Abstract
A computer algorithm, p53MH, was developed, which identifies putative p53 transcription factor DNA-binding sites on a genomewide scale with high power and versatility. With the sequences from the human and mouse genomes, putative p53 DNA-binding elements were identified in a scan of 2,583 human genes and 1,713 mouse orthologs based on the experimental data of el-Deiry et al. [el-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W. & Vogelstein, B. (1992) Nat. Genet. 1, 45–49] and Funk et al. [Funk, W. D., Pak, D. T., Karas, R. H., Wright, W. E. & Shay, J. W. (1992) Mol. Cell. Biol. 12, 2866–2871] (http://linkage.rockefeller.edu/p53). The p53 DNA-binding motif consists of a 10-bp palindrome and most commonly a second related palindrome linked by a spacer region. By scanning from the 5′ to 3′ end of each gene with an additional 10-kb nucleotide sequence appended at each end (most regulatory DNA elements characterized in the literature are in these regions), p53MH computes the binding likelihood for each site under a discrete discriminant model and then outputs ordered scores, corresponding site positions, sequences, and related information. About 300 genes receiving scores greater than a theoretical cut-off value were identified as potential p53 targets. Semiquantitative reverse transcription–PCR experiments were performed in 2 cell lines on 16 genes that were previously unknown regarding their functional relationship to p53 and were found to have high scores in either proximal promoter or possible distal enhancer regions. Ten (∼63%) of these genes responded to the presence of p53.
The p53 protein plays a central role in cancer surveillance (1, 2) and functions as a sequence-specific transcription factor. Although progress has been made in identifying numerous downstream effector genes regulated by p53, to date only about 20 genes have been confirmed to contain p53 DNA-responsive elements that bind to the p53 protein and are clearly transcriptionally regulated by p53. The presence of a p53 consensus DNA-binding sequence near or in a gene does not necessarily imply that it is regulated by p53 in vivo. However, such sequences, particularly when found in the regulatory region of a gene, can guide an experimental test of its functional relationship to p53.
The tetrameric p53 protein binds to two repeats of a consensus DNA sequence 5′-PuPuPuC(A/T)-3′, where (T/A)GPyPyPy is its inverted sequence. The sequence is commonly repeated in two pairs, each arranged as inverted repeats, → ← … → ←, where “→ ←” is PuPuPuC(A/T)(T/A)GPyPyPy and “…” is the spacer region (1, 3). The degenerate nature of the p53 DNA-binding consensus sequence might be critical for regulatory control, since it allows for diversity and flexibility in timing and levels in response to cellular signals. However, this variability or degeneracy complicates the identification of binding sites. There are three attributes that are implemented in the computational scheme presented here but are missing in other approaches (see ref. 4). First, although binding patterns can deviate from the consensus, the reiteration of a number of PuPuPuC(A/T) sites into a cluster recognized by p53 has been shown to stabilize binding and mediate expression (5), for example, in the MDM2 (6) and p21 (7) genes. Second, the spacer region located between the two members of the pair of 10-bp palindromes (i.e., → ←) consists of a number of nucleotides that can vary from 0 to approximately 14 bp. Third, an unbiased criterion for judging the overall binding likelihood is needed for a given gene of arbitrary size. Three features have been implemented in an algorithm to meet the above requirements: binding propensity plots, weighted scores, and statistical significance for most likely binding sites.
This computer algorithm has been used to identify putative binding elements on a genomewide scale. The algorithm, p53MH, uses an optimal scoring system as an indication of the percent similarity to the consensus. It can simultaneously screen thousands of genes for degenerate consensus sequences in the course of only a few minutes. With this, based on the available annotated human sequence databases, a White Page-like directory is created (hereafter referred to as the Directory). At the current stage, the Directory consists of binding information for over 4,000 genes loosely classified into 14 different biological pathways (http://linkage.rockefeller.edu/p53). These genes were classified by a complete survey of the pathways from Santa Cruz Biotechnology; sequence data for each gene have been obtained from the Celera database (http://cds.celera.com/), with an additional 10 kb of sequence included at each end for most genes. Every gene has been screened by p53MH for putative p53 binding elements. The sites and sequences for the 10 highest scores have been listed in the Directory, where the spacer region has been allowed to vary from 0 to 14 bp. The Directory is posted on the World Wide Web at http://linkage.rockefeller.edu/p53.
To test how well this algorithm predicts p53-responsive genes, semiquantitative real-time (RT)-PCR experiments were performed with 16 genes using two different cell lines in culture. These genes were chosen because the orthologs of both the human and mouse genomic sequences had high scores in either the proximal promoter or distal enhancer regions, and none of them have previously been reported in the literature as being p53-responsive genes.
Materials and Methods
p53MH Algorithm.
As described above, p53 DNA-binding sites tend to be more or less faithful to the consensus sequence. This variability or degeneracy may be captured in a weight matrix with rows corresponding to the four bases and entries in each column representing the relative base frequencies of a given position in known binding sites (8). In p53MH, the weight matrix has been derived from the combined data of 20 clones in el-Deiry et al. (3) and 17 clones in Funk et al. (9) (Table 1). With the weight as the input information, one can compute binding probabilities, or binding scores, for any given site. Methods based on statistical mechanics theory or artificial neural networks have been described and applied to other transcription factors (10). Depending on the pattern of the motif, different scoring systems furnish different degrees of accuracy. Here, a method based on discrete discriminant analysis, which is conceptually straightforward and has excellent properties in the p53 case, was used. In addition, previous experiments have shown that certain bases, for example, A, T, or C at positions 8 and 18 [X in (T/A)XPyPyPy], are incompatible with p53 binding (3, 11). Such offending bases were captured in a filtering matrix (Table 1), and sequences containing offending bases were assigned the minimum score. The algorithm can thus be summarized by three basic elements: weighting, scoring, and filtering. We then used experimentally known p53-responsive genes to test the effectiveness of the algorithm (Table 3) and developed several fine-tuning schemes to improve the power of detection (see Results and Discussion). Note that the clone sequences used to develop the weight matrix are different from those of the known p53-responsive genes that make up our test data set. What follows is a description of the scoring method.
Table 1.
Weight and filtering matrices
| 5′—R | R | R | C | W | W | G | Y | Y | Y | R | R | R | C | W | W | G | Y | Y | Y—3′ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Weight | ||||||||||||||||||||
| A | 14 | 11 | 26 | 0 | 28 | 2.5 | 0 | 0.5 | 0 | 3 | 6 | 2 | 11.5 | 0 | 27 | 4 | 0 | 0.5 | 1 | 2 |
| C | 3 | 1 | 1 | 36 | 1 | 0.5 | 0 | 24.5 | 33 | 23 | 2 | 0 | 0.5 | 36 | 2 | 0 | 0 | 9.5 | 24 | 15 |
| G | 16 | 24 | 10 | 0 | 0 | 0 | 37 | 0 | 0 | 0 | 23.5 | 34 | 25 | 0 | 2 | 1 | 37 | 0 | 0 | 3 |
| T | 4 | 1 | 0 | 1 | 7 | 34 | 0 | 12 | 4 | 10 | 5.5 | 1 | 0 | 1 | 5 | 32 | 0 | 27 | 12 | 16 |
| Filter* | ||||||||||||||||||||
| A | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| C | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| G | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| T | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
0 = filtered; 1 = nonfiltered.
Table 3.
An example of p53MH output for known p53-inducible genes
| Gene | Length | Site | Score | Sequence |
|---|---|---|---|---|
| Snk | 26283 | 7968 | 100 | AAACATGCCT.GGACTTGCCC |
| p48-ddb2 | 44746 | 10478 | 100 | GAACAAGCCC T GGGCATGTTT |
| gadd45 | 23161 | 16636 | 100 | AGGCATGTTT G GAGCTAGCTT |
| serpinb5-maspin | 48573 | 7263 | 100 | AGGCATGTTC TCCAG AAACTAGTTT |
| pten-10 | 123338 | 9522 | 100 | GAGCAAGCCC CAGGCAGCTACACT GGGCATGCTC |
| pigpc1-perp | 35853 | 13319 | 95.95 | AGGCAAGCTC.CAGCTTGTTC |
| p21-cdkn1a | 28622 | 15111 | 95.83 | AGACTTGCCT TTGTTGACAT TAGCTTGCCC |
| apaf1 | 110193 | 9397 | 95.69 | AGACATGTCT GGAGACCCTAGGA CGACAAGCCC |
| p53aip1 | 27599 | 19189 | 94.86 | GAGCAAGCTG TAGATCCA AGGCTTGCTT |
| Cycling-G | 26555 | 10309 | 93.62 | GCACAAGCCC.AGGCTAGTCC |
| pig1 | 40531 | 13335 | 93.6 | GCACAAGCCT TTTAAGTCAT GAGCTAGTCC |
| p53r2 | 54618 | 18129 | 93.16 | AACCTTGTTC ATACA AAACAAGTCT |
| sfn (14-3-3-σ) | 21312 | 11242 | 93.12 | AAGCATGTCT GCTGGGTGT GACCATGTTT |
| p53dinp1 | 35023 | 14871 | 92.79 | GAGCTTGTGT TTCATGGCTGAC AGACAAGTTT |
| p53r2 | 54618 | 16981 | 92.14 | TGGCATGTTT TACATACCTACAGT TAGCAAGTTC |
| fas-tnfrsf6 | 45151 | 10653 | 90.92 | GGACAAGCCC.TGACAAGCCA |
| igfbp3 | 29032 | 13172 | 90.9 | AAACAAGCCA C CAACATGCTT |
| siah1 | 47108 | 18036 | 90.89 | TGGCTAGCTA TAGTC GAGCATGTTC |
| serpine1-pai | 31893 | 9774 | 90.36 | ACACATGCCT.CAGCAAGTCC |
| mdm2 | 52293 | 10733 | 79.68 | GGTCAAGTTC.AGACACGTTC |
The first column shows the gene symbol. Length = the total length of the gene; site = first base position of the putative binding site identified by the p53MH algorithm (most of these correspond to the known binding sites), score, and sequence of the identified sites. Translation start site is at position 10,001.
Let x = (x1 … xL) denote the nucleotide sequence of length L including a spacer region of some length after element 10. For a given x, the objective is to decide whether it is a binding site. Discrete discriminant analysis theory shows that an optimal assignment is based on the probability ratio, P(x(b))/P(x(r)), where P(x(b)) and P(x(r)) stand for the multinomial distributions of sequences that do and do not bind to p53, respectively (12). This treatment of putative binding sites as dichotomous quantities is an approximation. In reality, the binding strength may be quantitative. Since the experimental observations with p53 DNA-binding are sparse, steps must be taken to reduce the dimensionality of the multinomial P(x). One possibility is to express the multinomial density in terms of a small number of orthogonal base functions (13, 14). In practice, independence models are often used and tend to work well; that is, P(x(b))/P(x(r)) is expressed as Πi(fi/gi), where, for simplicity, fi and gi represent the frequencies of the ith nucleotide in x (12). Estimates of fi and gi are obtained, in principle, from sequences that do and do not bind to p53, respectively. Because p53 DNA-binding sites are presumably relatively rare in comparison with the total sequence of a gene, gi may be estimated simply by the probability that the ith nucleotide conforms to the consensus by chance. For example, gi is estimated by P(A) + P(G), for P(A) and P(G) are the respective frequencies of occurrence of A and G nucleotides in the sequence involved.
These considerations justify the current practice of scoring a candidate site, x, as follows. For the ith base position in x, a score is defined as
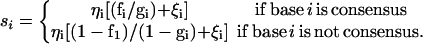 |
Eq. 1 is built on the formula given in Stormo and Hartzell (15). Summing the logarithm of si over the 20 bases in x while skipping the spacers, we get the site score, S(x; w(l)) = w(l) × Σilog(si), where w(l) is the weight for spacer length l as determined by its genomic frequency (unpublished results).
The two parameters, ξ and η in Eq. 1, are critical for obtaining optimal results. A smoothing factor ξ is imposed so that the score is not too drastically affected by inaccurate estimation of marginal frequencies, fi, due to limited experimental evidence. In the Directory, the value of ξ is chosen to make the maximum and minimum score symmetric about zero, |Smin| = Smax. This equation amounts to numerically solving the equation Σlog{(fi/gi + ξ)[(1 − fi)/(1 − gi) + ξ]} = 0 for ξ, where the maximum score, Smax, is the score for all sites being consensus, and the minimum score, Smin, for all sites not being consensus. Of the genes investigated here, ξ is in the neighborhood of 0.25. The core factor η serves to emphasize the biological importance of the eight nucleotides (CWWG in both palindromes) that most closely interact with the p53 protein residues (16). In the absence of the core factor η, the score treats each position as being equally likely for p53 binding. It is clear from the crystallographic results that one or two mutations in the core sequence prevent p53 binding even though the noncore sequences adhere to the consensus. In the Directory, η is set equal to 2 and 1 for the core and noncore regions, respectively. The ratio 2 to 1 has been determined simply by counting the frequency of zeros between the core and noncore nucleotide cells in the weight matrix.
The final site score was expressed as a percentage of the maximum possible score, i.e., S(x;w(l))/Smax, so that it ranges between 0 and 100.
Cell Culture, RNA Extraction, and Semiquantitative RT-PCR.
Murine F9 cells (ATCC no. CRL 1720) and Vm10 cells (17) were grown in 100-mm culture dishes in DMEM supplemented with 10% FCS, 10 units/ml of penicillin, and 10 μg/ml of streptomycin. F9 cells were cultured at 37°C, and Vm10 cells at 39°C. In F9 cells p53 was activated by etoposide treatment. F9 cells were grown to 50% confluence and then treated with etoposide (final concentration 10 μM) for 12 h. To activate p53 in Vm10 cells, the cells were grown to 70% confluence at 39°C then transferred to 32°C and cultured for another 24 h. The cells were then lysed and the total RNA was extracted with Trizol Reagent (Life Technologies, Rockville, MD) following the vendor's manual. RNA concentration was determined by spectrophotometry.
To detect the mRNA of the genes of interest, semiquantitative RT-PCR was performed with SuperScript One-Step RT-PCR System (Invitrogen) as recommended by the vendor. Briefly, a pair of oligonucleotides were designed for amplifying a ∼500-bp sequence from each gene of interest (Table 2). Total RNA (1 μg) and 0.025 μl of [α-32P]dCTP (3000 Ci/mmol, Amersham Pharmacia Biotech) were added to each 25 μl of RT-PCR reaction mix. The reaction mix was incubated in a thermocycler programmed as follows: 50°C for 30 min, 94°C for 2 min; 3-segment amplification cycles: 94°C for 30 sec, 55°C for 30 sec, and 72°C for 45 seconds, followed by final extension at 72°C for 5 min. The number of amplification cycles that gave linear cDNA amplification was determined experimentally. Twenty-three cycles were used for the Mdm2 gene and all test genes, 18 cycles for Gapdh and Ran genes.
Table 2.
Semiquantitative RT-PCR experiment and results, where the mdm2 gene is the positive control and mGAPDH and mRan are negative controls
| Gene | Left oligo sequence | Right oligo sequence | p53 Responsiveness*
|
|||
|---|---|---|---|---|---|---|
| F9
|
Vm10
|
|||||
| Basal | Fold | Basal | Fold | |||
| mBace2 | tgaagttggaaattggaggc | gtagaagccttccatcacgg | 774.8 | 4.26 Y (+) | ND | |
| mCdh13 | cagtgtgctgctgacagtga | atgggcaggttgtagtttgc | 12978.1 | 0.84 N | 231265.0 | 1.05 N |
| mCdk2 | cattcctcttcccctcatca | cgataacaagctccgtccat | 19984.1 | 5.82 Y (+) | 138612.5 | 0.84 N |
| mCradd | tcacacatcctcagcagctc | gccacaaagttccaaaccat | 30385.3 | 0.21 Y (−) | 38326.7 | 1.05 N |
| mEgfr | atgtcctcattgccctcaac | ggaacttttggcagaccaga | 9962.1 | 0.91 N | 10740.4 | 5.58 Y (+) |
| mEts2 | cttccaaaaggagcaacgac | gtcctggctgatggaacagt | 19678.5 | 0.67 N | 91144.4 | 0.96 N |
| mGrb10 | cctgattgctggaaagaagc | cacgagaccttgttgcttga | 14599.5 | 3.98 Y (+) | 4653.2 | 4.43 Y (+) |
| mMnat1 | gagttggaggaagcattgga | gttatctgggctgccagaag | 4243.6 | 0.7 N | 13820.0 | 0.84 N |
| mPer2 | attagacggtgctcggaaga | atgctccaaaccacgtaagg | 7975.0 | 0.81 N | 1963.4 | 4.66 Y (+) |
| mRab10 | ctgcttttcaagctgctcct | tttcggaggatgtcttcagc | 21075.9 | 0.29 Y (−) | 188298.2 | 0.32 Y (−) |
| mRab7 | tgaacccatcaaactggaca | caaggagggaggggtaaaag | 128765.0 | 0.48 N | 537400.2 | 1.14 N |
| mTyro3 | aaggccccctaggaccttat | tgaactgctgctctggaatg | ND | 26559.8 | 0.32 Y (−) | |
| mWisp2 | tagccacccgagtatccaac | gaagggacctggatgtttca | 21075.9 | 0.3 Y (−) | 188298.4 | 0.32 Y (−) |
| mWnt3 | tgtggaggcaggtctcttct | agcgggaaaggacaaattct | 16569.0 | 1.69 N | 83899.2 | 0.82 N |
| mTead1 | ctcagatctgcaacccacaa | tgaggggtgatgtcttcctc | 23336.9 | 0.78 N | 66787.8 | 0.78 N |
| mTshr | ctctcttacccgagccactg | ggctggttagcagaatgagc | 2336.9 | 8.62 Y (+) | 19939.6 | 0.52 N |
| mdm2 | tgtgtgagctgagggagatg | gcacatccaagccttcttct | 2688.1 | 29.68 | 436654.4 | 16.31 |
| mGAPDH | acccagaagactgtggatgg | cttgctcagtgtccttgctg | 468614.5 | 0.6 | 73008.5 | 1.17 |
| mRAN | aggacccatcaagttcaacg | ggcatccagcttcactttct | 193022.9 | 0.74 | 11437.3 | 1.31 |
Y (+), induction; Y (−), repression; N, no response; ND, nondetectable.
Representative results from two independent experiments.
After RT-PCR amplification, 5 μl of loading buffer (100% formamide/0.01% bromophenol blue and Xylene cyanole FF) was added to 5 μl of the reaction mix. The samples were denatured at 95°C for 3 min, loaded to 6% denaturing polyacrylamide gel, and separated by electrophoresis. The gel was then fixed and dried. Autoradiography was performed to visualize the amplified products and a PhosphorImager (Molecular Dynamics) was used to directly quantify the radioactive bands.
Results and Discussion
Comparison with Available Algorithms.
Although there are numerable computer programs for searching transcription factors binding sites, results have been disappointing when applied to p53. The programs have failed because (i) while a scoring system may work well for other transcription factors, it mostly likely needs to be modified, refined, or even exchanged for a new model to effectively deal with the highly degenerate p53 consensus sequence and the variable-length spacer region. (ii) Existing computer programs focus on the proximal promoters, whereas for many genes their p53-binding sites were found in introns, 3′ untranslated regions, or other distal enhancer regions. To our knowledge, the matinspector computer program (4) is the only publicly available algorithm that can detect putative p53 DNA-binding sites, albeit for sequences of limited lengths (<1,000 bp is recommended). Based on 17 relatively homogeneous clone sequences from Funk et al. (9), matinspector searches for the motif 5′-GGACATGCCCGGGCATGTCC-3′ and assumes complete absence of a spacer region. That is, it misses all binding sites with a spacer between the two palindromes. The merit of matinspector is that it can search for transcription factor-binding sites other than p53 while p53MH focuses on p53. However, given the versatility of p53MH, it may easily be expanded to accommodate other binding site sequences. The versatility and power of p53MH can be seen in the following subsections.
Asymmetry in the Weight and Filtering Matrices.
Both the weight and the filtering matrices built from experimental data (3, 9) are not symmetric with respect to either the two palindromes “→ ← … → ←” or within a palindrome “→ ←.” For example, of the two “→ ←” in “→ ← … → ←,” the left palindrome (5′ of the spacer region) seems to be more faithful to the consensus than the right palindrome, and within “→ ←”, “←” is more faithful than “→.” However, these trends are rather weak and not statistically significant and may merely be the chance consequence of small sample size. Further investigation is required to address this issue.
Binding Propensity Plots to Identify Putative Binding Clusters.
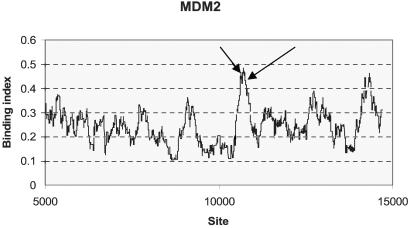
As mentioned in the introduction, many experimentally confirmed p53 DNA-binding sites tend to cluster together. Here we present a moving average approach to visualizing such clusters in a given gene, which is motivated by a scan statistics method (18). First, a binding index is calculated for each position in the sequence. The binding index is the average score (Eq. 1) within a “window” 100-bp long, for example. Thus, if the binding index is high for a number of consecutive positions in a gene, it tends to indicate a higher propensity of p53 binding than the signal at a single site. This cluster can be identified from the area with high peaks when binding indices are plotted against nucleotide numbers. An illustration of such a plot, conveniently called the binding propensity plot, is made with the promoter region of the MDM2 gene shown in Fig. 2. As one can see, from 3 kb upstream and downstream from the translation start site, the two experimentally proven p53-binding sites are situated in the window of the highest peak, whereas inside that window, single site scores are only in the range of 70s (see one of the true binding site scores in Table 3).
Figure 2.
Binding propensity plot for the MDM2 gene from 5 kb upstream and 5 kb downstream from the translation start site (at 10,001 on x axis) with a window size of 100 bp. Arrows point to the experimentally confirmed p53-binding sites at 10,733 and 10,771.
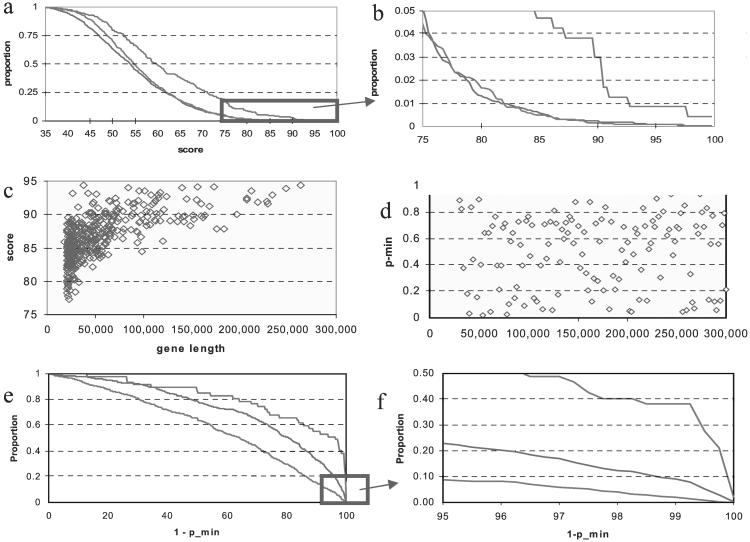
Length Bias in Site-Based Scores but Not in Gene-Based Binding Probability.
For a given gene, it is desirable to build an overall score that will predict its responsiveness to p53. Fig. 1 a and b shows that the sum of the 5 highest site scores in a gene are significantly larger in known p53-inducible genes (approximately 40 of these have been ascertained from refs. 19–21, and references therein) than in random sequences of comparable lengths (30 kb on average). In the graph, two kinds of random sequences are used, one is taken from segments on chromosome 6 and the other is obtained by computer simulations; their scores are indistinguishable and smaller than those of the p53-inducible genes. Results are unaltered when the p53-binding sequences in the known genes are incorporated into the weight matrix. Therefore, the ability to predict p53 responsiveness with this scoring system is valid and effective for these 40 genes. Table 3 lists p53MH output for some of these genes.
Figure 1.
Frequency distributions of the sum of the five highest scores for three gene groups displayed in a and b. The right line represents the known p53-inducible genes. The middle line represents sequences of 30 kb in length randomly picked from the P arm of chromosome 6. The left line represents computer-simulated random sequences of 30 kb in length. c shows the score vs. gene length for 400 genes arbitrarily chosen from the Directory. (d) Plot of the P values for the same 400 genes. The (1 − pmin) distributions are displayed in e and f, where pmin is the smallest significance level of consecutive sums of the 5 highest scores.
Fig. 1c examines ≈400 genes arbitrarily chosen from the Directory and plots the sum of the five largest scores in each gene against its total length. It is evident that shorter genes consistently correlate with lower scores—a length-bias phenomenon; that is, longer genes simply have more chance to experience higher scores at random. While there are many ways to minimize such an undesirable effect, the most intuitive one would be to determine “null” probabilities for the scores; that is, probabilities of having the scores given that the associated sequences are not the binding sites. Specifically, we compute the 5 highest scores and, with 5,000 bootstrap samples, obtain significance levels (P values) for the sums of ordered scores (i.e., P3 = significance level for the sum of the three highest scores). Then we take the minimum P value, Pmin, for i ranging from 1 through 5 as the gene-specific statistic (22), which automatically corrects for length-bias and is applicable to genes of any length (otherwise one could examine sequences in windows of fixed length before calculating site scores). This approach transforms Fig. 1c into Fig. 1d. Similarly, Fig. 1 a and b becomes Fig. 1 e and f with the proportions of (1 − Pmin) plotted for the three groups. It also captures subtle differences in base pair composition between chromosome 6 and the random sequences as seen in Fig. 1 e and f but not in a and b. However, the downside of this method is that it is time-consuming because of the need for bootstrap samples. For this reason, we implement a method based on fixed lengths as described below.
Potential p53 Target Genes.
Of the 4,296 genes in the Directory, 25 are found to contain perfect-match consensus sequences in both human and mouse (Table 4), although not every site is orthologous to each other in the two species. Among these 25 genes, BclII has been shown to be a downstream target of p53 transcriptional repression (23) and Pten is a p53-inducible gene (24). The ability of p53 to function as an activator or repressor may depend on the binding sequence as in the case of transcription factor Pit1, which can switch from activator to repressor by a two-base pair change in its binding site (25). One needs more experimental evidence to assess this issue with p53. In the present article, experimental tests are provided for some of these 25 genes.
Table 4.
Genes with a perfect score of 100 both in human and mouse orthologs
| Gene | Brief description |
|---|---|
| Bcl2 | B-cell CLL/lymphoma |
| Cradd* | CASP2; death domain |
| Egfr* | Avian erythroblastic leukemia viral oncogene homolog |
| Fbn1 | Fibrillin; Marfan syndrome |
| Fetub | Fetuin |
| Gabrb2 | GABA(A) receptor |
| Mnat1 | CDK-activating kinase assembly factor MAT1 |
| Mrvi1 | Murine retrovirus integration site 1 |
| Neo1 | Neogenin, a DCC-related protein |
| Pten | Phosphatase and tensin |
| Rab10* | RAS oncogene family |
| Rab7 | RAS oncogene family |
| Syn3 | Synapsin |
| Tlk2 | Tousled gene code for cell-cycle-regulated kinases |
| Top1 | Topoisomerase 1; DNA repair and transcription |
| Tshb | Thyroid-stimulating hormone |
| Tshr* | Thyroid-stimulating hormone receptor |
| Ucn | Urocortin; cognition-enhancing property |
| Wisp2* | WNT1 inducible signaling pathway protein 2 |
| Cacna1a | Calcium channel |
| Cryz | Crystallin, zeta (quinone reductase) |
| Lox11 | Lysyl oxidase-like |
| Pscd4 | Pleckstrin homology |
| Rhag | Rhesus blood group-associated glycoprotein |
| Ywhah | 14-3-3 eta chain gene |
In addition, a theoretical cut-off score was derived from the distribution of the 3 highest scores in each of 13-kb-long 10,000 reference sequences generated randomly according to the base frequencies in the human genome. Note that a fixed length was imposed to avoid a length bias as discussed previously. Of the 30,000 scores in the random sequences, less than 750 (<2.5%) exceeded 93 and less than 1,500 (<5%) exceeded 90. We take 93 as a cut-off score and classify 304 genes, each restricted in a 13-kb region (3 kb upstream and 10 kb downstream of the translation start site), in the Directory as potential p53 target genes (see http://linkage.rockefeller.edu/p53). A score of 90 is also a reasonable cutoff in light of the scores observed in known p53-responsive genes (see Table 3). On the basis of either cutoff (93 or 90), only MDM2 would have been missed among the genes with known p53-binding sites. On the other hand, as outlined in the previous subsection, the p53-binding sites of MDM2 are picked up by p53MH via the binding propensity plot (Fig. 2).
Semiquantitative RT-PCR.
The ability of p53 to activate the transcription of 16 different genes was examined with two different cell lines. One is a murine teratocarcinoma F9 cell line, which harbors the wild type but silent p53 (26). Etoposide treatment can promptly activate p53 in these cells (26). Another system uses the murine Vm10 cell line, which expresses a temperature-sensitive mutant p53 with alanine 135 changed to valine (17). Shifting the growth temperature from 39°C to 32°C changes the mutant p53 conformation to a wild-type conformation, therefore activating it. In both experiments, mdm2 gene was chosen as a positive control, while Gapdh and Ran genes were used as negative control. As shown in Table 2, the mRNA of 15 of 16 genes was detectable in F9 cells. Among these 15 genes, transcripts of 7 genes were changed 3-fold or more after p53 activation. Four transcripts went up and three went down. Similarly, the mRNA in 15 of 16 genes was detected in Vm10 cells. Transcripts of 6 of 15 genes were altered three or more fold after p53 activation. Three transcripts went up and three went down. Altogether, transcripts of a total of 10 genes with detectable mRNA were altered in response to p53 activation. The p53 protein did not activate the same set of genes in these two cell lines. Only three genes (Grb10, Wisp2, and Rab10) were similarly regulated in both F9 and Vm10 cell lines. It has been shown that the nature of the cell type and “stress” inducer can alter the type of p53-responsive gene that is regulated (20). To fully test the p53MH algorithm, a more thorough analysis of p53-responsive genes will need to be carried out in a variety of cell or tissue types stimulated by a variety of stress signals.
Acknowledgments
We thank Dr. Jenyue Tsai for many invaluable suggestions and discussions. Our appreciation also extends to the reviewers for their constructive comments. This work was supported by Human Genome Institute Grants K25HG00060-01A1 and R01HG00008, and by National Cancer Institute of the National Institutes of Health Grant P01CA87497.
Abbreviation
- RT
real time
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Tyner S D, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. Nature (London) 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 4.Quandt K, Frech K, Karas H, Wingender E, Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ptashne M, Gann A. Genes & Signals. Plainview, NY: Cold Spring Harbor Lab. Press; 2002. [Google Scholar]
- 6.Kaku S, Iwahashi Y, Kuraishi A, Albor A, Yamagishi T, Nakaike S, Kulesz-Martin M. Nucleic Acids Res. 2001;29:1989–1993. doi: 10.1093/nar/29.9.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.el-Deiry W S, Tokino T, Waldman T, Oliner J D, Velculescu V E, Burrell M, Hill D E, Healy E, Rees J L, Hamilton S R, et al. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- 8.Waterman M S. In: Mathematical Methods for DNA Sequences. Waterman M S, editor. Boca Raton, FL: CRC Press; 1989. pp. 93–115. [Google Scholar]
- 9.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stormo G D, Fields D S. Trends Biochem Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 11.Bian J, Sun Y. Proc Natl Acad Sci USA. 1997;94:14753–14758. doi: 10.1073/pnas.94.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon W R, Goldstein M. J Am Stat Assoc. 1978;73:305–313. [Google Scholar]
- 13.Ripley B D. Pattern Recognition and Neural Networks. Cambridge, U.K.: Cambridge Univ. Press; 1997. [Google Scholar]
- 14.Ott J, Kronmal R A. J Am Stat Assoc. 1976;71:391–399. [Google Scholar]
- 15.Stormo G D, Hartzell G W. Proc Natl Acad Sci USA. 1989;86:1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoh J, Ott J. Proc Natl Acad Sci USA. 2000;97:9615–9617. doi: 10.1073/pnas.170179197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin S, Levine A J. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 20.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman W H, Tom E, Mack D H, Levine A J. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Zhang L, Hwang P M, Rago C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoh J, Wille A, Ott J. Genome Res. 2001;11:2115–2119. doi: 10.1101/gr.204001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Shenk T. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak T W. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 25.Scully K M, Jacobson E M, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose D W, Hooshmand F, Aggarwal A K, Rosenfeld M G. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 26.Lutzker S G, Levine A J. Nat Med. 1996;2:804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]