room temperature were measured using a pump and probe FL100 setup (Photon Systems) for cyanobacteria and pulse amplitude modulated PAM101 (Walz) for plants.
Steady state oxygen evolution was measured using a Clark-type electrode (Yellow Springs Instruments). Measurements were performed at 30°C, and illumination was provided by a UV and heat filtered xenon lamp. Flash-induced oxygen evolution was measured using a bare platinum electrode setup (Artisan Scientific).
Accession Numbers
NCBI nonredundant database accession numbers for proteins discussed in this manuscript are as follows: NP_441343.1, Synechocystis sp PCC 6803, Sll1414; ZP_00178220.2, Crocosphaera watsonii WH 8501; ZP_00159115.1, Anabaena variabilis ATCC 29413; NP_484690.1, Nostoc sp PCC 7120; NP_484690.1, Nostoc sp PCC 7120; ZP_00164614.1, Synechococcus elongatus PCC 7942; ZP_00106172.1, Nostoc punctiforme PCC 73102; ZP_00324609.1, Trichodesmium erythraeum IMS101; NP_681924.1, Thermosynechococcus elongatus BP-1; NP_924346.1, Prochlorococcus marinus strain MIT 9313; gb|AAQ19850.1, Solanum tuberosum; XP_478693.1, Oryza sativa (japonica cultivar group); gb|AAD20906.1, Arabidopsis thaliana At2g20890; NP_897395.1, Synechococcus sp WH 8102; NP_875311.1, Prochlorococcus marinus subsp marinus strain CCMP1375; NP_892859.1, Prochlorococcus marinus subsp pastoris strain CCMP1986; NP_048481.1, Paramecium bursaria Chlorella virus; 166503, Chlamydomonas reinhardtii (from www.chalmy.org database)
Supplementary Material
Acknowledgments
This work was supported by funding from the National Science Foundation (MCB 0215359) to H.B.P. We thank Howard Berg of the Integrated Microscopy Facility at the Donald Danforth Plant Science Center for access to the electron microscopy equipment used in this study and the National Science Foundation MRI program for funding the purchase of this equipment. We thank the Salk Institute Genomic Analysis Laboratory for providing sequence-indexed Arabidopsis T-DNA insertion mutants.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Himadri B. Pakrasi (pakrasi@wustl.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035048.
References
- Adam, Z., and Ostersetzer, O. (2001). Degradation of unassembled and damaged thylakoid proteins. Biochem. Soc. Trans. 29, 427–430. [DOI] [PubMed] [Google Scholar]
- Adir, N., Shochat, S., and Ohad, I. (1990). Light-dependent D1 protein synthesis and translocation is regulated by reaction center II. Reaction center II serves as an acceptor for the D1 precursor. J. Biol. Chem. 265, 12563–12568. [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Blankenship, R.E., and Hartman, H. (1998). The origin and evolution of oxygenic photosynthesis. Trends Biochem. Sci. 23, 94–97. [DOI] [PubMed] [Google Scholar]
- Bricker, T.M., Morvant, J., Masri, N., Sutton, H.M., and Frankel, L.K. (1998). Isolation of a highly active photosystem II preparation from Synechocystis 6803 using a histidine-tagged mutant of CP 47. Biochim. Biophys. Acta 1409, 50–57. [DOI] [PubMed] [Google Scholar]
- Claros, M.G., and von Heijne, G. (1994). TopPred II: An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686. [DOI] [PubMed] [Google Scholar]
- Depege, N., Bellafiore, S., and Rochaix, J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–1575. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Ferreira, K.N., Iverson, T.M., Maghlaoui, K., Barber, J., and Iwata, S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838. [DOI] [PubMed] [Google Scholar]
- Grossman, A.R., Bhaya, D., Apt, K.E., and Kehoe, D.M. (1995). Light-harvesting complexes in oxygenic photosynthesis: Diversity, control, and evolution. Annu. Rev. Genet. 29, 231–288. [DOI] [PubMed] [Google Scholar]
- Hall, D.O., and Rao, K.K. (1995). Photosynthesis. (Cambridge, UK: Cambridge University Press).
- Hundal, T., Forsmark-Andree, P., Ernster, L., and Andersson, B. (1995). Antioxidant activity of reduced plastoquinone in chloroplast thylakoid membranes. Arch. Biochem. Biophys. 324, 117–122. [DOI] [PubMed] [Google Scholar]
- Joliot, P. (1972). Modulated light source use with the oxygen electrode. Methods Enzymol. 24, 123–134. [DOI] [PubMed] [Google Scholar]
- Kashino, Y., Lauber, W.M., Carroll, J.A., Wang, Q., Whitmarsh, J., Satoh, K., and Pakrasi, H.B. (2002). Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012. [DOI] [PubMed] [Google Scholar]
- Keren, N., Kidd, M.J., Penner-Hahn, J.E., and Pakrasi, H.B. (2002). A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry 41, 15085–15092. [DOI] [PubMed] [Google Scholar]
- Keren, N., Liberton, M., and Pakrasi, H.B. (2005). Photochemical competence of assembled photosystem II core complex in cyanobacterial plasma membrane. J. Biol. Chem. 280, 6548–6553. [DOI] [PubMed] [Google Scholar]
- Keren, N., and Ohad, I. (1998). State transition and photoinhibition. In The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas, J.D. Rocaix, M. Goldschmidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Press), pp. 569–596.
- Kramer, D.M., Johnson, G., Kiirats, O., and Edwards, G.E. (2004). New fluorescence parameters for the determination of Q(A) redox state and excitation energy fluxes. Photosynth. Res. 79, 209–218. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Gilmore, A.M., and Niyogi, K.K. (2002). Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J. Biol. Chem. 277, 33590–33597. [DOI] [PubMed] [Google Scholar]
- Maxwell, K., and Johnson, G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Mullet, J.E. (1988). Chloroplast development and gene expression. Annu. Rev. Plant Physiol. 39, 475–502. [Google Scholar]
- Nakamura, Y., et al. (2003). Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids (supplement). DNA Res. 10, 181–201. [DOI] [PubMed] [Google Scholar]
- Ossenbuhl, F., Gohre, V., Meurer, J., Krieger-Liszkay, A., Rochaix, J.D., and Eichacker, L.A. (2004). Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16, 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J.B., Ytterberg, A.J., Sun, Q., and van Wijk, K.J. (2004). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279, 49367–49383. [DOI] [PubMed] [Google Scholar]
- Rokka, A., Suorsa, M., Saleem, A., Battchikova, N., and Aro, E.M. (2005). Synthesis and assembly of thylakoid protein complexes: Multiple assembly steps of photosystem II. Biochem. J. 388, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, A.W. (1989). Photosystem II: The water-splitting enzyme. Trends Biochem. Sci. 14, 227–232. [DOI] [PubMed] [Google Scholar]
- Rutherford, A.W., and Boussac, A. (2004). Water photolysis in biology. Science 303, 1782–1784. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W. (2003). Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet. Syst. 78, 1–9. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15, 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, H.P. (1993). Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15, 831–834. [PubMed] [Google Scholar]
- Shikanai, T., Munekage, Y., Shimizu, K., Endo, T., and Hashimoto, T. (1999). Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol. 40, 1134–1142. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, L.E., Ohkawa, H., Roose, J.L., Kashino, Y., Keren, N., and Pakrasi, H.B. (2004). Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. Plant Cell 16, 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Best, J.A., and Duysens, L.N. (1975). Reactions between primary and secondary acceptors of photosystem II in Chlorella pyrenoidosa under anaerobic conditions as studied by chlorophyll fluorescence. Biochim. Biophys. Acta 408, 154–163. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J., Roobol-Boza, M., Kettunen, R., Andersson, B., and Aro, E.M. (1997). Synthesis and assembly of the D1 protein into photosystem II: Processing of the C-terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry 36, 6178–6186. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Sullivan, R.W., Kight, A., Henry, R.L., Huang, J., Jones, A.M., and Korth, K.L. (2004). Deletion of the chloroplast-localized thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136, 3594–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal, S., Soll, J., and Vothknecht, U.C. (2003). Evolution of chloroplast vesicle transport. Plant Cell Physiol. 44, 217–222. [DOI] [PubMed] [Google Scholar]
- Yoon, H.S., Hackett, J.D., Pinto, G., and Bhattacharya, D. (2002). The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. USA 99, 15507–15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak, E., Norling, B., Maitra, R., Huang, F., Andersson, B., and Pakrasi, H.B. (2001). The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. USA 98, 13443–13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.