Abstract
Genome-wide comparisons between enteric bacteria yield large sets of conserved putative regulatory sites on a gene-by-gene basis that need to be clustered into regulons. Using the assumption that regulatory sites can be represented as samples from weight matrices (WMs), we derive a unique probability distribution for assignments of sites into clusters. Our algorithm, “PROCSE” (probabilistic clustering of sequences), uses Monte Carlo sampling of this distribution to partition and align thousands of short DNA sequences into clusters. The algorithm internally determines the number of clusters from the data and assigns significance to the resulting clusters. We place theoretical limits on the ability of any algorithm to correctly cluster sequences drawn from WMs when these WMs are unknown. Our analysis suggests that the set of all putative sites for a single genome (e.g., Escherichia coli) is largely inadequate for clustering. When sites from different genomes are combined and all the homologous sites from the various species are used as a block, clustering becomes feasible. We predict 50–100 new regulons as well as many new members of existing regulons, potentially doubling the number of known regulatory sites in E. coli.
New microbial genomes are sequenced almost daily, and the first step in their annotation is the elucidation of their protein-coding regions. The noncoding regions of the genome can provide clues about gene regulation, because they contain various regulatory elements. These elements generally are much smaller and more variable than typical coding regions and thus harder to identify. Computational methods are needed, because even for Escherichia coli there are only 60–80 genes for which binding sites and regulated genes are known (1, 2), whereas protein sequence homology suggests there are ≈300 DNA-binding proteins (3). Binding sites have been identified experimentally in only 300 of the 2,400 regulatory regions of E. coli (2). For important pathogens such as Vibrio cholerae, Yersinia pestis, or Mycobacterium tuberculosis very little is known about gene regulation from direct experimentation.
Computational strategies for the discovery of regulatory sites began with algorithms (4–6) that identified sets of similar sequences in the regulatory regions of functionally related groups of genes. More recently, algorithms were proposed to identify repetitive patterns within an entire genome (7). Here we develop methods for partitioning a large set of putative regulatory sites into clusters based on sequence similarity, with the goal of identifying regulons. That is, we aim to partition the set of sites such that each cluster corresponds to those targeted by the same transcription factor (TF).
Many authors have noted the potential of interspecies comparisons to elucidate regulatory motifs (e.g., ref. 8). Generally, a group of functionally related genes in bacteria is pooled to extract common sites within the regulatory regions of these genes (e.g., refs. 9 and 10). More recent studies (11, 12) have shown that when upstream regions of orthologous genes from several suitably related species are compared at once, there is sufficient signal for regulatory sites to be inferred on a gene-by-gene basis, yielding thousands of potentially new sites. These sites form the data sets on which our algorithm operates.
Previous algorithms that fit weight matrices (WMs) cannot process genome scale data representing sites from hundreds of TFs simultaneously. Other schemes (7), not based on WM representations of regulatory sites, are not well suited for processing sites that were inferred from interspecies comparison. Our algorithm partitions the entire set of sites at once, infers the number of clusters internally, and assigns probabilities to all partitions of sequences into clusters. Within this framework, we also derive theoretical limits on the clusterability of sets of regulatory sites.
A set of sites, sampled from a set of unknown WMs, is said to be clusterable if it is possible to infer which sites were sampled from the same WM. If the WMs from which the sites were sampled are known, we have the much simpler classification problem: determining which sites were sampled from which WM. It is important to realize that the cell is performing a classification task because it knows the WMs of the TFs, i.e. the chemistry of the DNA–protein interaction automatically assigns a binding energy to each site just as a WM assigns a score to each site. However, since we cannot infer binding specificities from a TF's protein sequence, we face the much harder clustering task. Our theoretical arguments and the available data for E. coli in fact suggest that the set of all regulatory sites in the E. coli genome is unclusterable by itself. However, we also show how this problem can be circumvented by taking into account information from interspecies comparison.
Model
Protein binding sites in bacterial genomes are commonly described by a WM, w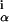 , which gives the probabilities of finding base α at position i of the binding site (13). The probabilities in different columns i are assumed independent, which accords well with existing compilations (1). Motif-finding algorithms (4–6) score the quality of an alignment of putative binding sites by the information score I of its (estimated) WM,
, which gives the probabilities of finding base α at position i of the binding site (13). The probabilities in different columns i are assumed independent, which accords well with existing compilations (1). Motif-finding algorithms (4–6) score the quality of an alignment of putative binding sites by the information score I of its (estimated) WM,
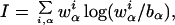 |
1 |
where bα is the background frequency of base α, and the w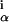 are the WM probabilities estimated from the sequences in the alignment. The rationale for this scoring function is that the probability of an n sequence alignment with frequencies w
are the WM probabilities estimated from the sequences in the alignment. The rationale for this scoring function is that the probability of an n sequence alignment with frequencies w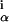 arising by chance from n independent samples of the background distribution of bases bα is given by P ≈ e−nI.
arising by chance from n independent samples of the background distribution of bases bα is given by P ≈ e−nI.
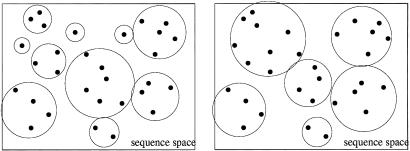
Instead of distinguishing sequence motifs for a single TF against a background distribution, our task is to cluster a set of binding sites of an unknown number of different TFs, i.e. a set of sequences sampled from an unknown number of unspecified WMs. To this end, we consider all ways of partitioning our data set into clusters and assign a probability to each partition. Fig. 1 depicts, schematically, two ways of partitioning a set of sequences into clusters. We will assign probabilities to all such partitions. The probability of a partition is the product of the probabilities, for each cluster, that all sequences within the cluster arose from a common WM.
Figure 1.
Two ways of partitioning the same set of sequences into clusters. The rectangle schematically represents the space of all possible DNA sequences of some particular length l. The dots denote the sequences in the data set, and the circles indicate which sequences are partitioned together into clusters.
To calculate these probabilities, consider first the conditional probability P(S|w) that a set of n length l sequences S was drawn from a given WM w,
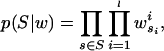 |
2 |
where si is the letter at position i in sequence s. The probability P(S) that all sequences in S came from some w can be obtained by integrating over all allowed w, namely over the simplex ∑αw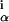 = 1 for each position i. Lacking any knowledge regarding w, we use a uniform prior over the simplex. We obtain
= 1 for each position i. Lacking any knowledge regarding w, we use a uniform prior over the simplex. We obtain
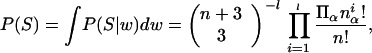 |
3 |
where n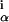 is the number of occurrences of base α in column i. The last factor in Eq. 3 is just the inverse of the multinomial factor that counts the number of ways of constructing a specific vector (na, nc, ng, nt) from n bases, which bears an obvious relation to Eq. 1. High probabilities thus are given to vectors, which can be realized in the least number of ways. The factor (
is the number of occurrences of base α in column i. The last factor in Eq. 3 is just the inverse of the multinomial factor that counts the number of ways of constructing a specific vector (na, nc, ng, nt) from n bases, which bears an obvious relation to Eq. 1. High probabilities thus are given to vectors, which can be realized in the least number of ways. The factor (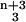 ) counts the number of distinct vectors (na, nc, ng, nt) that can be obtained from n samples.
) counts the number of distinct vectors (na, nc, ng, nt) that can be obtained from n samples.
We now can define for any partition C of a data set of sequences D into clusters Sc the likelihood P(D|C) that all sequences in each Sc were drawn from a single WM: P(D|C) = ∏cP(Sc), with P(Sc) given by Eq. 3. Then the posterior probability P(C|D) for partition C given the data D is
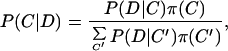 |
4 |
where π(C) is the prior distribution over partitions, which we will assume to be uniform.
Consider the simplest example of a data set of only two sequences with matching bases in b of their l positions. We have P = 2b(1/20)l for the probability that the sequences came from the same WM, whereas P = (1/16)l for the probability that they came from different WMs. P(C|D) thus will prefer to either cluster or separate the two sequences depending on b. In general, the probability distribution P(C|D) will prefer partitions in which similar sequences are coclustered. The state space of all partitions (the number of which grows nearly as rapidly as n!; ref. 14) acts as an “entropy,” which opposes (stable) clustering of similar sequences.
The probability distribution Eq. 4 allows us to calculate any statistic of interest by summing over the appropriate partitions C. For instance, to calculate the probability that the data set separates into n clusters, one sums P(C|D) over all partitions that contain n clusters. Analogously, we can calculate the probability that any particular subset of sequences forms a cluster by summing P(C|D) over all partitions in which this occurs. Note that our clustering framework thus allows for direct calculations of these quantities. In the implementation section below we describe how we sample P(C|D) and identify significant clusters by finding subsets of sequences that cluster consistently.
Generalizations to data arising from WMs of different lengths and sequences that are not aligned consistently are straightforward and considered below. It is also trivial to incorporate prior information on the number of clusters (e.g., that it should equal the number of TFs).
Classifiability vs. Clusterability
Correct regulation of gene expression requires that TFs should bind preferentially to their own sites. Associating TFs with WMs, P(s|w) commonly is taken to be the probability that w binds to s. Correct regulation thus implies that for a sample s from w, we have that P(s|w) > P(s|w′) for all other TFs w′ ≠ w, which we call a classification task. Formally, we are given a set of WMs and a set of sequences sampled from them and assign each sequence s to the WM from the set that maximizes P(s|w). We define the data to be classifiable when, in at least half of the cases, the WM w that maximizes P(s|w) is the WM from which s was sampled. As mentioned in the Introduction, classification is much simpler than clustering a set of sites in the absence of knowledge of the set of WMs from which they were sampled.
To quantify clusterability, assume we are clustering nG sequences that were obtained by sampling n times from each of G different WMs. For each of these WMs we can calculate the probability that m of its n samples cocluster by summing the probabilities P(C|D) over all partitions C in which m, and no more than m, samples of w occur together in any of the clusters. We will define the set to be “clusterable” if for more than half of the G WMs the average of m, 〈m〉 > n/2.
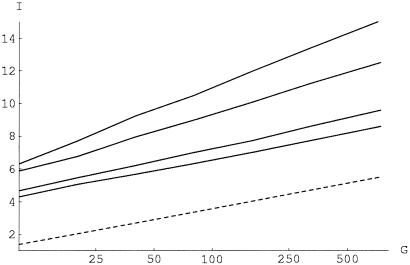
We have performed analytical and numerical calculations that identify under what conditions a data set is classifiable and clusterable. This theory is beyond the scope of this paper and will be reported elsewhere. The results are summarized in Fig. 2. Given the information score I (Eq. 1) of a WM, the fraction of the space of 4l sequences filled by the binding sites for this WM is e−I. One thus can regard I as a measure of the specificity of a WM. Fig. 2 shows the minimal WM specificity necessary to cluster (solid lines) or classify (dashed line) as a function of the number of WMs G and the number of samples n per WM. Fig. 2 shows that exp(−I) ∝ 1/G for classification and exp(−I) ∝ 1/G2 for clustering a set of n = 3 binding sites, with fractional exponents in between these extremes. Thus, all G WMs together consume a fixed fraction of sequence space at the classification threshold (independent of G), while it decreases as a function of G at the clusterability threshold. Moreover, there is a significant gap between the requirements for classification vs. clustering even for large numbers of samples. Thus, clustering is impossible for data sets close to the classification threshold. The results presented below suggest that the collection of E. coli binding sites may well be in this unclusterable regime, where few regulons can be inferred correctly.
Figure 2.
The critical information score I for clusterability (solid lines) or classifiability (dashed line) as a function of the number of clusters G (shown on a log scale). The solid lines correspond, from top to bottom, to sets of n = 3, 5, 10, and 15 samples per cluster. The WM length is l = 27.
However, comparative genomic information can salvage this situation. The putative binding sites of our data sets were extracted by finding conserved sequences upstream of orthologous genes of different bacteria (see below). Such conserved sequence sets are likely to contain binding sites for the same TF and should be clustered together. Therefore, we can reduce the size of the state space significantly by preclustering these conserved sites into so-called mini-WMs, and instead of clustering single sequences we will be clustering these mini-WMs with the same probabilities shown in Eq. 3, which improves clusterability dramatically.
Implementation
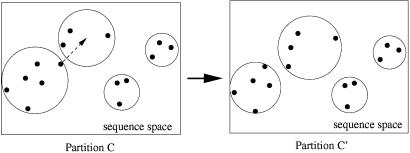
We have implemented a Monte Carlo random walk to sample the distribution P(C|D). At every “time step” we choose a mini-WM at random and consider reassigning it to a randomly chosen cluster (or empty box). These moves are accepted according to the Metropolis–Hastings scheme (15): moves that increase the probability P(C|D) are always accepted, and moves that lower P(C|D) are accepted with probability P(C′|D)/P(C|D). Fig. 3 shows an example of a move from a partition C to a partition C′. This sampling scheme thus generates “dynamic” clusters, the membership of which fluctuates over time. Clusters may evaporate altogether, and new clusters may form when a pair of mini-WMs is moved together. We wish to identify “significant” clusters by finding sets of mini-WMs that are grouped together persistently during the Monte Carlo sampling. Ideally, we would find a set of clusters, each with stable “core” members that are present at all times, while the remaining mini-WMs move about between different clusters. Reality unfortunately is more complicated. One finds clusters that are drifting constantly such that their membership is uncorrelated on long time scales. Other clusters, with stable membership, may evaporate and reform many times. Although we can sample P(C|D) easily to obtain significance measures for any given “candidate cluster,” the rich dynamics of drifting, fusing, and evaporating clusters makes it nontrivial to identify good candidate clusters.
Figure 3.
Monte Carlo sampling of partitions: example of a move from partition C to partition C′. The dots are sequences, and the circles delineate the clusters.
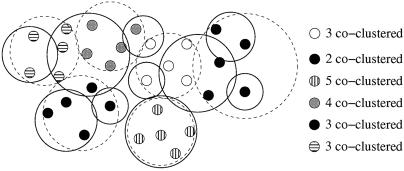
We have experimented with a number of schemes for identifying candidate clusters (see supporting information, which is published on the PNAS website, www.pnas.org). One approach is to search for the maximum likelihood (ML) partition that maximizes Eq. 4, which can be done by simulated annealing: we raise P(D|C) to the power β, increasing β over time (in practice β = 3 is large enough). The ML partition gives us a set of candidate clusters. The significance of the ML clusters then are tested by sampling P(C|D). Fig. 4 illustrates this procedure. For each partition encountered during the sampling, we define the number of coclustering members of an ML cluster as the maximum number of mini-WMs from the ML cluster that co-occur in a single cluster (see Fig. 4). In this way we measure, for each ML cluster, the probabilities p(k) that k of its members cocluster. The mean size of the cluster thus is ∑k k p(k). Finally, we calculate the minimal length interval [kmin, kmax] for which ∑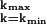 p(k) > 0.95. All clusters for which kmin ≥ 2 are deemed significant.
p(k) > 0.95. All clusters for which kmin ≥ 2 are deemed significant.
Figure 4.
The ML partition obtained by annealing is indicated by the thin, dashed circles and the fill patterns of the dots. The thick lines show an alternative partition that may arise during sampling. The number of coclustering members in this partition are shown on the right for each of the ML clusters.
This method is computationally prohibitive for large data sets (because we cannot run long enough to converge all cluster statistics). For larger data sets we measure, using several Monte Carlo random walks, the probability that each pair of mini-WMs coclusters (note that these pair statistics cannot be calculated in terms of the sequences in the pair of mini-WMs themselves; they depend on the full data set). We then construct a graph in which nodes correspond to mini-WMs, and edges between mini-WMs i and j exist if and only if their coclustering probability pij > ½. Candidate clusters now are given by the connected components of this graph. The pairwise statistics are then processed further to obtain probabilistic cluster membership, which yields for each mini-WM i the probabilities p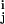 that mini-WM i belongs to cluster j (see supporting information). We also calculate, for each cluster, the probability distribution p(k) of k of its members coclustering. Cluster significance is judged from p(k) as described above. Fortunately, there is substantial agreement on the significant clusters among these ways of extracting significant clusters from P(C|D).
that mini-WM i belongs to cluster j (see supporting information). We also calculate, for each cluster, the probability distribution p(k) of k of its members coclustering. Cluster significance is judged from p(k) as described above. Fortunately, there is substantial agreement on the significant clusters among these ways of extracting significant clusters from P(C|D).
After we have inferred the clusters and their members, we can estimate a WM for each cluster. We then classify all mini-WMs in the full data set in terms of these cluster WMs. Finally, we search for additional matching motifs to the cluster WMs in all the regulatory regions of the E. coli genome. Details for all these procedures are described in the supporting information.
Data Sets
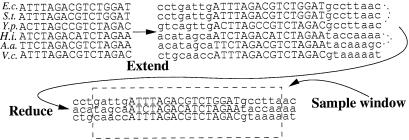
Our primary data sets (11, 12) consist of alignments of relatively short sequences, i.e. typically 15–25 bases, that where extracted from upstream regions of orthologous genes in different prokaryotic genomes. Data set (11) uses the genomes of E. coli, Actinobacillus actinomycetemcomitans, Haemophilus influenzae, Pseudomonas aeruginosa, Shewanella putrefaciens, Salmonella typhimurium, Thiobacillus ferrooxidans, V. cholerae, and Y. pestis. Data set (12) uses E. coli, Klebsiella pneumoniae, S. typhimurium, V. cholerae, and Y. pestis. An example alignment is shown in Fig. 5. The available evidence suggests that these alignments either include or substantially overlap a set of binding sites for a TF (or another kind of regulatory site). Our algorithm will have to decide which stretch of bases in each alignment corresponds to the regulatory site. Known binding sites (1) are between 11 and 50 bases long with a mean of 24.5 and a standard deviation of just under 10. We will assume that all binding sites are exactly 27 bases long, compromising between diluting the signal in the small binding sites and missing some of the signal in long binding sites. We symmetrically expand the alignments in our data set to length 32, padding bases from the genomes (see Fig. 5). We would like to treat these sequences as independent samples of a single WM, but for closely related species this assumption probably is untenable. For alignments from data set (11) we therefore replace sites from the triplet E. coli, Y. pestis, and S. typhimurium, and from the duplet H. influenzae and A. actinomycetemcomitans by their respective consensi. For the data set (12) we only replace the triplet E. coli, K. pneumoniae, and S. typhimurium by their consensus. The mini-WMs thus obtained are the objects that our algorithm clusters. Finally, every time the Monte Carlo algorithm reassigns a mini-WM to a cluster, it samples over the six different ways of picking a length 27 window out of the length 32 alignment and over both strands (see supporting information).
Figure 5.
Operations on the data sets. Starting from an alignment of variable length, we extend the alignment to length 32 by padding bases from the genome and then replace sequences of closely related species by their consensus. This yields so-called mini-WMs, which are the objects that our algorithm clusters. When moved between clusters, a window of length 27 is sampled from the alignment.
Before clustering these primary data sets we tested the algorithm on a set of experimentally determined TF binding sites in E. coli that was collected in ref. 1. We again extended (or cropped) these sequences symmetrically to length 32. After excluding σ factor sites and sites that overlap one another by 27 or more bases, there are 397 binding sites representing 53 TFs remaining in this test set. See the supporting information for comments on the preprocessing of this and our other data sets.
For data set (11) we removed all alignments that overlap known binding sites or repetitive elements and then took the top 2,000 nonoverlapping alignments ordered by their score. For data set (12) we also took the top 2,000 nonoverlapping sites based on significance, but we left sites overlapping known binding sites in this set. Finally, in order to separate new regulons from new sites for TFs with sites in the collection (1), we aligned all known E. coli sites for each TF into its own mini-WM and added these 56 mini-WMs to sets (11) and (12) [3 out of the 53 TFs (argR, metJ, and phoB) have two different types of sites, which we align separately into mini-WMs]. Both these sets thus contain 2,056 mini-WMs.
We created an additional test set consisting of the 397 known binding sites from ref. 1 and the E. coli sequences of the top 2,000 unannotated mini-WMs from (11). As described below, this test verified our prediction that by embedding the 397 known sites in a larger set of sites, many clusters will fail to be inferred correctly.
Results
We used the test set of 397 known binding sites in several ways. First, we sampled P(C|D) and measured, for each factor, how well its sites cluster. That is, we measured the coclustering distribution p(k) for each TF. Using the significance threshold described above, we found significant clusters for 24 of the 53 TFs. Twenty two TFs have three or fewer sites in the test set, and with the exception of trpR their sites did not cluster significantly. As a better test of our algorithm, we compared the clusters inferred from annealing this data set with the site annotation. We performed two annealing runs to identify an ML partition and then performed sampling runs to test the significance of these ML clusters. We found that, in general, there is good agreement between the annotation and the clusters inferred by annealing. For 17 of the 24 TFs that form significant clusters there was an analogous significant cluster obtained by the annealing. The full results are in supporting information. We have found also that the likelihood P(C|D) for the partition obtained in all annealing runs is significantly higher than that obtained when the sites are partitioned according to their annotation. Thus we feel that the clustering for this data set cannot be improved within our scoring scheme. In short, our algorithm recovers almost half of all regulons for which binding sites are known and the large majority of regulons for which there are more than three sites known.
We sampled P(C|D) for the 2,397-site test set and found that, as predicted, many clusters are lost (only 9 of 24 significant clusters remain). Several of those that remain where reinforced by the presence of additional unannotated sites in the supplemental set of 2,000. (Using more samples improves clusterability as we have seen in Classifiability vs. Clusterability.) For this larger data set, the total number of clusters fluctuates around 350 during the run, but only ≈5% of them are significant, which suggests that most E. coli binding sites are in the unclusterable regime, and that comparative genomic information is essential to effectively cluster. We also performed simulations with “surrogate” data sets that support this claim further. For each cluster of known binding sites, we calculated the information score I of its WM and created four random WMs with equal I. By drawing samples from each of these, we “scaled up” the set of known binding sites and clusters by a factor of 5 to correspond to the estimated number of TFs in E. coli. In sampling P(C|D) for this set, we found that less than 10% of the clusters are inferred correctly.
For the larger data sets from (11) and (12), which are our main interest, repeated annealing and sampling runs indicated that both the annealed state and the significance statistics are not converged fully within our running times (1010 steps, taking a week on a workstation per run). We therefore extracted significant clusters via pair statistics as described above, which did converge and allowed us to assign error bars to all pair statistics. For the data set (11) there were 365 ± 5 clusters on average, and the connectivity graph gave 274 components containing 1,139 out of 2,056 mini-WMs. Thus, about half of the data set clusters stably, whereas the other half moves in and out of the ≈100 unstable clusters. There were 115 significant clusters comprising 645 mini-WMs. Of the 115 significant clusters, 21 contained as one of its member mini-WMs the alignment of a set of known binding sites for a TF from ref. 1. These clusters thus contain new sites for known regulons. The other 94 clusters correspond to new putative regulons, some examples of which are described below.
It is interesting to calculate the cluster information scores, I, to compute the fractions, e−I, of sequence space occupied by our clusters. Summing these volumes, we find that ≈1% of the space is filled by the top 45 clusters, the top 80 clusters fill 10% of the space, and all our 115 significant clusters fill 39% of the space, which again supports the idea that the set of all WMs is close to the classification boundary; their binding sites fill almost the entire sequence space.
For the data set (12) there are 275 ± 4 clusters on average during the sampling. The connectivity graph has 176 clusters containing 726 mini-WMs. There were 65 significant clusters (containing 398 mini-WMs), of which 25 correspond to known regulons. With respect to the sequence space volume filled by the WMs of these clusters, 1% of the space is filled by the first 30 clusters, 50 clusters fill 10% of the space, and the full set of 65 WMs fills ≈50% of the sequence space.
Examples
Table 1 contains a synopsis of some of predicted new regulons we have examined in detail from the data set 11. Primary cluster membership is noted along with additional sites that can be found by scanning the cluster WM over the full data set and all regulatory regions of E. coli. The complete lists are on our web site (www.physics.rockefeller.edu/∼erik/website.html).
Table 1.
Sample clusters from data set 11
| Cluster name | Rank | Defining operons |
|---|---|---|
| Thiamin biosynthesis | 0 | thiCEFGH tpbA/yabKJ thiMD thiL |
| gntR/idnR regulon | 1 | idnK,idnDOTR gntKU gntT b2740 edd/eda |
| Elongation factor | 2 | tufB |
| Ribonucleotide reductase | 3 | nrdAB nrdDG nrdHIEF |
| ? | 4 | coaA tgt/yajCD/secDF yegQ b3975 tpr yeeO |
| Stem–loop/attenuator repair ? | 5 | yhbc/nusA/infB mutM arsRBC yhdNM nadA/pnuC lig ptsHI/crr rbfA/truB/rpsO |
| ntrC regulon | 11 | glnK/amtB cmk/rpsA glnALG glnHPQ narGHJI hisJQMP |
| Ribosomal protein attenuation | 15 | thdF fabF recQ tsf pnp pyrE himD |
| Anaerobic oxidation | 16 | cydAB appCB yhhK,livKHMGF torCAD,torR ansB/yggM ybbQ yiiE |
| Fatty acid biosynthesis | 17 | fabA b2899(yqfA) fabB fabHDG |
| Cell envelope replication ? | 25 | pcnB/folK pssA dksA/yadB yaeS mreCD/yhdE/cafA sanA cmk/rpsA |
| Alkaline phosphatase peptidoglycan | 26 | yaiB/phoA/psiF, ddlA dnaB/alr creABCD iap avtA |
| Transport | 37 | abc,yaeD cadBA araFGH,yecI celABCDF citAB,citCDEF agaBCD tauABCD |
| fruR regulon | 71 | fruR fruBKA epd yggR |
| Fe-S radicals | 85 | metK,yqgD ftn pykA yheA/bfr |
The cluster rank is by WM information score. The defining operons come in three categories: those with member sites in the data set on which the algorithm was run (bold), those with sites in data set (11) that match the WM (normal font), and those that were found by scanning the regulatory regions of E. coli (italics). Multiple genes within an operon are separated by a /or by multiple capitals at the end of the gene name. Operons separated by a comma indicate that the site fell between divergently transcribed genes.
Our thiamin cluster is an example of a predicted regulon that recently has been confirmed experimentally. A comprehensive review of thiamin biosynthesis in prokaryotes (16) places the genes from the three operons of our thiamin cluster (thiBPQ is also called tbpA/yabJK) into a single pathway, together with the four single genes: thiL, thiK, dxs (yajP), and thiI (yajK). A recent paper (17) shows that the three thiamin operons share a common RNA stem–loop motif that is responsible for posttranscriptional regulation. It is precisely a portion of this motif that we cluster. A fragment of this structure also occurs just upstream of translation start in thiL. For the remaining genes, thiK, dxs, and thiI, there are no putative sites in data set (11).
Besides the main gluconate metabolism pathway, a second pathway that utilizes input from the catabolism of l-idonic acid has been reported recently (18) and corresponds to our second cluster. The first two operons (idnK and idnDOTR) code for the enzymes that import l-idonate and convert it to 6-P-gluconate. The operon gntKU contains a gluconokinase, which catalyzes the same reaction as the idnK protein, and a low-affinity gluconate permease. b2740 is a gene of unknown function that belongs to the family of gluconate transporters. Finally, gntT is a high-affinity gluconate permease. Additional sites were found upstream of the edd/eda operon that encode the key enzymes of the Entner–Doudoroff pathway (19). Ref. 18 suggests that idnR both up-regulates the l-idonate catabolism genes and represses gntKU and gntT when growing on l-idonate, suggesting that our sites may bind indR. However, there are two sites upstream of gntT that are annotated as gntR sites (20), which are also part of our cluster.
The pathway for ribonucleotide reduction to deoxyribonucleotides is pictured on page 591 of ref. 21 and includes the first two operons of our like-named cluster. We did not find sites in the regulatory regions of the other two genes in this pathway (ndk, dcd). Scanning of the genome with the WM inferred from the nrdAB and nrdDG sites reveals an additional three (weaker) sites upstream of the nrdHIEF operon. The nrdEF genes are annotated as a cryptic ribonucleotide reductase. The regulation of our two primary operons (nrdAB and nrdDG) is known to be complex and includes an fnr site upstream of nrdD (which we correctly clustered with other fnr sites) and additional fis, dnaA, and unattributed sites upstream of nrdA (22). The nrdA site in our cluster is located downstream of transcription start. Because nrdA is down-regulated during anaerobiosis and nrdD is essential for anaerobic growth, we would guess that our sites are involved in the switch.
The estimated WM of cluster 5 has a prominent inverted repeat sequence as its consensus (AAAAacCC***TT***GGGGgTTTTTT) and has over 20 matches in the genome. These sites may correspond to an RNA secondary structure, possibly involved in attenuation. There is no clear predominant functional theme to the genes in our cluster 5. Noteworthy are sites upstream of the arsenic resistance operon (arsRBC), the crr regulator of a multidrug efflux pump, and the ydnM (zntR) regulator for Pb(II), Cd(II), and Zn(II) efflux. Also, two genes involved in DNA repair occur (MutM and lig).
The sites in cluster 15 occur upstream of genes whose proteins are involved in RNA modification (thdF and pnp), recombination (recQ and himD), and translation (tsf). More strikingly, 6 of 7 of these sites occur downstream of genes coding for ribosomal protein subunits and one RNase. For five of these genes, there is evidence (see the ecocyc database, ecocyc.org:1555/server.html/) that our site falls within a transcription unit, i.e. that the genes upstream and downstream of our site are cotranscribed. It seems likely that these sites are involved in either attenuation or translational regulation.
E. coli has a rich repertory of respiratory chains that are built from a variety of electron donors and acceptors (see ref. 21, page 218). One of our clusters (16) involves two homologous cytochrome operons cydAB and appCB (cyxAB), which transfer electrons to oxygen and are active mainly during anaerobic conditions. The torACD operon (divergently transcribed with its regulator torR) transfers electrons to trimethylamine N-oxide. There is a third cytochrome complex, cyoABCD, with different specificity that is not linked to this cluster. Other operons in this cluster such as livKHMGF, which is involved in amino acid import, and ansB, which catalyzes asparagine to aspartate conversion, seem unrelated but are divergently transcribed with genes of unknown function. However, refs. 23 and 21 (page 366) suggest that ansB also can provide fumarate as a terminal electron acceptor. AnsB is up-regulated strongly during anaerobic conditions and has known crp and fnr sites. The ansB site in our cluster is different from these sites.
Cluster number 17 corresponds to the fatty acid biosynthesis regulon with TF yijC (fabR) that was identified in ref. 11. Our cluster contains the sites they found upstream of fabA and b2899. Additionally, we found WM matches upstream of the related genes fabB and fabHDG. Other operons with lower quality sites in the cluster include the mglBAC operon (methyl-galactoside transport), clpX (component of clpP serine protease), and the putative peptidase b2324.
We are unable to guess the functional role of the binding sites clustered in cluster number 25. Some of the genes have functionalities related to the cell envelope and membrane (pssA, yaeS, mreCD, and sanA), and some seem involved in replication (dskA, cafE). However, these functions seem rather diverse.
For cluster 26, we find sites upstream of genes involved in peptidoglycan biosynthesis (alr, ddlA, avtA, and mrcB) and genes that are known to be regulated in response to phosphate starvation (creABC, iap, and phoA/psiF). In particular, alkaline phosphatase (phoA) is upregulated more than 1,000-fold and accounts for as much as 6% of the protein content of the cell during phosphate starvation (see ref. 21, page 1,361). Because alkaline phosphatase is active in the periplasm, it seems conceivable that peptidoglycan synthesis is down-regulated when phoA is expressed at such high levels.
Additional clusters with obvious common functionality include cluster 85 for Fe-S radical synthesis (24) and the large cluster 37, which contains several phosphotransferase system and other transport systems. Cluster 71 contains sites that overlap binding sites for the fructose repressor fruR. These sites were clustered separately from the known fruR sites because of a systematic shift, larger than the range our algorithm scans, between how they were given in data set (11) and the annotated fruR sites. Similarly, cluster 11 contains sites that overlap binding sites for the nitrogen fixation regulator ntrC (glnG).
Apart from the 94 putative regulons, our web site has an additional 270 sites that cluster with WMs of known TFs. Summing their membership probabilities, this corresponds to an expected 135 binding sites. The web site also provides information for each E. coli gene separately: inferred regulatory sites upstream of the gene and the cluster memberships of these sites.
The clusters inferred from data set (12) are also on our web site. We have not evaluated their functional significance yet, but some of them correspond to clusters that we also found in the data of data set (11), e.g., the thiamin cluster reappears.
Discussion
We introduced a new inference procedure for probabilistically partitioning a set of DNA sequences into clusters. Currently, the algorithm assumes all WMs to be of a fixed length, but prior information about site lengths, their dimeric nature, and the length of spacers between dimeric sites could be included easily. One also could extend the hypothesis space on which the algorithm operates; one may assume that only some fraction, rather than all, of the sequences are WM samples, whereas the rest should described by a background model, which would, for instance, be appropriate for analyzing entire upstream regions. In all these generalizations, the algorithm would still assign probabilities to sets of sequences belonging to a single TF. This essentially Bayesian approach should be contrasted with approaches (e.g., refs. 4 and 7), in which “promising” motifs are selected based on how unlikely it is for them to occur under some null hypothesis of randomness.
By applying our algorithm to data sets (11, 12) of putative regulatory sites extracted from enteric bacteria, we predicted ≈100 new regulons in E. coli, containing ≈500 binding sites, and ≈150 binding sites for known TFs. The functionality of many of the predicted regulons is supported by the fact that their sites are found upstream of genes that are clearly related functionally. Even if there is no common theme in the annotation of the genes controlled by the sites, our significance measures suggest that a large fraction of the clusters is functional; the data sets contain only conserved sites upstream of orthologous genes in different organisms, and a highly significant association of groups of such sites was found. We note that our set is a considerable augmentation of the ≈400 non-σ sites that are known experimentally. Analysis of some of our clusters shows that included in our predicted regulons in addition to TF binding sites are RNA stems controlling translation and even termination motifs.
The clusters and sites resulting from our genome-wide analysis of regulatory motifs allows for a more quantitative evaluation of the global structure of regulatory networks in bacteria. The regulatory network is often imagined as a rather loosely coupled collection of “modules” where each regulon controls a set of genes with closely linked functionality (although of course many exceptions exist such as the structural TFs fis, ihf, etc.). Our predicted regulons are often much less orderly. In several cases, some but not all genes of a well studied pathway entered the regulon. In other cases, a regulon contains sets of sites for genes of two or three clearly distinct functionalities for which no regulatory connection is known. Our overall impression is of a more haphazard regulatory network than traditionally imagined.
Finally, we have emphasized the distinction between classifying and clustering a set of binding sites. We have argued that the TFs of a cell are essentially solving a classification task, and that inferring regulons from the set of binding sites of a single genome may well be impossible in principle. There are also evolutionary arguments that support this claim. Like any piece of DNA, binding sites are subject to random mutations. The more specific binding sites are, the more likely they are to be disrupted by mutations. Evolution thus will naturally drive TFs and their binding sites to become as unspecific as possible (25, 26) within the constraints set by their function. That is, evolution will drive the set of binding sites toward the “classification threshold” where they become unclusterable. The situation is reminiscent of the situation in communication theory, where optimally coded messages look entirely random to receivers that are not in possession of the code. Information from comparative genomics thus is essential for the inference of regulons from genomic data, and as the number of sequenced genomes grows, so will our algorithm's ability to discover new regulons.
Supplementary Material
Acknowledgments
The support of National Science Foundation Grant DMR-0129848 is acknowledged.
Abbreviations
- TF
transcription factor
- WM
weight matrix
- ML
maximum likelihood
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Robison K, McGuire A M, Church G M. J Mol Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- 2.Salgado H, Santos-Zavaleta A, Gama-Castro S, Millan-Zarate D, Blattner F, Collado-Vides J. Nucleic Acids Res. 2000;28:65–7. doi: 10.1093/nar/28.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salgado H, Moreno-Hagelsieb G, Smith T, Collado-Vides J. Proc Natl Acad Sci USA. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stormo G D, Hartzell G W. Proc Natl Acad Sci USA. 1989;86:1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence C E, Altschul S F, Boguski M S, Liu J S, Neuwald A F, Wootton J C. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 6.Bailey T, Elkan C. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 7.Bussemaker H J, Li H, Siggia E D. Proc Natl Acad Sci USA. 2000;97:10096–10100. doi: 10.1073/pnas.180265397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardison R, Oeltjen J, Miller W. Genome Res. 1997;10:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- 9.Gelfand M, Koonin E, Mironov A. Nucleic Acids Res. 2000;28:695–705. doi: 10.1093/nar/28.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGuire A M, Hughes J D, Church G M. Genome Res. 2000;10:744–757. doi: 10.1101/gr.10.6.744. [DOI] [PubMed] [Google Scholar]
- 11.McCue L A, Thompson W, Carmack C S, Ryan M P, Liu J S, Derbyshire V, Lawrence C E. Nucleic Acids Res. 2001;29:774–782. doi: 10.1093/nar/29.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajewsky N, Socci N D, Zapotocky M, Siggia E D. Genome Res. 2002;12:298–308. doi: 10.1101/gr.207502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg O G, von Hippel P H. J Mol Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 14.de Bruijn N G. Asymptotic Methods in Analysis. New York: Dover; 1958. [Google Scholar]
- 15.Metropolis N, Rosenbluth A W, Rosenbluth M N, Teller A H, Teller E. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- 16.Begley T, Downs D, Ealick S, McLafferty F, van Loon A, Taylor S, Campobasso N, Chiu H J, Kinsland C, Reddick J J, Xi J. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 17.Miranda-Rios J, Navarro M, Soberón M. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bausch C, Peekhaus N, Utz C, Blais T, Murray E, Lowary T, Conway T. J Bacteriol. 1998;180:3704–3710. doi: 10.1128/jb.180.14.3704-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peekhaus N, Conway T. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peekhaus N, Conway T. J Bacteriol. 1998;180:1777–1785. doi: 10.1128/jb.180.7.1777-1785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidhardt F C, editor. Escherichia coli and Salmonella Typhimurium: Cellular and Molecular Biology. Washington DC: Am. Soc. Microbiol.; 1996. [Google Scholar]
- 22.Jacobson B A, Fuchs J A. Mol Microbiol. 1998;28:1315–1322. doi: 10.1046/j.1365-2958.1998.00897.x. [DOI] [PubMed] [Google Scholar]
- 23.Jennings M, Beacham I. J Bacteriol. 1990;172:1491–1498. doi: 10.1128/jb.172.3.1491-1498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheek J, Broderick J. J Biol Inorg Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 25.van Nimwegen E, Crutchfield J P, Huynen M. Proc Natl Acad Sci USA. 1999;96:9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengupta A M, Djordjevic M, Shraiman B I. Proc Natl Acad Sci USA. 2002;99:2072–2077. doi: 10.1073/pnas.022388499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.