Abstract
Frequently, mtDNA with pathogenic mutations coexist with wild-type genomes (mtDNA heteroplasmy). Mitochondrial dysfunction and disease ensue only when the proportion of mutated mtDNAs is high, thus a reduction in this proportion should provide an effective therapy for these disorders. We developed a system to decrease specific mtDNA haplotypes by expressing a mitochondrially targeted restriction endonuclease, ApaLI, in cells of heteroplasmic mice. These mice have two mtDNA haplotypes, of which only one contains an ApaLI site. After transfection of cultured hepatocytes with mitochondrially targeted ApaLI, we found a rapid, directional, and complete shift in mtDNA heteroplasmy (2-6 h). We tested the efficacy of this approach in vivo, by using recombinant viral vectors expressing the mitochondrially targeted ApaLI. We observed a significant shift in mtDNA heteroplasmy in muscle and brain transduced with recombinant viruses. This strategy could prevent disease onset or reverse clinical symptoms in patients harboring certain heteroplasmic pathogenic mutations in mtDNA.
Keywords: gene therapy, mitochondria
Mammalian mtDNA is a maternally inherited circular genome present in hundreds to thousands of copies in most cells. mtDNA, the only extrachromosomal DNA in the cell, encodes for components of the oxidative phosphorylation system (OXPHOS), and it is located within the mitochondrial matrix in discrete protein-DNA complexes called nucleoids (1-4). Most pathogenic mtDNA mutations exist in a heteroplasmic state, and several studies have shown that a high percentage of mutated mtDNA is necessary to trigger a metabolic defect in specific tissues and organs (5, 6).
When the pathogenic nucleotide change creates a novel site for the restriction endonuclease, the expression of a mitochondrially targeted restriction endonuclease has the potential to reduce the proportion of mutated mtDNA in heteroplasmic mitochondria. This is the case for the T8399G transversion in the mitochondrial ATP6 gene, which creates a unique SmaI-XmaI site in the human mtDNA. This mutation is associated with neuropathy ataxia and retinitis pigmentosa (NARP), or when present at high (>90%) levels, with a more severe syndrome termed maternally inherited Leigh syndrome (6, 7).
We have previously shown that a mitochondrially targeted PstI is able to degrade mtDNA harboring PstI sites, leading to a loss of mitochondrial genomes in cultured human cells. The same construct was able to shift mtDNA heteroplasmy in cultured cells containing both mouse and rat mtDNA, because the latter lacks PstI sites (8). In a similar study, targeting the restriction endonuclease SmaI to mitochondria of cultured cells harboring the T8399G NARP mutation prompted specific elimination of mutated mtDNA, followed by repopulation by the wild-type mtDNA. This resulted in restoration of normal intracellular ATP level and mitochondrial membrane potential (9).
Here, we developed a system to define the kinetics of mtDNA heteroplasmy change as well as test whether this therapeutic approach could be effective in vivo. We took advantage of a well characterized heteroplasmic mouse model containing two polymorphic mtDNA sequence variants (NZB and BALB). This mouse line was created by cytoplasmic fusion in single cell embryos (10). In these mice, both mtDNA haplotypes appear to behave as neutral variants except for one characteristic: there is an age-related selection of different mtDNA haplotypes in the same animal (10-12). The BALB mtDNA variant harbors a unique ApaLI site that is not present in the NZB variant, so we developed a mitochondrially targeted ApaLI gene construct to specifically decrease BALB mtDNA levels in the tissues of these animals.
Methods
Mito-ApaLI-Hemagglutinin (HA) Constructs. A synthetic gene coding for the mammalian codon usage-optimized version of ApaLI-HA was purchased from Integrated DNA Technologies (Coralville, IA) and cloned downstream and in-frame with a mitochondrial targeting sequence from the cytochrome oxidase subunit VIII (13). For inducible expression, a Mifepristone-(RU486) inducible system was used (see Supporting Text, which is published as supporting information on the PNAS web site).
Cell Lines, Transfections, Enzyme Assays, and Immunodetection. All cell lines used, including variants of the HP202B mouse heteroplasmic hepatocyte cell line and their characterization, are described in detail as Supporting Text.
Restriction Fragment Length Polymorphism (RFLP) and Southern Blot. Total DNA from cellular clones, tail biopsies, or microdissected tissues samples were purified and a region of mtDNA PCR was amplified and digested with ApaLI or with HindIII, which differentiate BALB and NZB mtDNAs. These analyses as well as the Southern blot procedures are described in Supporting Text.
Animal Handling and Virus Administration. NZB/BALB heteroplasmic female founders were produced as described (10), and a colony was maintained by breeding with C57BL/6J males. Replication-deficient, Mito-ApaLI-HA encoding adenovirus (Ad5-Mito-ApaLI-HA 3.4 × 1012 viral particles per ml based in OD260) and a hybrid adeno-associated viral (AAV) vector derived from serotypes 1 and 2 (14, 15) encoding Mito-ApaLI-HA (AAV1,2-Mito-ApaLI-HA; 2.4 × 1012 genomic particles per ml) was injected in brain and muscle, and these tissues were analyzed by laser capture microscopy as described (see Supporting Text).
Results
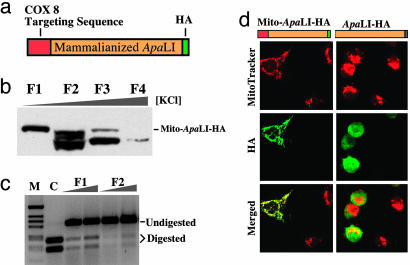
Expressing ApaLI in Mitochondria Promotes a Shift in mtDNA Heteroplasmy. We have optimized the codon usage of the gene coding for the Acetobacter pasterianus restriction endonuclease ApaLI to enhance its expression in mammalian cells (16). For immunological detection of ApaLI, a HA epitope tag was added to its C terminus (Fig. 1a). We tested whether ApaLI containing the HA tag was enzymatically active by expressing a cytosolic version of the enzyme (without the mitochondrial targeting sequence, ApaLI-HA) in transiently transfected human embryonic kidney (HEK) 293T cells. The cytosolic S-100 fraction of transfected cells showed specific cleavage of a test DNA fragment, demonstrating that the HA tag did not abrogate ApaLI activity (Fig. 1c). Next, we showed that when the mitochondrial targeting sequence from the nuclear-encoded cytochrome c oxidase subunit VIII (COX8) was added in-frame to the N terminus of the construct, ApaLI-HA colocalized with the mitochondrial dye Mitotracker (Invitrogen) in transiently transfected COS-1 cells (Fig. 1d).
Fig. 1.
Construction and characterization of a Mito-ApaLI-HA. (a) The genetic code of a synthetic ApaLI gene was optimized for mammalian translation and a HA-tag coding sequence added to the 3′ end. The gene was cloned downstream of the cytochrome oxidase subunit 8 (COX 8) mitochondrial targeting sequence in a pcDNA3 vector. (b) Mito-ApaLI-HA containing the COX 8 targeting sequence was expressed in human embryonic kidney 293T cells and after 48 h, an S-100 fraction was purified by heparin Sepharose binding. Fractions eluted with increasing salt concentrations (F1-F4) were analyzed by immunoblot using an anti-HA antibody. Fraction 1 shows mostly full length protein, whereas the other lanes have degradation products. (c) Fraction F1 was able to digest a DNA fragment at the ApaLI cleavage sites. Control (C) is the same PCR fragment digested with commercial ApaLI. (d) pcDNA3-based plasmids harboring either Mito-ApaLI-HA or ApaLI-HA constructs were transiently transfected into COS-7 cells. After 24 h, cells were stained with the mitochondrial specific dye Mitotracker red, fixed, and immunostained for the HA epitope. In the merged image, note the mitochondrial colocalization of HA immunofluorescence (in green) and Mitotracker (in red) in cells expressing Mito-ApaLI-HA.
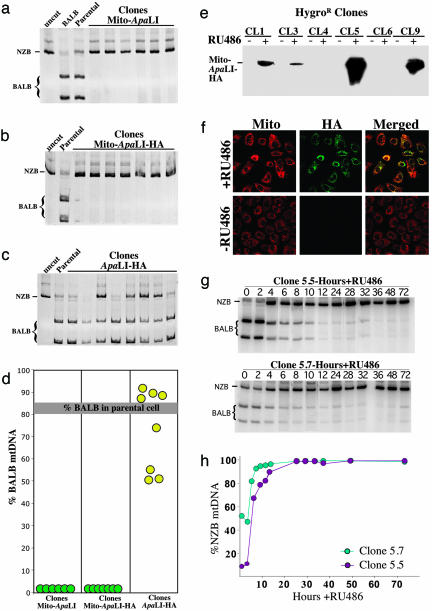
To test the ability of the Mito-ApaLI-HA to promote a shift in mtDNA heteroplasmy, we stably transfected ApaLI-HA, Mito-ApaLI, and Mito-ApaLI-HA into hepatocytes derived from heteroplasmic mice harboring both BALB and NZB mtDNA haplotypes. The stable expression of Mito-ApaLI in these hepatocytes resulted in a complete shift in heteroplasmy toward NZB mtDNA, regardless of the presence of the HA tag (Fig. 2 a and b). Cytosolic expression of ApaLI-HA did not alter mtDNA heteroplasmy (Fig. 2c). No apparent toxicity, as judged by cell morphology and growth rates (not shown), was observed in cells expressing either the cytosolic or the mitochondrial versions of the construct. These results demonstrate that this system is highly efficient in promoting a shift in mtDNA heteroplasmy in cultured hepatocytes (Fig. 2d).
Fig. 2.
Rapid and complete shift in mtDNA heteroplasmy by Mito-ApaLI-HA. Cultured hepatocyte clones from heteroplasmic BALB/NZB mice stably transfected with the different ApaLI constructs were analyzed for their mtDNA haplotypes. Cells were collected after selection in G418. DNA was purified and used to amplify a PCR fragment spanning an ApaLI site. After digestion with ApaLI, the samples were analyzed by PAGE (“uncut,” PCR fragment not digested; “BALB,” PCR fragment from a BALB mouse digested with ApaLI; “Parental,” PCR fragment from untransfected hepatocytes digested with ApaLI). a-c show autoradiographies of PAGE/RFLP of amplicons from clones expressing the different ApaLI constructs. Mitochondrially targeted ApaLI promoted a complete shift in mtDNA heteroplasmy toward the NZB haplotype (no ApaLI sites), irrespective of the presence of the HA tag (a and b). c shows that in the absence of a mitochondrial targeting sequence, no mtDNA heteroplasmy shift took place. d summarizes the data from the individual hepatocyte clones. e shows the immunoblot (anti-HA) analysis of clones expressing an inducible form of mito-ApaLI-HA. A strong induction of mito-ApaLI-HA expression can be observed in some hepatocyte clones after the addition of 10 nM RU486. f shows the RU486 induction of Mito-ApaLI-HA by immunocytochemistry in clone 5. g shows the time course of the shift in mtDNA heteroplasmy after induction of Mito-ApaLI-HA expression in clones 5.5 and 5.7. h shows a similar analysis, after digestion of PCR fragments with HindIIII (also discriminates BALB and NZB mtDNA), in graphic format.
Mito-ApaLI Promotes a Rapid Shift in mtDNA Heteroplasmy. To investigate the kinetics of the shift in mtDNA heteroplasmy, we used a RU486-inducible system derived from the Gene Switch system (Invitrogen) to express the Mito-ApaLI-HA construct. The isolation of heteroplasmic hepatocyte lines with this vector integrated in the genome yielded clones with undetectable levels of basal expression of Mito-ApaLI-HA (Fig. 2e). In two of them (clones 5 and 9), high levels of Mito-ApaLI-HA expression were observed after RU486 induction (Fig. 2e). By immunocytochemistry, no Mito-ApaLI-HA expression was detected before induction, and the protein colocalized with the mitochondria after induction (Fig. 2f).
We produced Mito-ApaLI-HA-inducible cell lines with different levels of mtDNA heteroplasmy by treating the highly inducible clone 5 with the mitochondrial toxin rhodamine-6G and replenishing the cell with mitochondria from chemically enucleated mouse cells harboring homoplasmic BALB mtDNA. Treated cells were fused and clones isolated as described in Supporting Text.
We determined the percentage of each mtDNA haplotype in clones 5.5 (9% NZB mtDNA) and 5.7 (52% NZB mtDNA) at different times after the induction of Mito-ApaLI-HA. The mtDNA haplotype rapidly shifted to essentially 100% NZB in both clones after RU486 induction (Fig. 2g). In clone 5.7, containing an initial proportion of NZB of 52%, this complete shift was achieved in ≈12 h. In clone 5.5, with a lower initial level of NZB mtDNA (9%), a virtually complete change in mtDNA haplotype was achieved in ≈24 h. To verify the mtDNA heteroplasmy levels by using an independent approach, we digested a PCR-amplified region of the mtDNA with HindIII, an enzyme capable of discriminating between BALB and NZB mtDNA haplotypes. The results were identical to those obtained by ApaLI digestion (Fig. 2h). These results showed that the kinetic of shift in mtDNA heteroplasmy by mitochondrially targeted ApaLI is extremely rapid.
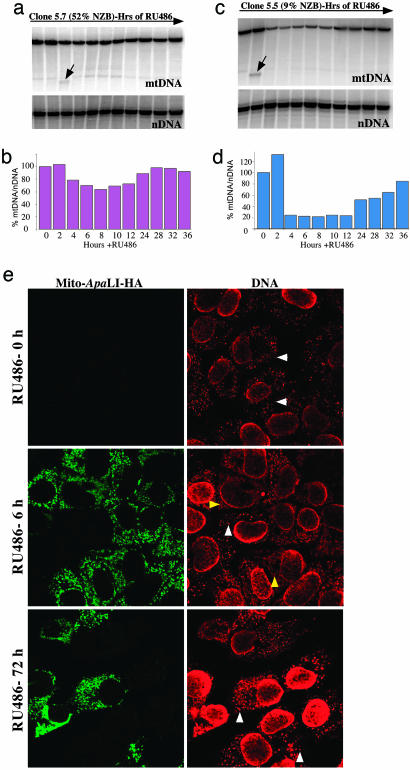
Activation of Mitochondrially Targeted Restriction Endonuclease Causes Transient mtDNA Depletion and Nucleoid Disorganization. We quantified mtDNA levels by Southern blot after Mito-ApaLI-HA induction in clones 5.5 and 5.7, respectively, at several time points after induction. When normalized to a nuclear DNA gene (18S rDNA), clone 5.5 (with 9% NZB mtDNA) exhibited a stronger transient mtDNA depletion than clone 5.7 (with 52% NZB mtDNA), and the recovery of the initial mtDNA levels took longer to approach the initial levels (36 and 24 h, respectively; Fig. 3 a-d). A Mito-ApaLI-digested mtDNA fragment could be detected by Southern blots 2-4 h after Mito-ApaLI-HA induction but not at later times (indicated by arrows in Fig. 3 a and c).
Fig. 3.
Transient mtDNA depletion associated with expression of Mito-ApaLI-HA. Cells were collected at different time points after expression of Mito-ApaLI-HA, and their DNA was isolated and analyzed by Southern blot. Total DNA was digested with SacI, which cuts both BALB and NZB mtDNA once at nucleotide 9047, probed to a mtDNA probe and subsequently to a nuclear 18SrDNA probe. a and c show the Southern blot, whereas b and d show the quantitation of mtDNA/nuclear DNA ratios. A fragment with the size predicted from a double-digestion SacI+ApaLI was observed only at 2-4 h of Mito-ApaLI-HA induction (arrow). (e) Cells with 9% NZB mtDNA were induced to express Mito-ApaLI-HA with RU486. The cells were fixed at different time points after induction and immunostained for HA (green) and DNA (red). Note that at 6-h induction, cells expressing Mito-ApaLI-HA lost the punctate nucleoid structures (white arrowhead) that were substituted by a diffuse crossreacting material (yellow arrowheads). However, nucleoids were reestablished in Mito-ApaLI-HA-expressing cells at 72-h induction.
To further characterize the effect of Mito-ApaLI-HA in heteroplasmic cells, we analyzed mtDNA-containing nucleoids at different time points after the Mito-ApaLI-HA induction. Nucleoids are DNA-protein structures that are important in mtDNA maintenance and stability (2-4). For this purpose, we used an antibody raised against double-stranded DNA that recognizes the mtDNA-enriched nucleoids (2). Nucleoids disappeared gradually when the cells were treated with a mtDNA-depleting agent, such as ethidium bromide (Fig. 6, which is published as supporting information on the PNAS web site). We monitored the nucleoid presence in a heteroplasmic clone (9% NZB). There was a decrease in the number of nucleoids (white arrowheads in Fig. 3e) at 6 h after Mito-ApaLI-HA induction, and only a diffuse staining for DNA was observed in mitochondria from Mito-ApaLI-HA expressing cells (yellow arrowheads). However, the nucleoids recovered their punctate appearance after 72 h (Fig. 3e). As shown in Fig. 3 a-d, this is due to an initial BALB mtDNA depletion followed by NZB mtDNA repopulation. Fig. 6 shows a similar analysis in two additional hepatocyte clones, homoplasmic for either NZB or BALB mtDNA, after RU486 induction. In the clone homoplasmic for BALB mtDNA, we observed progressive nucleoid elimination after 6, 18, and 72 h of Mito-ApaLI-HA expression (Fig. 6). In the clone homoplasmic for NZB mtDNA, because of the absence of ApaLI sites, nucleoids were unchanged by Mito-ApaLI-HA expression (Fig. 6).
Mitochondrially Targeted Restriction Endonuclease as a Gene Therapy Approach. We evaluated the potential of the mitochondrially targeted restriction endonuclease to induce a shift in heteroplasmy in vivo in both skeletal muscle and brain of BALB/NZB heteroplasmic mice. We used either recombinant adenovirus vector (rAd5) or AAV vector (rAAV1,2) to deliver the Mito-ApaLI-HA gene.
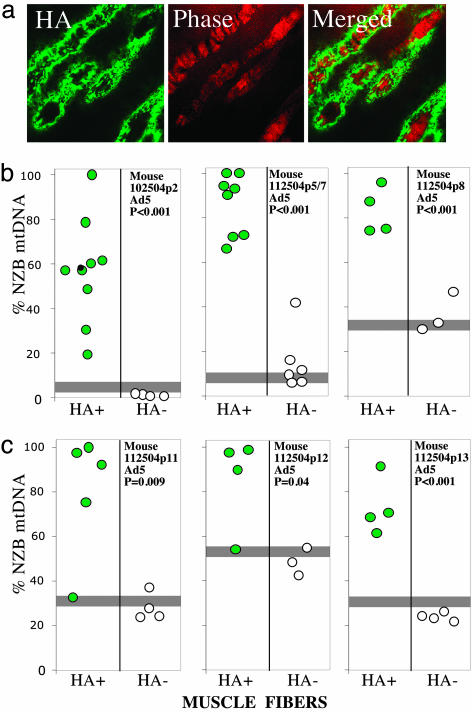
rAd5-Mito-ApaLI transduction resulted in the expression of the transgene in differentiated muscle fibers of 5-day-old mice, as shown in HA-immunostained tissue sections (Fig. 4a). To estimate the shift in heteroplasmy in muscle transduced with rAd5-Mito-ApaLI-HA, we microdissected individual muscle fibers from HA-immunostained muscle sections by using a laser capture microscope followed by PCR/RFLP analyses. We analyzed muscle from three animals 1 week after viral injection (Fig. 4b) and from three additional animals 2 weeks after the injections (Fig. 4c). In all of the animals tested, mtDNA heteroplasmy of the fibers expressing Mito-ApaLI-HA showed a statistically significant higher proportion of NZB mtDNA when compared with the HA-negative ones (Figs. 4 b and c).
Fig. 4.
Expression of Mito-ApaLI-HA in muscle leads to efficient shift in mtDNA heteroplasmy. Skeletal muscle (gastrocnemius) of 5-day-old mice was injected with Ad5-Mito-ApaLI-HA, as described in Methods. After 1 (b) or 2 (c) weeks, animals were killed and muscle frozen in liquid nitrogen-cooled isopentane. Twenty-micrometer muscle sections were immunostained for HA (a) and microdissected using laser capture microscopy. Microdissected samples were treated with an alkaline lysis solution and subjected to PCR/RFLP for mtDNA haplotype determination. The proportions of NZB/BALB mtDNA were calculated after “last cycle hot” PCR, digestion with ApaLI, PAGE, and phosphorimaging (b and c). Horizontal gray bars represent the percent NZB mtDNA in the mouse tail.
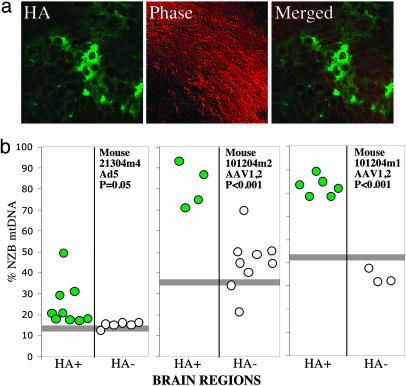
To investigate whether the same viral approach could cause a shift in mtDNA heteroplasmy in the brain, we injected rAd5-Mito-ApaLI-HA in the right cerebral hemisphere of 2- to 4-mo-old BALB/NZB heteroplasmic mice. rAd5 transduces mostly neurons but also astrocytes and other glial cells (17). Injections targeted the putamen of adult mice. One week after rAd5 injection, HA-immunostained brain sections were laser-capture microscopy-microdissected and their mtDNA heteroplasmy levels determined by RFLP as described above. The transduced areas expressing Mito-ApaLI-HA were relatively small (Fig. 5a). Nevertheless, we observed a shift in mtDNA heteroplasmy, albeit the change was lower in magnitude than the one observed in muscle (Fig. 5b Left).
Fig. 5.
Expression of Mito-ApaLI-HA in brain leads to a shift in mtDNA heteroplasmy. The putamen of anesthetized 2- to 4-mo-old mice was stereotactically injected with 2 μl of virus suspensions (Ad5-Mito-ApaLI-HA or AAV1,2-Mito-ApaLI-HA). After 1 (Ad5) or 2 weeks (AAV1,2), the animals were killed and the brains snap-frozen in liquid nitrogen. Twenty-micrometer sections were screened for GFP expression (not shown) and adjacent sections stained for HA expression (a). Regions of positive and negative staining were microdissected by laser capture microscopy and subjected to mtDNA haplotype analysis as described in the legend to Fig. 4b. Horizontal gray bars represent the percent NZB mtDNA in the mouse tail.
The rAAV1,2-Mito-ApaLI-Ha vector, which transduces mostly neurons (18), was also used to transduce the same area of the brain. We obtained better transduction efficiency with this vector when compared with rAd5, with larger areas of the brain expressing Mito-ApaLI-HA (not shown). RFLP of microdissected regions showed a stronger shift in heteroplasmy than the one observed with rAd5 (Fig. 5b Right).
Discussion
The modulation of mtDNA heteroplasmy has been recognized as a promising approach for the treatment of mitochondrial disorders (19). Earlier approaches using antisense technology directed against mutated molecules showed high specificity in vitro (20), but because of the requirement to deliver antisense nucleic acids to mitochondria, this approach has not yet been able to shift mtDNA heteroplasmy in vivo (21).
Muscle satellite cells usually have a lower percentage of mutated mtDNA when compared with mature muscle. Incorporation of satellite cells into existing myofibers in patients by tissue damage/regeneration (22, 23), or exercise (24), has been used to lower the mutant load, but the effect on mtDNA heteroplasmy was modest (25). Ketogenic treatment of cultured cells also triggered a small shift in heteroplasmy toward the wild-type mtDNA (26). Here we show that a mitochondrially targeted restriction endonuclease can promote rapid, specific, and robust shifts in mtDNA heteroplasmy in vivo, and that this approach could be used for gene therapy in patients harboring mtDNA mutations that create restriction sites (e.g., NARP syndrome).
The use of the RU486 inducible activation vector gave us the ability to turn the expression of the mitochondrial restriction endonuclease on and off. This allowed us to determine the kinetics of the shift in mtDNA heteroplasmy and to quantify the restriction endonuclease-driven mtDNA depletion. The shift in heteroplasmy was efficient, and mtDNA molecules were quickly degraded once they had been cleaved by the endonuclease. This was shown by the presence of predicted DNA fragments in Southern blots at 2-4 h after the restriction endonuclease was turned on but not at later times. After specific mtDNA cleavage, the mitochondria were repopulated with the mtDNA haplotype lacking the restriction site. The number of nucleoids decreased with ApaLI induction but recovered along with the mtDNA levels. These observations are similar to those reported by Tanaka et al. (9), who showed a reduction in mtDNA copy number upon expression of SmaI or EcoRI in mitochondria of cultured cells (9). Taken together, our results indicate that the shift in mtDNA heteroplasmy by mitochondrially targeted restriction endonucleases is rapid and specific.
Transient mtDNA depletion is probably not severe enough to compromise the oxidative phosphorylation system (OXPHOS), which would not be surprising considering OXPHOS complexes are likely to have a slow turnover rate. Additional data from the literature support this concept. Using an inducible dominant negative DNA polymerase γ, Jazayeri et al. (27) showed that decreasing mtDNA levels by 99% resulted in a reduction of the activity of the respiratory complexes to 18-30% of controls (27). In mice with a brain-specific Tfam knockout, which leads to lack of mtDNA replication, there was a 1- to 2-mo interval between severe mtDNA depletion and the detection of a biochemical defect (28). More importantly, we show that, in heteroplasmic cells treated with mitochondrially targeted restriction endonucleases, mtDNA levels decreases are rapidly compensated by increases in the nonsusceptible mtDNA. Despite these reassuring data, care may be required in treating patients with extremely high levels of the mutated mtDNA to avoid an abrupt mtDNA depletion of the mutated mtDNA (that retains partial function), which could compromise tissue viability. The administration of a mitochondrially targeted restriction endonuclease could be done step-wise, avoiding infecting the complete organ in one session. Still, if a substantial number of postmitotic cells are suspected of being homoplasmic (or near homoplasmic) in a particular patient, this approach may not be indicated.
We did not detect evidence of nuclear DNA damage by the TUNEL assay in transfected cultured hepatocytes (data not shown), suggesting that because of the specific mitochondrial localization of the Mito-ApaLI-HA, nuclear DNA digestion does not occur. Similarly, nuclear DNA breakdown was not observed in the studies of Tanaka et al. (9) expressing mitochondrially targeted restriction endonucleases in NARP cells. Therefore, nuclear DNA damage is unlikely to be a concern in the administration of mitochondrially targeted restriction endonucleases in vivo.
We tested the efficacy of this approach in the NZB/BALB mtDNA heteroplasmic mouse using viral vectors encoding the Mito-ApaLI-HA gene. In this animal model, heteroplasmic mtDNA in muscle fibers expressing the restriction enzyme consistently shows a shift toward the NZB haplotype. This was observed if they were analyzed 1 or 2 weeks after Ad5-Mito-ApaLI-HA transduction. Adenoviral vectors can transduce a variety of tissues in vivo, including skeletal and cardiac muscle, liver, kidney, and bronchial epithelial and nervous system tissues (29). Transduction does not require active cell division or integration of the adenovirus genome for Ad genome-derived gene expression, because the viral genome is normally maintained in an episomal state. However, Ad-driven transgene expression is transient, and the vector elicits a strong immune response (30). Therefore, immunosuppression therapy may be required to avoid strong antibody responses. This could be a serious disadvantage for many gene therapy approaches. However, because only transient expression of the restriction endonuclease is necessary to produce a fast and long-lasting shift in heteroplasmy, adenoviral vectors may be suitable for gene therapy of mitochondrial disorders with mitochondrially targeted restriction endonucleases.
When brain from NZB/BALB heteroplasmic mouse was transduced with Ad5 containing Mito-ApaLI-HA, the shift in heteroplasmy was modest. We think this was due to the analyses of a mixture of transduced and nontransduced cells in the microdissected sections. With the use of AAV1,2-based viral vector, the efficiency of transduction improved, resulting in a more dramatic shift in heteroplasmy.
AAV-based vectors can be maintained in persistent episomal form or integrate in the host genome (31). An inducible expression system such as GeneSwitch, which results in high levels of gene expression only in the presence of the inductor molecule (32), could be used with AAV to avoid unnecessary long-term expression of the restriction endonuclease and control the expression levels to modulate the ensuing mtDNA depletion.
The T8399G mtDNA mutation, commonly associated with Leigh syndrome, when the mutation is present in >90-95%, and with the NARP syndrome, when the proportion of mutated mtDNA is 70-90%, would be a suitable candidate for this therapeutic approach. Tanaka et al. (9) showed that cultured cells with the T8399G mutation underwent a shift in heteroplasmy when a mitochondrially targeted SmaI was expressed. However, it took five cycles of transfection to completely eliminate the mutated mtDNA. This may have resulted from the fact that the SmaI construct used had the bacterial codon usage, and expression could have been low in human cells (16).
One potential concern with the mitochondrial restriction endonuclease approach is the recent observation that double-strand breaks in the mtDNA can lead to the formation of low levels of mtDNA deletions in muscle of mice expressing a mitochondrially targeted PstI (33). However, in these mice, all mtDNA molecules were homoplasmic for the restriction site and susceptible to PstI digestion, and mtDNA deletions are unlikely to accumulate to high levels in a heteroplasmic environment. In fact, we were unable to detect mtDNA deletions, even by PCR amplification using primers flanking the ApaLI site and the mtDNA D-loop region, in hepatocytes expressing Mito-ApaLI-HA (not shown).
Conclusion
We have shown that the manipulation of mtDNA heteroplasmy by a mitochondrially targeted restriction endonuclease is a powerful approach to modulate mtDNA heteroplasmy in vivo. This approach could be suitable as a genetic treatment of patients with mitochondrial diseases caused by mutations that create a unique restriction endonuclease cleavage site. It is also important to emphasize that even a small shift in mtDNA heteroplasmy would have a dramatic effect in the bioenergetic state of the cells. Although the need for a unique restriction site is an obvious limitation, this approach may also modulate mtDNA heteroplasmy in the context of multiple cleavage sites, if expression of the restriction endonuclease could be tightly regulated.
Supplementary Material
Acknowledgments
We are grateful to Ms. Dayami Hernandez and Ms. Sofia Garcia for help with mouse maintenance, perfusions, and typing. We also thank Dr. R. Voellmy (University of Miami, Miami) for vectors. This work was supported by grants from the National Eye Institute (EY10804 and EY014801) and the National Institute of Neurological Disorders and Stroke (NS41777) (to C.T.M.). E.A.S. is an international scholar of the Howard Hughes Medical Institute and a Senior Investigator of the Canadian Institutes of Health Research. B.J.B. was supported by a Natural Sciences and Engineering Research Council postdoctoral fellowship.
Author contributions: M.P.B.-B. and C.T.M. designed research; M.P.B.-B., B.B., and C.T.M. performed research; B.B., B.J.B., and E.A.S. contributed new reagents/analytic tools; M.P.B.-B. and C.T.M. analyzed data; and M.P.B.-B. and C.T.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NZB, New Zealand black; AAV, adeno-associated virus; NARP, neuropathy ataxia and retinitis pigmentosa; RFLP, restriction fragment length polymorphism; HA, hemagglutinin; rAd5, recombinant adenovirus vector.
References
- 1.Kaufman, B. A., Newman, S. M., Hallberg, R. L., Slaughter, C. A., Perlman, P. S. & Butow, R. A. (2000) Proc. Natl. Acad. Sci. USA 97, 7772-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legros, F., Malka, F., Frachon, P., Lombes, A. & Rojo, M. (2004) J. Cell Sci. 117, 2653-2662. [DOI] [PubMed] [Google Scholar]
- 3.Bogenhagen, D. F., Wang, Y., Shen, E. L. & Kobayashi, R. (2003) Mol. Cell Proteomics 2, 1205-1216. [DOI] [PubMed] [Google Scholar]
- 4.Garrido, N., Griparic, L., Jokitalo, E., Wartiovaara, J., van der Bliek, A. M. & Spelbrink, J. N. (2003) Mol. Biol. Cell 14, 1583-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorburn, D. R. & Dahl, H. H. (2001) Am. J. Med. Genet. 106, 102-114. [DOI] [PubMed] [Google Scholar]
- 6.DiMauro, S. & Schon, E. A. (2003) N. Engl. J. Med. 348, 2656-2668. [DOI] [PubMed] [Google Scholar]
- 7.Holt, I. J., Harding, A. E., Petty, R. K. & Morgan-Hughes, J. A. (1990) Am. J. Hum. Genet. 46, 428-433. [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava, S. & Moraes, C. T. (2001) Hum. Mol. Genet. 10, 3093-3099. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka, M., Borgeld, H. J., Zhang, J., Muramatsu, S., Gong, J. S., Yoneda, M., Maruyama, W., Naoi, M., Ibi, T., Sahashi, K., et al. (2002) J. Biomed. Sci. 9, 534-541. [DOI] [PubMed] [Google Scholar]
- 10.Jenuth, J. P., Peterson, A. C. & Shoubridge, E. A. (1997) Nat. Genet. 16, 93-95. [DOI] [PubMed] [Google Scholar]
- 11.Battersby, B. J. & Shoubridge, E. A. (2001) Hum. Mol. Genet. 10, 2469-2479. [DOI] [PubMed] [Google Scholar]
- 12.Battersby, B. J., Loredo-Osti, J. C. & Shoubridge, E. A. (2003) Nat. Genet. 33, 183-186. [DOI] [PubMed] [Google Scholar]
- 13.De Giorgi, F., Ahmed, Z., Bastianutto, C., Brini, M., Jouaville, L. S., Marsault, R., Murgia, M., Pinton, P., Pozzan, T. & Rizzuto, R. (1999) Methods Cell Biol. 58, 75-85. [DOI] [PubMed] [Google Scholar]
- 14.Burger, C., Gorbatyuk, O. S., Velardo, M. J., Peden, C. S., Williams, P., Zolotukhin, S., Reier, P. J., Mandel, R. J. & Muzyczka, N. (2004) Mol. Ther. 10, 302-317. [DOI] [PubMed] [Google Scholar]
- 15.Rabinowitz, J. E., Rolling, F., Li, C., Conrath, H., Xiao, W., Xiao, X. & Samulski, R. J. (2002) J. Virol. 76, 791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos, M. A., Moura, G., Massey, S. E. & Tuite, M. F. (2004) Trends Genet. 20, 95-102. [DOI] [PubMed] [Google Scholar]
- 17.Davidson, B. L., Stein, C. S., Heth, J. A., Martins, I., Kotin, R. M., Derksen, T. A., Zabner, J., Ghodsi, A. & Chiorini, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passini, M. A., Watson, D. J., Vite, C. H., Landsburg, D. J., Feigenbaum, A. L. & Wolfe, J. H. (2003) J. Virol. 77, 7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor, R. W., Wardell, T. M., Lightowlers, R. N. & Turnbull, D. M. (2000) Neurol. Sci. 21, S909-S912. [DOI] [PubMed] [Google Scholar]
- 20.Taylor, R. W., Chinnery, P. F., Turnbull, D. M. & Lightowlers, R. N. (1997) Nat. Genet. 15, 212-215. [DOI] [PubMed] [Google Scholar]
- 21.Muratovska, A., Lightowlers, R. N., Taylor, R. W., Turnbull, D. M., Smith, R. A., Wilce, J. A., Martin, S. W. & Murphy, M. P. (2001) Nucleic Acids Res. 29, 1852-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoubridge, E. A., Johns, T. & Karpati, G. (1997) Hum. Mol. Genet. 6, 2239-2242. [DOI] [PubMed] [Google Scholar]
- 23.Clark, K. M., Bindoff, L. A., Lightowlers, R. N., Andrews, R. M., Griffiths, P. G., Johnson, M. A., Brierley, E. J. & Turnbull, D. M. (1997) Nat. Genet. 16, 222-224. [DOI] [PubMed] [Google Scholar]
- 24.Taivassalo, T., Fu, K., Johns, T., Arnold, D., Karpati, G. & Shoubridge, E. A. (1999) Hum. Mol. Genet. 8, 1047-1052. [DOI] [PubMed] [Google Scholar]
- 25.Taivassalo, T. & Haller, R. G. (2004) Biochim. Biophys. Acta 1659, 221-231. [DOI] [PubMed] [Google Scholar]
- 26.Santra, S., Gilkerson, R. W., Davidson, M. & Schon, E. A. (2004) Ann. Neurol. 56, 662-669. [DOI] [PubMed] [Google Scholar]
- 27.Jazayeri, M., Andreyev, A., Will, Y., Ward, M., Anderson, C. M. & Clevenger, W. (2003) J. Biol. Chem. 278, 9823-9830. [DOI] [PubMed] [Google Scholar]
- 28.Sorensen, L., Ekstrand, M., Silva, J. P., Lindqvist, E., Xu, B., Rustin, P., Olson, L. & Larsson, N. G. (2001) J. Neurosci. 21, 8082-8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amalfitano, A. (2004) Methods 33, 173-178. [DOI] [PubMed] [Google Scholar]
- 30.Cao, H., Koehler, D. R. & Hu, J. (2004) Viral Immunol. 17, 327-333. [DOI] [PubMed] [Google Scholar]
- 31.Monahan, P. E. & Samulski, R. J. (2000) Mol. Med. Today 6, 433-440. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Y., Tsai, S. Y. & O'Malley, B. W. (2000) Adv. Pharmacol. 47, 343-355. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava, S. & Moraes, C. T. (2005) Hum. Mol. Genet. 14, 893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.