Abstract
The eukaryotic intra-S-phase checkpoint, which slows DNA synthesis in response to DNA damage, is poorly understood. Is DNA damage recognized directly, or indirectly through its effects on replication forks? Is the slowing of S phase in part because of competition between DNA synthesis and recombination/repair processes? The results of our genetic analyses of the intra-S-phase checkpoint in the fission yeast, Schizosaccharomyces pombe, suggest that the slowing of S phase depends weakly on the helicases Rqh1 and Srs2 but not on other recombination/repair pathways. The slowing of S phase depends strongly on the six checkpoint-Rad proteins, on Cds1, and on Rad4/Cut5 (similar to budding yeast Dpb11, which interacts with DNA polymerase ɛ) but not on Rhp9 (similar to budding yeast Rad9, necessary for direct damage recognition). These results suggest that, in fission yeast, the signal activating the intra-S-phase checkpoint is generated only when replication forks encounter DNA damage.
Eukaryotic organisms reduce their rate of DNA synthesis when DNA is damaged during S phase. This is a consequence of the combined effects of checkpoint-independent, damage-dependent inhibition of replication fork movement (1) and checkpoint- and damage-dependent inhibition of origin firing (2, 3).
Checkpoint- and damage-dependent origin inhibition is frequently referred to as the intra-S-phase checkpoint. It is likely that this response enhances genomic stability by providing time for cells to repair DNA damage. Indeed, human patients suffering from the genomic instability cancer-prone diseases ataxia telangiectasia (2), ataxia-telangiectasia-like disorder (4), and Nijmegen breakage syndrome (5) are all deficient in suppressing origin firing in response to DNA damage by ionizing radiation. Cancer patients with mutations in the CHK2/CDS1 gene are also deficient in this response (6, 7).
The fission yeast, Schizosaccharomyces pombe, has proved particularly useful for study of DNA-damage and replication checkpoints. Several distinct checkpoint pathways have been defined in fission yeast. In addition to the intra-S-phase checkpoint, these include the SdNTP-M checkpoint, which prevents mitosis when replication is inhibited by lack of dNTPs and the G2-M checkpoint, which prevents mitosis when DNA is damaged during G2 phase (reviewed in ref. 8).
These checkpoint pathways resemble each other in using a common set of proteins for signal transduction. The first six of the genes listed in Table 1 encode a group of proteins that frequently cooperate with each other in checkpoint signaling (9, 10). In fission yeast, these proteins are called the “checkpoint-Rad” proteins, and their functions are usually so interdependent and so central to DNA-damage and replication checkpoint signaling that deletion of the gene encoding any one of them usually completely eliminates checkpoint signaling (11, 12). There is one known exception to this generalization: in G2 phase of the cell cycle, damage-induced Rad3-dependent phosphorylation of Rad26 does not require the proliferating cell nuclear antigen-like and RFC-like checkpoint-Rad proteins (13).
Table 1.
Conserved damage and replication checkpoint genes
| S. pombe | S. cerevisiae | Homo sapiens | Protein function |
|---|---|---|---|
| rad1 | RAD17 | RAD1 | PCNA-like protein |
| rad9 | DDC1 | RAD9 | PCNA-like protein |
| hus1 | MEC3 | HUS1 | PCNA-like protein |
| rad17 | RAD24 | RAD17 | RFC-like protein |
| rad26 | DDC2/LCD1 | ATRIP | PIK-related kinase binding protein |
| rad3 | MEC1 | ATR | PIK-related kinase |
| tel1 | TEL1 | ATM | PIK-related kinase |
| chk1 | CHK1 | CHK1 | Downstream protein kinase |
| cds1 | RAD53 | CHK2/CDS1 | Downstream protein kinase with FHA domain |
This table summarizes work from many laboratories, reviewed in refs. 9 and 10. The first six S. pombe proteins (above the heavy line) are frequently called “checkpoint-Rad” proteins.
PCNA, proliferating cell nuclear antigen; RFC, replication factor C; PIK, phosphoinositide-3-kinase; FHA, fork-head-associated.
In all eukaryotic organisms, each characterized DNA-damage and replication checkpoint pathway depends completely on one or more members of the phosphoinositide-3-kinase-related family (9, 14) (Table 1). These proteins (called Rad3 and Tel1 in fission yeast) are protein kinases, and in many cases they act by phosphorylating and thus activating one or both of the downstream protein kinases, Chk1 or Cds1 (Table 1). In fission yeast, most or all phosphorylation of Chk1 and Cds1 is carried out by Rad3. In contrast to budding yeast and mammals, where direct phosphorylation of Rad53/Chk2 by Tel1/ATM has been described, there is not yet evidence in fission yeast for Chk1 or Cds1 phosphorylation by Tel1 (15).
In fission yeast, the activation and function of Chk1 and Cds1 are cell-cycle-dependent. Cds1 is responsible for DNA-damage and replication checkpoints in early S phase, whereas Chk1 is responsible for checkpoints in late S and G2 (16, 17).
Preliminary studies of the intra-S-phase checkpoint in fission yeast have already been reported. Lindsay et al. (16) demonstrated that the slowing of S phase in response to the alkylating agent methyl methanesulfonate (MMS) depends on Rad3, Rad26, and Cds1, but not on Chk1. Hartsuiker et al. (18) found that MMS-induced slowing of S phase does not require the recombination/repair protein Rad50. Christensen et al. (19) demonstrated that ionizing-radiation-induced retardation of S phase requires Rad3 but not Chk1. Other investigators have detected slowing of S phase in response to UV light (20). These previous investigations left important questions unanswered. Does the intra-S-phase checkpoint require all six checkpoint-Rad proteins or are Rad3 and Rad26 sufficient—a possibility suggested by the results of Edwards et al. (13)? Does the full intra-S-phase checkpoint response require two parallel upstream signaling pathways (one to detect stalled replication forks and another to directly detect damaged sites) as in budding yeast (21–24)? What proteins, if any, are required to mediate checkpoint-independent slowing of replication forks (1)?
With regard to the latter question, several investigators have recently suggested that damage-inducible recombination and/or repair processes may compete with and thus slow down replication (23, 25, 26). This possibility seems consistent with the fact that the helicases Sgs1 and Srs2, which may help to regulate recombinational repair and recombinational damage bypass, contribute to the MMS-induced intra-S-phase checkpoint in budding yeast (22, 23).
Here we report a detailed investigation of the proteins required to retard S phase in response to MMS damage in fission yeast. We chose MMS because both mammalian cells (27) and budding yeast (28) also retard S phase in response to MMS damage, and the proteins required for this response have been well studied in budding yeast (21–24, 28).
We found that all six checkpoint-Rad proteins are needed for the MMS-induced intra-S-phase checkpoint. Surprisingly, of the additional checkpoint proteins whose mutant genes we tested, only Rad4/Cut5, which may be needed for recognition of stalled replication forks, proved to be important for the upstream signaling portion of the intra-S-phase checkpoint pathway. In addition, the helicases Rqh1 and Srs2 contribute slightly to the slowing of S phase.
Methods
Cell Culture and Synchronization by Nitrogen Starvation.
The strains used in this study are listed in Table 2, which is published as supporting information on the PNAS web site (www.pnas.org). S. pombe cells were grown in rich yeast extract medium (YES) (29) and were synchronized in G1 by nitrogen starvation. A preculture of log-phase cells was washed twice with water and then resuspended in Edinburgh Minimal Medium (29) containing supplements but no nitrogen source. Cells were starved for 36 h at 25°C. After starvation, most cells were small and round. However, the rad50Δ, rad32Δ, srs2Δ, and srs2Δ rqh1Δ populations also contained long, dead cells. Because these cells interfered with FACS data quality, they were largely eliminated from the starved culture by brief centrifugation in a table-top centrifuge (TJ-6; Beckman Coulter). The rotor was accelerated for 2 sec at a speed setting of 8 and then allowed to coast to a stop. The supernatant, enriched with small cells, was recovered and used for further analysis. In all cases, the cells were released from starvation by a 1:10 dilution with YES. In the case of temperature-sensitive strains, cells were released from the G1 block at the permissive, semipermissive, and restrictive temperatures, whereas in the case of deletion strains, the cells were released from the G1 block at 25°C. To compensate for occasional variability in the kinetics of S-phase progression, 0.03% MMS (Sigma) was added to portions of the cultures at 2, 3, and 4 h post-release. Samples were collected at 1-h intervals, and the cells were fixed in 70% ethanol. The results shown in Figs. 1–3 are those in which MMS addition was closest to the beginning of S phase in the majority of cells, and this time (2 or 3 h postrelease) is indicated in the figure legends.
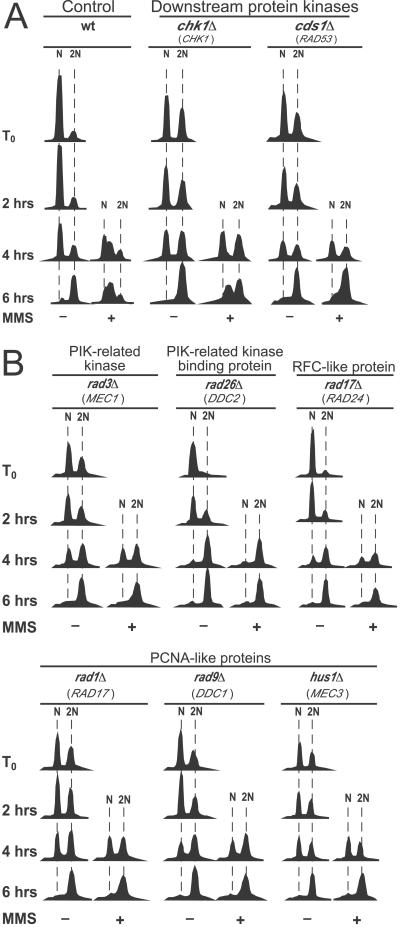
Figure 1.
Effects of conserved checkpoint proteins on MMS-induced slowing of DNA synthesis in fission yeast. Cell synchronization, MMS treatment, and flow cytometric analysis are described in Methods. N and 2N refer to number of haploid genomes. MMS was added 2 h after release from nitrogen starvation. The gene names in parentheses below the S. pombe gene names are the names of the corresponding genes in Saccharomyces cerevisiae. (A) Wild-type and chk1Δ cells, but not cds1Δ cells, proceed through S phase more slowly in the presence of MMS than in its absence. (B) Strains bearing single deletions in each of the six checkpoint-Rad genes (Table 1) are unable to retard S phase in the presence of MMS.
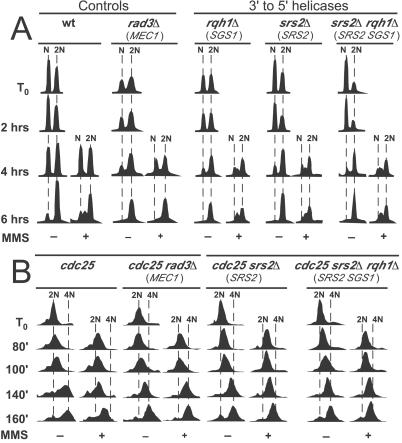
Figure 3.
Effects of the Rqh1 and Srs2 helicases on MMS-induced slowing of DNA synthesis. (A) Analyses were as in Methods and Fig. 1, except that MMS was added 3 h after release. (B) The srs2Δ, rqh1Δ, and rad3Δ mutations were introduced by mating into a cdc25 strain. Strains bearing the indicated mutations were synchronized to G2 by incubation at 36°C for 4 h, then released by shift to 25°C (at T0). MMS was added at 60 min, and samples were taken for flow cytometric analysis at the indicated times (see Methods).
G2 Synchronization by cdc25 Temperature Block.
Log-phase cells were arrested in G2 by incubation at 36°C for 4 h. Cells were then released into the cell cycle at 25°C. MMS (0.03%) was added after 60 min, and then samples were collected at 20-min intervals.
Flow Cytometry.
Cells, fixed in 70% ethanol, were stored at 4°C until needed. Before sorting, cells were washed once in 50 mM sodium citrate, pH 7.0, then resuspended in the same buffer supplemented with 100 μg/ml of RNaseA (Sigma) and incubated for 1–18 h at 37°C. Cells were then stained in the same buffer supplemented with 1 μM Sytox Green (Molecular Probes) and immediately analyzed on a FACScan (Becton Dickinson).
Results
The Six Checkpoint-Rad Proteins and Cds1 Are Essential for the Intra-S-Phase Checkpoint.
To confirm Lindsay et al.'s earlier observation that Rad3, Rad26, and Cds1 (but not Chk1) are required for the S. pombe intra-S-phase checkpoint (16), and to test whether the other four checkpoint-Rad proteins are also required, we synchronized cells in G1 by nitrogen starvation, then released them into the cell cycle in rich medium. At 2 and 3 h after release, we added MMS to a part of each culture, and we used flow cytometry to follow progress through S phase. In the absence of MMS, wild-type cells completed S phase within 6 h of release (Fig. 1A). In the presence of MMS, the rate of DNA replication was much slower, and most cells were still in early S phase at 6 h after release. Cells lacking Chk1 behaved similarly (Fig. 1A). However, cells lacking Cds1 (Fig. 1A) or any of the six checkpoint-Rad proteins (Fig. 1B) completed S phase in the presence of MMS nearly as rapidly as in the absence of MMS.
One BRCT-Domain-Containing Protein (Rad4/Cut5) Is Important, but Another (Rhp9/Crb2) Is Not Important, for the Intra-S-Phase Checkpoint.
The Brca1 carboxyl terminal (BRCT) repeat motif of about 95 aa is commonly found in eukaryotic proteins involved in DNA repair and in DNA-damage checkpoints (30). For example, two previously studied fission yeast proteins, Rad4/Cut5 (similar to Dpb11 in budding yeast) and Rhp9/Crb2 (similar to Rad9 in budding yeast), which respectively contain four and two BRCT domains, are both required for the G2-M damage checkpoint (31–33). However, only Rad4/Cut5 is important for the SdNTP-M checkpoint (31–34). Because the Rad9 protein in budding yeast is important for the intra-S-phase checkpoint (21), we expected that its fission yeast counterpart, Rhp9/Crb2, would also prove important.
To evaluate the roles of these BRCT-domain-containing proteins in the intra-S-phase checkpoint, we tested strains bearing mutations in each of the corresponding genes (Fig. 2). Surprisingly, we found that deletion of rhp9/crb2 did not diminish the MMS-dependent slowing of S phase compared with wild type [Fig. 2A; see also Fig. 5, which is published as supporting information on the PNAS web site (www.PNAS.org)]. Therefore, unlike its budding yeast relative, Rad9, Rhp9/Crb2 in fission yeast does not contribute to the intra-S-phase checkpoint.
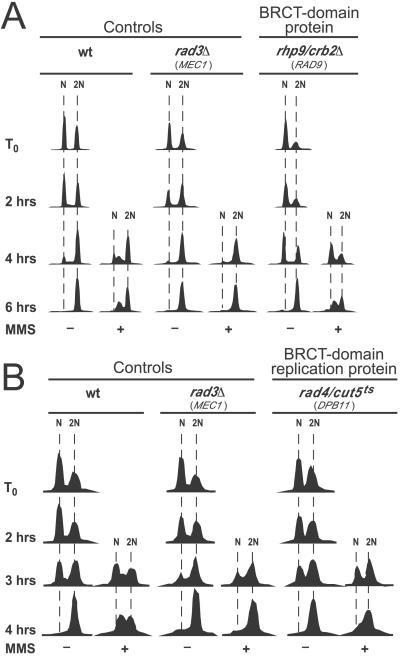
Figure 2.
Effects of BRCT-domain-containing proteins on MMS-induced slowing of DNA synthesis. Unless otherwise noted, analyses were as in Methods and in Fig. 1. (A) The Rhp9/Crb2 protein is not required to slow DNA synthesis. (B) The Rad4/Cut5 protein is required to slow bulk synthesis. Nitrogen starvation was at 25°C, but incubation after release from nitrogen starvation was at 30°C.
Because the Rad4/Cut5 protein is essential for initiation of replication, we tested a strain with a mutant temperature-sensitive allele, rad4–116, that had previously been shown to be functional for replication but defective in the SdNTP-M and G2-M checkpoints at the semipermissive temperature of 32°C (31) (Fig. 2B). We found that at 30°C (Fig. 2B) and at 32°C (data not shown), the rad4–116 strain was able to synthesize DNA in the presence of MMS nearly as rapidly as in the absence of MMS. Thus Rad4/Cut5 is important for the intra-S-phase checkpoint in fission yeast. Its budding yeast relative, Dpb11, has not yet been tested for a role in the intra-S-phase checkpoint. It seems likely that Dpb11 will prove to be important, because DNA polymerase ɛ is important for the intra-S-phase checkpoint in budding yeast (22), and Dpb11 cooperates with DNA polymerase ɛ in the SdNTP-M checkpoint (35, 36).
MMS-Damage-Dependent Slowing of S Phase Depends Slightly on Rqh1 and Srs2.
MMS-damage-dependent slowing of S phase in budding yeast depends partially on the Srs2 and possibly on the Sgs1 3′-5′ DNA helicases (22, 23). These helicases are thought to have overlapping functions in regulating homologous recombination in response to DNA-damage and replication abnormalities (reviewed in ref. 37). The homologous fission yeast proteins are called Srs2 and Rqh1, respectively. Rqh1 is important for maintenance of cell viability when DNA is damaged or replication forks are blocked (38–40). Srs2 in fission yeast is required for regulation of homologous recombination and resistance to DNA damaging agents (41).
We tested the effects of mutations in the rqh1 and srs2 genes on the rate of passage through S phase of fission yeast cells that had been synchronized to G1 by nitrogen starvation. We found (Fig. 3A) that cells bearing complete deletions of the rqh1 and/or srs2 genes appeared to behave like wild-type cells and not like checkpoint-deficient rad3Δ cells (Fig. 3A).
However, the nitrogen starvation and release procedure does not provide as good synchrony as does the α-factor block and release procedure used for the corresponding studies in budding yeast (22, 23). In fact, the time required for fission yeast cells to complete S phase during recovery from nitrogen starvation is much less than the time span over which individual cells within the population enter S phase. Thus the 1N peak diminishes while the 2N peak increases, but at no time (in the absence of MMS) is there a discrete S-phase peak (Figs. 1–3A). Under these circumstances, mutations having only small effects on the time required for passage through S phase might not detectably affect the rate at which the 1N peak decreases and the 2N peak increases.
Because deletions of the SRS2 and (especially) SGS1 genes had only small effects on the rate of passage through S phase in MMS-treated budding yeast cells (22, 23), we decided to test the effects of deletions of the corresponding fission yeast genes with an alternative synchronization procedure, cdc25 block and release. When cdc25 mutant cells are incubated at 36°C, they accumulate in late G2 with a 2N DNA content. When the temperature is dropped to 25°C, they complete mitosis and then proceed synchronously (as a single peak by flow cytometry) through G1 and S phases. Because cytokinesis does not take place until late S or early G2, DNA content per cell increases from 2N to 4N. The results in Fig. 3B show that in the absence of MMS the majority of wild-type (cdc25) and mutant cells passed through S phase—as indicated by movement of the major peak from 2N to 4N—between 80 and 160 min after temperature downshift to 25°C. In the presence of MMS, wild-type (cdc25) cells completed only about 2/3 of S phase during the same time interval, but rad3Δ cells synthesized DNA in the presence of MMS as rapidly as in its absence. Interestingly, srs2Δ, rqh1Δ (not shown), and srs2Δ rqh1Δ cells synthesized DNA more rapidly than wild type but more slowly than rad3Δ cells, indicating that the function of these helicases is required for maximal slowing of S phase in response to MMS damage.
MMS-Damage-Dependent Slowing of S Phase Does Not Depend on the Rad50 Complex, Nonhomologous End-Joining, Homologous Recombination, or Nucleotide Excision Repair.
The Rad50 complex usually consists of three interacting proteins. Two of these, Rad50 and Mre11 (Rad32 in fission yeast), are structurally conserved. The third protein (Nbs1 in mammalian cells; Xrs2 in budding yeast) has not yet been identified in fission yeast. In addition to its roles in homologous recombination and nonhomologous end joining, the Rad50 complex is also important for the intra-S-phase checkpoint in mammals and in budding yeast (4, 5, 42, 43). Therefore, we were surprised to find that deletions of either rad50 (18) or rad32 (44) had no detectable effect on the slowing of S phase in response to MMS damage in fission yeast using either the nitrogen starvation or the cdc25 synchronization procedures (see Figs. 5 and 6A, which are published as supporting information on the PNAS web site).
We have also tested the effects of deleting the genes encoding Rad13 (the fission yeast homologue of mammalian XPG and budding yeast Rad2; necessary for 3′ incision during nucleotide excision repair; ref. 45), pKu70 and Lig4 (the fission yeast homologues of Ku70 and DNA ligase IV, which are essential for nonhomologous end joining; refs. 46, 47), and Rhp51 (a homologue of budding yeast and mammalian Rad51, essential for homologous recombination; ref. 48). No effect on the intra-S-phase checkpoint was observed in cells deleted for the genes encoding Rad13, pKu70, Lig4, and Rhp51 (see Fig. 6 B and C, which is published as supporting information on the PNAS web site).
Discussion
DNA-Repair and Recombination Processes Contribute Minimally to the Slowing of S Phase in Fission Yeast.
We have attempted to test the hypothesis that inhibition of DNA synthesis in response to MMS damage is mediated in part by competition between replication and recombination/repair processes (23, 25, 26). We measured the effects of deleting several genes involved in DNA repair or recombination. Our results indicate that individual deletions of most of these genes had no detectable effect on MMS-induced slowing of S phase in fission yeast. However, deletion of the genes encoding Rqh1 and Srs2 (repair and recombination helicases similar to budding yeast Sgs1 and Srs2) does have a small effect on MMS-induced slowing of S phase. The effect of a double deletion of the rqh1 and srs2 genes was not significantly different from the effects of the single deletions. Our observations are consistent with the earlier demonstration by Paulovich et al. (21) that deletions of genes essential for base excision repair, nucleotide excision repair, mismatch repair, homologous recombination, and mutagenic bypass synthesis all fail to affect MMS-dependent slowing of S phase in budding yeast. Thus, of the tested yeast mutations in repair or recombination genes, the only ones that affect damage-induced slowing of S phase are rqh1 and srs2 in fission yeast, the corresponding genes (SGS1 and SRS2) in budding yeast, and the Rad50 complex in budding yeast. These results suggest that damage-induced, checkpoint-independent slowing of replication forks (1) is probably not a consequence of DNA repair processes interfering with fork movement. However, these results do not permit distinction between several other possibilities: (i) direct repair-independent stalling at damaged sites may be responsible for most or all damage-induced slowing of replication forks; (ii) processes requiring the Rqh1/Sgs1 and Srs2 helicases (such as recombinational damage bypass) may contribute to fork slowing; and/or (iii) in budding yeast, action of the Rad50 complex at damaged sites may contribute to fork slowing.
The available data also do not permit distinction between roles for Rqh1/Sgs1 and Srs2 in directly slowing replication forks and in checkpoint signaling, which would lead to inhibition of origin firing. Evidence from budding yeast suggests that Sgs1 (22) and Srs2 (23) do indeed play important roles in checkpoint signaling. However, this does not exclude the possibility that they may also contribute, in checkpoint-independent fashion, to replication fork slowing.
Considering the importance of the Rad50 complex to intra-S-phase checkpoint signaling in mammals (4, 5) and budding yeast (42), it may seem surprising that we do not detect a role for the Rad50 complex in fission yeast. There are two possible explanations for this apparent discrepancy. First, previous studies demonstrating the importance of the Rad50 complex for the intra-S-phase checkpoint have used DNA damaging agents that produce double-strand breaks (4, 5, 42), which are the preferred substrate of the Rad50 complex (49). In contrast, we used MMS as a damaging agent. MMS leads to significant double-strand breakage only at high concentration (50). Second, Rad50-complex-dependent checkpoint signaling in mammals is initiated by ATM and in budding yeast by ATM's homolog, Tel1 (Table 1). Budding yeast Tel1 (42, 43) appears to play a more important role in cellular responses to DNA damage than does fission yeast Tel1 (15), whose function appears largely confined to telomeres. It is possible that contribution of the fission yeast Rad50 complex (and Tel1) to damage-induced slowing of S phase may become detectable when a larger portion of the damage is due to double-strand breaks.
The Fission Yeast Intra-S-Phase Checkpoint Is Mediated by a Single Signaling Pathway, Which Resembles the SdNTP-M Checkpoint.
Our results indicate that in fission yeast all six checkpoint-Rad proteins, the Cds1 kinase, the Rad4/Cut5 protein, and to a lesser extent the repair and recombination helicases, Srs2 and Rqh1, are required for slowing S phase in response to DNA damage (Fig. 4). With the exception of Srs2 and Rqh1, these are the same proteins that are required for the SdNTP-M checkpoint (which inhibits mitosis when replication forks are blocked by dNTP starvation), and they are somewhat different from the proteins required for the G2-M checkpoint, where Rhp9/Crb2 is required and Chk1 is required instead of Cds1 (reviewed in ref. 8).
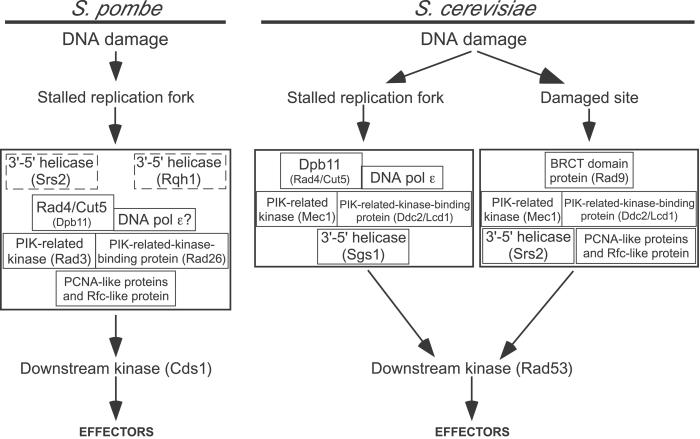
Figure 4.
Models for the S. pombe and S. cerevisiae intra-S-phase checkpoint pathways. The S. pombe model is based on the protein requirements for the fission yeast MMS-induced intra-S-phase checkpoint, as demonstrated previously (16) and in this paper. The boxes representing the Srs2 and Rqh1 helicases are shown with dashed outlines both to indicate that there is only partial dependence of S-phase slowing on these helicases and to emphasize that it is not yet known whether these helicases are in fact part of the intra-S-phase checkpoint (as suggested here) or contribute to checkpoint-independent replication fork slowing. The S. cerevisiae model is based on studies in other laboratories of the proteins required for the budding yeast MMS-induced intra-S-phase checkpoint and of the genetic interactions between them (21–24, 28, 35, 51, 57, 58). The S. cerevisiae model shows only proteins homologous to those tested by us in S. pombe. In both cases, the upstream proteins are grouped in large boxes to indicate that their order of action is still uncertain (13, 24). We infer that the fission yeast pathway and the Dpb11 branch of the budding yeast pathway are activated by stalled replication forks, because replication proteins are involved in both cases (Rad4/Cut5 in fission yeast, Dpb11 and DNA polymerase ɛ in budding yeast).
The evidence for a single intra-S-phase checkpoint pathway in fission yeast is surprising, because in budding yeast at least two signaling pathways contribute to the intra-S-phase checkpoint (21–24) (Fig. 4). One of these resembles the budding yeast SdNTP-M signaling pathway. This pathway requires DNA polymerase ɛ (51) and Dpb11 (35), the homologue of Rad4/Cut5. The other budding yeast intra-S-phase checkpoint signaling pathway resembles the budding yeast G2-M pathway and requires Rad9 (21, 24, 52), the homologue of Rhp9/Crb2. Our results indicate that an Rhp9/Crb2-dependent pathway is not used for the intra-S-phase checkpoint in fission yeast.
The single fission yeast intra-S-phase signaling pathway suggested by our data partially resembles the Dpb11-dependent branch of the budding yeast signaling pathway (Fig. 4). Because Dpb11 interacts and cooperates with budding yeast DNA polymerase ɛ both during DNA synthesis and in checkpoint signaling (35), it seems likely that fission yeast polymerase ɛ will also prove to be important for signaling in both the SdNTP-M and intra-S-phase checkpoints.
In an attempt to evaluate this hypothesis, we have tested all available DNA polymerase ɛ alleles (53, 54), none of which precisely matches the checkpoint-deficient alleles of budding yeast DNA polymerase ɛ (51). In all cases, the rate of DNA replication in the presence of MMS was slowed to the same extent as in wild type (data not shown). Further testing with mutant alleles that match the checkpoint-deficient polymerase ɛ alleles from budding yeast is needed to more rigorously evaluate this hypothesis.
The apparent existence of a single replication-fork-dependent signaling pathway in S-phase fission yeast cells is consistent with the cell-cycle specificity of the downstream kinases in fission yeast: Cds1 is activated by DNA damage or replication fork blockage only in S phase, whereas Chk1 is activated by DNA damage only in G2 (16, 17, 55, 56). In contrast, the budding yeast homologue of Cds1, Rad53, can be activated in G1, S, and G2 (25). Because there are no replication forks present during G1 or G2, it follows that Rad53 must be capable of being activated by fork-independent as well as fork-dependent pathways. During S phase, a significant portion of Rad53 associates with replication forks (22). It is likely that the same will prove to be true for Cds1 in fission yeast.
Supplementary Material
Acknowledgments
We are grateful to Deborah Mahoney, who initiated these studies in the Huberman lab, and to Marius Poitelea and Yetunde Fatunmbi for important contributions. We thank Hiroshi Murakami and Hiroto Okayama (University of Tokyo) for the cds1Δ strain, Clive Price (University of Sheffield) for the rad4–116 strain, and Teresa Wang (Stanford University), Gennaro D'Urso (University of Miami), and anonymous referees for constructive comments on the manuscript. This research was supported by grants from the National Institutes of Health to J.A.H. (GM49294 and CA84302) and G.A.F. (ES07940) and from the Human Frontiers Science Program (RGO178/2000M) to A.M.C. E.H. was supported by a Marie Curie Fellowship from the European Union (MCF1-1999-01425). We are grateful for support of the Roswell Park Cancer Institute Flow Cytometry Facility by the Roswell Park Cancer Center Support Grant (P30 CA16056-26).
Abbreviations
- MMS
methyl methanesulfonate
- BRCT
Brca1 carboxyl terminal
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tercero J A, Diffley J F. Nature (London) 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 2.Painter R B, Young B R. Proc Natl Acad Sci USA. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Nature (London) 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 4.Stewart G S, Maser R S, Stankovic T, Bressan D A, Kaplan M I, Jaspers N G J, Raams A, Byrd P J, Petrini J H J, Taylor A M R. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 5.Carney J P, Maser R S, Olivares H, Davis E M, Beau M L, III, J. R Y, Hays L, Morgan W F, Petrini J H J. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 6.Bell D W, Varley J M, Szydlo T E, Kang D H, Wahrer D C, Shannon K E, Lubratovich M, Verselis S J, Isselbacher K J, Fraumeni J F, et al. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 7.Falck J, Mailand N, Syljuåsen R G, Bartek J, Lukas J. Nature (London) 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 8.Huberman J A. Prog Nucleic Acid Res Mol Biol. 1999;62:369–395. doi: 10.1016/s0079-6603(08)60513-9. [DOI] [PubMed] [Google Scholar]
- 9.Zhou B-B S, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 10.Cortez D, Guntuku S, Qin J, Elledge S J. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 11.Al-Khodairy F, Carr A M. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J F, Lehmann A R, Carr A M. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards R J, Bentley N J, Carr A M. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 14.Bosotti R, Isacchi A, Sonnhammer E L L. Trends Biochem Sci. 2000;25:225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura A, Naito T, Ishikawa F. Genetics. 1999;152:1501–1512. doi: 10.1093/genetics/152.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindsay H D, Griffiths D J F, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brondello J-M, Boddy M N, Furnari B, Russell P. Mol Cell Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartsuiker E, Vaessen E, Carr A M, Kohli J. EMBO J. 2001;20:6660–6671. doi: 10.1093/emboj/20.23.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen P U, Bentley N J, Martinho R G, Nielsen O, Carr A M. Proc Natl Acad Sci USA. 2000;97:2579–2584. doi: 10.1073/pnas.97.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhind N, Russell P. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frei C, Gasser S M. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 23.Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foss E J. Genetics. 2001;157:567–577. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foiani M, Pellicioli A, Lopes M, Lucca C, Ferrari M, Liberi G, Falconi M M, Plevani P. Mutat Res. 2000;451:187–196. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- 26.Rhind N, Russell P. Curr Biol. 2000;10:R908–R911. doi: 10.1016/s0960-9822(00)00849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahle D B, Griffiths T D, Carpenter J G. Mol Pharmacol. 1978;14:278–289. [PubMed] [Google Scholar]
- 28.Paulovich A G, Hartwell L H. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 29.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 30.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 31.McFarlane R J, Carr A M, Price C. Mol Gen Genet. 1997;255:332–340. doi: 10.1007/s004380050504. [DOI] [PubMed] [Google Scholar]
- 32.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verkade H M, O'Connell M J. Mol Gen Genet. 1998;260:426–433. doi: 10.1007/s004380050913. [DOI] [PubMed] [Google Scholar]
- 34.Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araki H, Leem S-H, Phongdara A, Sugino A. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Elledge S J. Proc Natl Acad Sci USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McVey M, Kaeberlein M, Tissenbaum H A, Guarente L. Genetics. 2001;157:1531–1542. doi: 10.1093/genetics/157.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray J M, Lindsay H D, Munday C A, Carr A M. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davey S, Han C S, Ramer S A, Klassen J C, Jacobson A, Eisenberger A, Hopkins K M, Lieberman H B, Freyer G A. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S-W, Goodwin A, Hickson I D, Norbury C J. Nucleic Acids Res. 2001;29:2963–2972. doi: 10.1093/nar/29.14.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Amours D, Jackson S P. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Usui T, Ogawa H, Petrini J H J. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 44.Wilson S, Warr N, Taylor D L, Watts F Z. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carr A M, Sheldrick K S, Murray J M, al-Harithy R, Watts F Z, Lehmann A R. Nucleic Acids Res. 1993;21:1345–1349. doi: 10.1093/nar/21.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baumann P, Cech T R. Mol Biol Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manolis K G, Nimmo E R, Hartsuiker E, Carr A M, Jeggo P A, Allshire R C. EMBO J. 2001;20:210–221. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muris D F, Vreeken K, Carr A M, Broughton B C, Lehmann A R, Lohman P H, Pastink A. Nucleic Acids Res. 1993;21:4586–4591. doi: 10.1093/nar/21.19.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haber J E. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 50.Chlebowicz E, Jachymczyk W J. Mol Gen Genet. 1979;167:279–286. doi: 10.1007/BF00267420. [DOI] [PubMed] [Google Scholar]
- 51.Navas T A, Zhou Z, Elledge S J. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 52.Navas T A, Sanchez Y, Elledge S J. Genes Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- 53.D'Urso G, Nurse P. Proc Natl Acad Sci USA. 1997;94:12491–12496. doi: 10.1073/pnas.94.23.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng W, D'Urso G. Mol Cell Biol. 2001;21:4495–4504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, Carr A M, Bentley N J. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhind N, Russell P. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paciotti V, Clerici M, Lucchini G, Longhese M P. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 58.Rouse J, Jackson S P. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.