Abstract
Protein–protein interactions are often mediated by the recognition of short continuous amino acid stretches on target proteins by specific binding domains. Affinity-based selection strategies have successfully been used to define recognition motifs for a large series of such protein domains. However, in many biological systems specificity of interaction may be of equal or greater importance than affinity. To address this issue we have developed a peptide library screening technology that can be used to directly define ligands for protein domains based on both affinity and specificity of interaction. We demonstrate the value of this approach by the selection of peptide ligands that are either highly specific for the Grb2 Src homology 2 (SH2) domain or that are cross-reactive between a group of related SH2 domains. Examination of previously identified physiological ligands for the Grb2 SH2 domain suggests that for these ligands regulation of the specificity of interaction may be an important factor for in vivo ligand selection.
The intracellular organization that enables a cell to respond to external stimuli consists of a complex web of signal transduction pathways. Key factors in the regulation of cellular signaling are the protein binding domains—small, conserved protein modules that mediate intracellular protein–protein interactions. Many of these domains, such as the families of SH2 (Src homology 2), SH3 (Src homology 3), PTB (phosphotyrosine binding), or PDZ (postsynaptic density-95/Discs large/zona occludens-1) domains, use short peptide sequences for ligand recognition (1). For example, the binding of SH2 domains to target proteins involves the recognition of a phosphorylated tyrosine residue, and specificity of individual SH2 domains is mediated by the recognition of amino acid residues immediately C-terminal to the phospho-tyrosine (2). The binding preferences of SH2 domains have been studied extensively through the use of peptide libraries, and predictions for the optimal binding motifs for a large number of SH2 domains have been obtained in this manner (3, 4). In addition, such binding motifs have been used extensively as lead structures for the design of selective small-molecular SH2 inhibitors (5, 6). Importantly, in traditional library screening strategies used to define SH2 ligand motifs the selection of ligands is exclusively based on the strength of the SH2–phospho-peptide interaction. Consequently, the motifs that are identified in this manner describe ligands with an optimal affinity for a given SH2 domain (here named affinity motifs). Because both affinity and specificity of protein interactions are controlled by the same thermodynamic factors (shape and charge complementarity in the ground state), the selection of high affinity ligands will often also result in the selection of highly specific ligands. However, it has previously been argued that for closely related targets (such as the families of SH2 domains and other signal transduction modules) affinity-based selections may result in the identification of ligands that cross-react with related molecules (7). To address this issue we set out to develop a library screening strategy that can be used to define both affinity and specificity motifs for protein–ligand interactions. We have used this strategy to identify highly specific phospho-tyrosine ligands for the SH2 domain of the Grb2 adaptor molecule. This ubiquitously expressed adaptor protein is composed of a single SH2 domain flanked by two SH3 domains (8). The SH2 domain of Grb2 directly recognizes phospho-tyrosine-containing sites on a number of tyrosine kinases and tyrosine kinase receptors. The Grb2 SH3 domains bind to the Ras guanine nucleotide exchange factor Sos, thereby linking Grb2 recruitment to Ras activation (8–10). Importantly, this Grb2-dependent Ras activation pathway has been shown to be essential for cellular transformation in a subset of human tumors. Approximately 40–50% of breast tumors display increased expression levels of members of the erbB family of receptor tyrosine kinases, and suppression of Grb2 function in these cells inhibits cell proliferation (11, 12). Because of the clear potential of Grb2 inhibitors as therapeutic agents, significant interest has grown in the development of inhibitors of the Grb2 SH2 domain (5, 13, 14). We show here that the conventional affinity-based library selections for Grb2 SH2 ligands result in phospho-peptides that display cross-reactivity toward related SH2 domains and we define ligands that express a desirable specificity profile. The value of specificity-based screening strategies for the prediction of protein interactions and for drug discovery is discussed.
Materials and Methods
Glutathione S-Transferase (GST)–SH2 Fusion Proteins.
cDNAs encoding mGrb2, mGrb2(R67H), and the SH2 domains of hAbl, Nck, bPI3Kp85-N, bPI3Kp85C, mSHP-2N, mSHP-2C, and vSrc were subcloned into the expression vector pGEX (Amersham Pharmacia). GST fusion proteins were produced in the Escherichia coli strain BL21DE3pLysS by isopropyl β-d-thiogalactopyranoside induction and purified with glutathione-Sepharose beads (Amersham Pharmacia). GST fusion proteins were biotinylated by using NHS-LC-biotin (Pierce). The identity and purity were checked by SDS/PAGE.
Peptide Library Synthesis.
Nα-N-(9-fluorenyl)methoxycarbonyl (Fmoc)-based peptide syntheses were conducted on a Syro II synthesizer (MultiSynTec, Bochum, Germany) by using benzotriazole-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate(Py BOP)/diisopropylethylamine (DIPEA) coupling protocols. Solid-phase peptide libraries were synthesized onto 10 μm Tentagel M beads (Rapp Polymere, Tübingen, Germany) by split and pool synthesis (15). Protected amino acids and coupling reagents were from Perkin–Elmer and Nova Biochem. Where indicated, the peptide density on beads was controlled by mixing Nα-Fmoc-alanine with Nα-acetylated alanine (Bachem) in the first cycle of synthesis.
Multiparameter Peptide Library Screening.
Anti-β-endorphin library screening.
An octameric peptide library (density: 0.29 μmol/g) was generated by using all naturally occurring amino acids as building blocks. A total of ≈0.5 × 106 library beads was incubated with 0.1 μg anti-β-endorphin 3E7 antibody (Gramsch Laboratories, Schwabhausen, Germany) in 100 μl PBS/1% BSA for 1 h at room temperature. Beads were washed three times in PBS and stained with both FITC and phycoerythrin (PE)-conjugated goat anti-mouse IgG2A antibodies (Southern Biotechnology Associates) to detect 3E7-bound beads. The percentage of FITC/PE double-positive beads was calculated.
Grb2 phospho-peptide library screening.
A phospho-peptide library was generated (density: 29 μmol/g) by using all naturally occurring amino acids as building blocks. A total of ≈0.5 × 106 library beads was incubated with 0.1 μM biotinylated Grb2(R67H)-GST and 1.0 μM each of hAbl, Nck, bPI3Kp85-N, bPI3Kp85-C, mSHP-2N, mSHP-2C, and vSrc SH2-GST fusion proteins in 100 μl PBS/1% BSA for 1 h. Beads were washed three times in PBS and stained with 1 μg PE-labeled Avidin (Molecular Probes) for 20 min. Beads were washed three times in PBS and incubated with 100 μl precoupled 0.1 μM Grb2-GST/1 μg FITC-labeled Avidin (Molecular Probes) in PBS/1% BSA/0.1 mM biotin for 1 h and washed three times in PBS. Reactions were performed in 100 μl PBS (pH 7.4)/1% BSA at room temperature. The percentages of beads that stained positive only for fluorescein (Grb2) were determined.
All samples were monitored on a FACS-Calibur flow cytometer (Becton Dickinson) and analyzed with cellquest software. Analysis was restricted to single beads by using forward and sideward scatter parameters. Autofluorescent beads were excluded by gating on FL4 parameter-negative beads.
Grb2 Binding Assay.
pYVNV tetramers were generated by incubation of 10 μg horseradish peroxidase-conjugated streptavidin (CLB, Amsterdam) with an excess (10 nmol) of biotin-conjugated SApYVNVSA peptide, and they were purified by gel filtration HPLC. Microtiter plates (Nunc) were coated with Grb2-GST at 5 μg per well in 0.1 M Na2CO3/NaHCO3, pH 9.6. Plates were washed with PBS and blocked with 2% milk powder/PBS for 1 h. Plates were washed five times with PBS/0.005% Tween 20 and incubated for 1 h with 3 nM pYVNV tetramers in the presence of the indicated concentrations of competing peptides in 100 μl of 1% milk powder/PBS. Plates were washed five times in PBS/0.005% Tween 20 and incubated with 1 mg/ml 3,5,3′,5′-tetramethylbenzidine/0.003% H2O2 in 100 μl of 0.1 M NaAc, pH 5.5. Reaction was stopped by addition of 100 μl of 1.8 M H2SO4, and absorption was measured at OD450.
CD28 Immunoprecipitation.
A variant of the mCD28 cDNA in which the YMNM motif is changed to YKNI was generated by PCR assembly (16) with the following primers: mCD28top/BamHI (GGGGGATCCTAAACCATGACACTCAGGCTGCTGTTC), mCD28KNItop (GGAACAGACTCCTTCAAAGTGACTACAAAAACATCACTCCCCGGAGG), and mCD28bottom/NotI (ATAAGAATGCGGCCGCTCAGGGGCGGTACGCTG). Both the wild-type CD28 (CD28wt) and mutant CD28KNI cDNAs were cloned into the pMX retroviral vector. The CD28-negative variant Jurkat cell line J.RT3-T3.5 was grown in Iscove's modified Dulbecco's medium (Life Technologies, Paisley, Scotland) with 5% FCS (BioWhittaker). Jurkat cells were transduced with pMX vectors encoding CD28wt or CD28KNI by using a transduction protocol as described (17) and sorted for CD28-expressing cells. A total of 2 × 107 Jurkat/CD28wt or Jurkat/CD28KNI cells (in 500 μl PBS) was stimulated with 10 μg/ml anti-CD28 mAb 37.51 in the presence of 0.1 mM pervanadate for 20 min on ice. CD28/antibody complexes were cross-linked with rabbit anti-Syrian hamster IgG (Jackson Immunoresearch) at 40 μg/ml for 10 min on ice and 15 min at 37°C. Cells were lysed by adding 500 μl ice-cold 2× lysis buffer [1× lysis buffer contains 50 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 μM MgCl2, 1 mM EGTA, 1 mM Na3VO4, and protein inhibitor mixture (Roche Diagnostics)] for 20 min on ice. As a control, cells were exposed to the same procedure, but with addition of the antibodies after cell lysis. The detergent-insoluble fraction was removed by centrifugation, and the supernatants were incubated with 20 μl protein A Sepharose beads (Amersham Pharmacia) in the presence of 30 nM biotinylated Grb2-GST and 70 nM biotinylated phosphatidylinositol 3-kinase-associated p85 (PI3Kp85-C) SH2-GST at 4°C for 1 h. The beads were washed five times in 1× lysis buffer, and immunoprecipitates were separated by SDS/PAGE and transferred to nitrocellulose membranes. GST fusion proteins were detected with horseradish peroxidase-coupled streptavidin (Amersham Pharmacia) plus SuperSignal Chemiluminescent Substrate (Pierce).
Results
We set out to develop a library screening strategy that allows the simultaneous measurement of the binding properties of ligands to multiple target proteins. Flow cytometry has been used extensively to simultaneously determine the binding of large sets of antibodies to individual cells and to isolate (rare) cell types that display a specific mAb-binding profile. In our experiments, we wanted to exploit the ability of flow cytometry analysis to measure multiple receptor–ligand interactions simultaneously for the identification of ligands with a desired specificity pattern. In this setup, a library of peptide ligands is synthesized on nanometer-sized beads, such that individual beads contain a single sequence. This library is then confronted with a set of differentially labeled target proteins and the library components are analyzed for their reactivities toward any of these targets by flow cytometry. Because of its ability to simultaneously measure the interaction of ligands with multiple targets, this multiple-parameter ligand identification strategy can be used for the one-step identification of ligands with a defined specificity pattern. In addition, the ability of flow cytometers to scan the binding pattern of individual beads with extremely high speed (more than 1 × 107 events per h) permits the rapid identification of optimal (peptide) ligands from libraries of extremely large size.
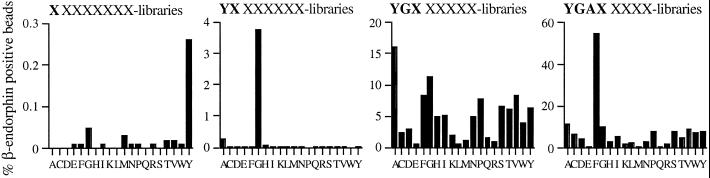
To first examine the feasibility of ligand selection by flow cytometry a synthetic peptide library was screened for peptide sequences that are recognized by the well-studied anti-β-endorphin mAb 3-E7. This antibody specifically binds with high affinity (Kd 7.1 nM) to the N-terminal sequence YGGF (single-letter amino acid designation) of the β-endorphin protein (18). An octa-peptide library containing all 20 naturally occurring amino acids as building blocks was synthesized and screened for binding to the 3-E7 antibody by recursive deconvolution (19, 20). In subsequent screening rounds the preferred amino acid at each position was determined and used to deduce an optimal binding motif. Screening of this peptide library by multiple-parameter ligand identification resulted in the definition of the minimal amino acid sequence YGAF as the optimal peptide ligand for the 3-E7 antibody (Fig. 1). This sequence is in full agreement with results previously obtained in phage display screens with the 3-E7 antibody (21, 22). In addition, the relative contribution of individual amino acid side chains in this motif to the total binding energy can directly be deduced from the bias toward one (or more) residue(s) in each individual screening round (Fig. 1).
Figure 1.
Selection of peptides that bind the anti-β-endorphin (3-E7) mAb. A degenerate octa-peptide library was screened for library beads that bind 3-E7 antibody, and it was analyzed by flow cytometry. The percentage of fluorochrome-labeled beads was determined for all amino acids on each position. From left to right, screening for first, second, third, and fourth positions.
Multiple-parameter ligand identification was subsequently used to identify binding motifs for the SH2 domain of the Grb2 adaptor protein. The target specificity of individual SH2 domains is based on the recognition of the three amino acids carboxyl terminal to the phospho-tyrosine within the target molecule. For the Grb2 SH2 domain the optimal binding motif has previously been identified as the sequence pY-E/Q-N-Ψ (where Ψ indicates a hydrophobic residue) (4, 13, 14, 23). In this motif, the asparagine at p+2 (position+2 respective to the phospho-tyrosine residue) is essential for Grb2 SH2 binding, whereas the selectivity at p+1 and p+3 is apparent, but less stringent. A phospho-peptide library was designed with the sequence ASpYXXXXSA, where X indicates positions of degeneracy in which all 20 aa are included (library diversity: 204 = 1.6 × 105). This library was first screened for high affinity binding motifs for Grb2 SH2, by selection of sequences that bind recombinant full-length Grb2 (SH3-SH2-SH3) but do not interact with a mutant Grb2, in which a R67H substitution in the SH2 domain abolishes phospho-tyrosine recognition (Fig. 2 A and B). Screening of the multiple-parameter ligand identification peptide library in this manner results in a Grb2 SH2 binding motif, pY-Q/E-N-L/I (Fig. 3 A–C), that is in perfect accordance with the optimal binding motif, as previously determined for Grb2 SH2 (4, 13, 14, 23). Furthermore, these library screening results confirm the strong preference for Asn at p+2 for Grb2 recognition and the somewhat smaller bias at p+1 and p+3. As previously established, amino acid residues at p+4 did not affect Grb2 SH2 binding (not shown).
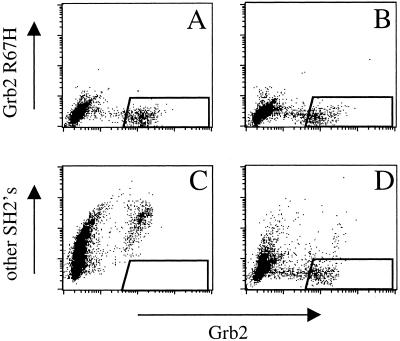
Figure 2.
Amino acids at the first position C-terminal from the phospho-tyrosine determine Grb2 SH2 specificity. ASpYEXXXSA (A and C) and ASpYKXXXSA (B and D) phospho-peptide sublibraries were stained with FITC-labeled Grb2-GST versus either PE-labeled Grb2(R67H)-GST (A and B) or PE-labeled Grb2(R67H)-GST, Abl, Nck, PI3Kp85-N, PI3Kp85-C, SHP-2N, SHP-2C, and Src SH2-GST fusion proteins (C and D). Sublibraries were analyzed by flow cytometry. Results are represented as flow cytometry dot plots where each bead is represented by a dot. The percentage of Grb2-specific sequences within each library was determined as the fraction of dots that fall within the quadrangle shown in the lower right corner of the plots.
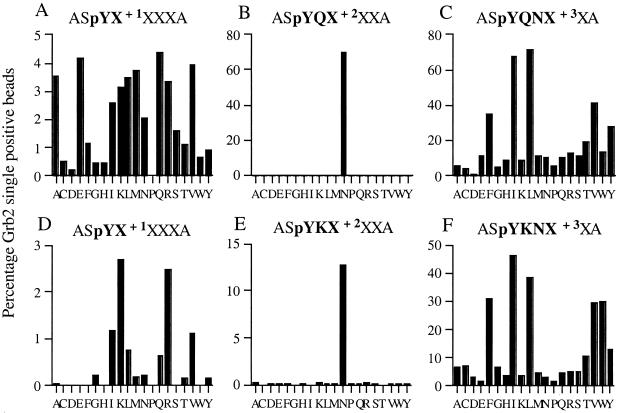
Figure 3.
Selection of affinity and specificity motifs for the Grb2 SH2 domain. A degenerate phospho-tyrosine library was screened for Grb2 SH2-specific sequences and analyzed by flow cytometry. Libraries were screened for Grb2 SH2 optimal binding motifs (A–C) and Grb2 SH2 specificity motifs (D–F). A/D, B/E, and C/F represent screenings for amino acids at p+1, p+2, and p+3, respectively. The percentages of Grb2 SH2-specific sequences were determined as the percentage of gated beads as shown in Fig. 2.
To identify phospho-peptide motifs that define ligands with an optimal specificity for the Grb2 SH2 domain, the same phospho-peptide library was subsequently screened for Grb2-reactive sequences that do not bind a series of other SH2 domains. All known SH2 domains can be placed into four different groups on the basis of the amino acid at position βD5. The side chain at the βD5 position contacts p+1 and p+3 of the phospho-tyrosine ligand and has been shown to have a major effect on SH2 selectivity (3, 24, 25). In our set of control SH2 domains we included three closely related SH2 domains from signaling molecules Abl, Src, and Nck that are derived from the same group as Grb2 (group 1). In addition, the two SH2 domains of PI3Kp85 (PI3Kp85-N and PI3Kp85-C; both group 3) and the two SH2 domains of the SHP-2 phosphatase (SHP-2-N, group 3; SHP-2-C, group 4) were used as competitors in this library screen, as both of these signaling molecules have been described to share ligands with Grb2 SH2 in vivo (26–29). The phospho-peptide library was screened for sequences that bind Grb2 but are not reactive toward any of the control set SH2 domains, by using FITC fluorescence to detect Grb2 binding and PE fluorescence to detect ligand binding within the control set (Fig. 2 C and D). These library beads that selectively interact with Grb2 fall in the quadrangle in the lower right corner as shown in Fig. 2. Competitor SH2 domains were used in a 10-fold higher concentration compared with Grb2 to identify peptide motifs with high selectivity for Grb2 SH2. These experiments reveal that the vast majority of sequences that avidly bind to the Grb2 SH2 domain are strongly cross-reactive with other SH2 domains, whereas only a subset of Grb2 binding sequences displays Grb2 SH2 specificity (Fig. 3 D–F). More specifically, whereas the Asn at p+2 functions as the main determinant for Grb2 affinity, the residue on p+1 appears to function as a prime determinant for Grb2 specificity. In affinity-based selections a large number of amino acid side chains are tolerated at this position with a small preference for Gln and Glu. However, only positively charged residues (Lys and Arg) at this position yield ligands that exclusively interact with the Grb2 SH2 domain (Figs. 2 and 3). For this class of high-specificity ligands a positive charge at p+1 is sufficient to obtain Grb2 specificity (motif: pY-K/R-N-I/L). A smaller group of Grb2-specific ligands is also found that harbor an aliphatic residue at p+1 (Fig. 3D). However, for these ligands Grb2 specificity also depends on the amino acid side chain at p+3. Specifically, whereas the naturally occurring Grb2 ligands pYVNV and pYVNI are highly cross-reactive, substitution of the amino acid at p+3 to pYVNQ reduced this cross-reactivity significantly (see Fig. 5).
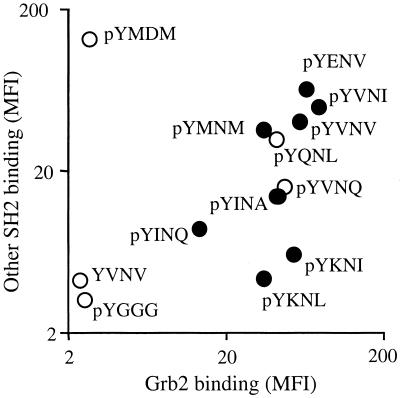
Figure 5.
Specificity of Grb2 SH2 binding to naturally occurring ligands. A set of naturally occurring binding motifs was synthesized onto beads and analyzed for their capacities to bind Grb2-GST (0.1 μM, x axis) or a panel of other SH2 domains (1.0 μM each, y axis) by measuring the mean fluorescence intensity (MFI) by flow cytometry. ●, Beads that contain naturally occurring Grb2-binding motifs. ○, Non-naturally occurring Grb2-binding motifs pYQNL and pYVNQ, and the p85-specific pYMDM, pYGGG and unphosphorylated YVNV as negative controls.
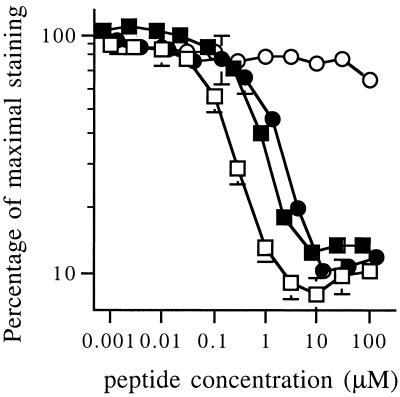
Although the pYKNI motif was selected for high specificity, it only has a minor decrease in binding affinity for Grb2 as compared with the optimal binding motif pYQNL (Fig. 4, IC50 of 0.9 μM and 0.5 μM, respectively). Notably, in these binding studies the pYVNV peptide has a 4-fold higher affinity for Grb2 than the pYQNL sequence identified in library screens. This discrepancy is consistently observed in different library selection procedures that seek optimal ligands for Grb2 SH2 (14, 23).
Figure 4.
Binding affinity of phospho-peptide motifs for Grb2. The capacity of the indicated soluble phospho-peptides to compete with horseradish peroxidase-conjugated ASpYVNVSA tetramers for binding to Grb2-GST was measured by ELISA. Competitor peptides used were: ASpYQNLSA (■), ASpYKNISA (●), ASpYVNVSA (□), and the Src SH2-specific sequence ASpYEEISA (○). Data shown are means of quadruplicates ± SD.
Having established that affinity and specificity-based selection strategies yield distinct motifs for the Grb2 SH2 domain we wanted to assess the relevance of these two motifs for in vivo ligand selection. To this purpose an array of naturally occurring phospho-peptide ligands previously shown to interact with the Grb2 SH2 domain was generated, including the pYKNI/L sequences identified here. Subsequently, these sequences were analyzed for both their capacity to bind the Grb2 SH2 domain and their reactivity toward the series of competitor SH2 domains (Fig. 5). These data indicate that Grb2 SH2 ligands can be defined into distinct classes, based on their selectivity. The first group of ligands appears to have evolved toward high specificity for Grb2 SH2 binding. This group of ligands is characterized by the presence of a specificity determinant at p+1 and/or p+3 and includes the pYKNI/L sequence that was identified here. This sequence has been shown to be a conserved Grb2 binding site in both the colony stimulating factor 1 receptor Fms and its viral homologue v-Fms (30, 31). In addition, the pYKNL sequence is the binding site for Grb2 SH2 in the protooncogene product c-kit. The Grb2 SH2 ligands that harbor an Ile at p+1 (present in EGFR and SHP-2) also can be regarded as part of this Grb2-specific class of phospho-peptides, although selectivity appears less stringent for these ligands. The second class of Grb2 SH2 ligands can be described as hybrid ligand motifs that contain residues at p+1 and p+3 that are permissive but not selective for Grb2 binding. For example, the pYMNM sequence derived from the CD28 molecule, a T cell-specific tyrosine kinase receptor, contains both the Grb2 SH2 binding motif pYXNX and the phosphatidylinositol 3-kinase (PI3-kinase) SH2 binding motif pYMXM (3). In line with these findings, this CD28 binding site has been shown to bind both Grb2 and PI3-kinase upon tyrosine phosphorylation (32).
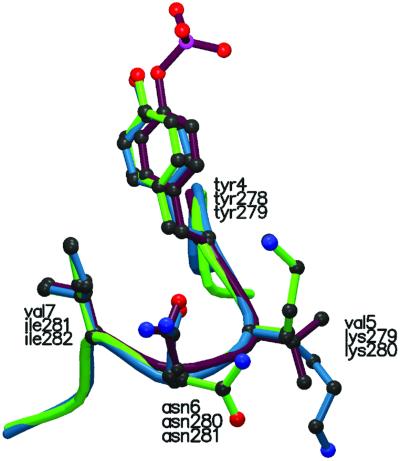
To reveal the potential use of specificity motifs for the prediction of novel physiologically relevant protein interactions a database search was performed for molecules containing YKNI/L sequences. Interestingly, the YKNI motif is present in both the SHP-1 (PTP-1C) and SHP-2 tyrosine phosphatases. Although both phosphatases are known to interact with the Grb2 SH2 domain upon phosphorylation, the known phosphorylation sites within these molecules lie within the C-terminal tail and do not involve the YKNI motif (33–35). Examination of the crystal structure of the SHP-1 and SHP-2 phosphatases reveals that in both molecules the YKNI sequence is surface-exposed close to the phosphatase active site of the molecule (36). Remarkably, the structure of the 4-aa YKNI loops in SHP-1 and SHP-2 overlay exceptionally well with the structure of a phospho-tyrosine peptide bound by the Grb2 SH2 domain (rms differences between Cα backbones of 0.286 Å and 0.134 Å, respectively; Fig. 6). This structural homology is particularly suggestive in view of the fact that phospho-peptide ligands can bind only the Grb2 SH2 domain in a unique β-turn conformation, which is not observed in phospho-peptide ligands for other SH2 domains (37, 38). However, the assignment of these YKNI loops within SHP-1 and SHP-2 as tyrosine phosphorylation sites and as putative Grb2 binding sites remains hypothetical in the absence of direct experimental confirmation.
Figure 6.
YKNI sites in SHP-1 and SHP-2 display a β-turn conformation similar to a Grb2-bound phospho-peptide ligand. The YKNI (278–281) loop from SHP-1 (blue) and the YKNI (279–282) loop from SHP-2 (green) are overlaid with the structure of a Grb2 SH2-bound phospho-peptide ligand pYVNV (purple) (37). The rms differences between Cα backbones of SHP-1 loop and Grb2 ligand and SHP-2 and Grb2 ligand are 0.286 Å and 0.134 Å, respectively.
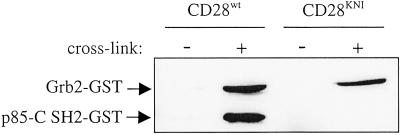
To directly assess the effects of hybrid versus specificity motifs on the functional properties of signaling molecules, the YMNM motif within the cytoplasmic tail of CD28 was changed to the Grb2-specific YKNI sequence identified here. Jurkat T cells that lack endogenous CD28 expression were transduced with either CD28wt or CD28KNI cDNA and stimulated with anti-CD28 mAbs. Subsequently, the capacity of both receptors to bind Grb2 and PI3Kp85-C SH2 GST fusion proteins was determined by immunoprecipitation and Western blotting, as described (39). In line with the peptide selection experiments, CD28wt containing the original YMNM motif associates with both Grb2 and PI3Kp85-C SH2, whereas CD28KNI has fully lost the ability to bind PI3Kp85-C SH2, but remains capable of recruiting Grb2 (Fig. 7). Tyrosine phosphorylation of CD28 is required for Grb2 and PI3Kp85-C SH2 binding, as no interaction was observed in cells that were left unstimulated. These results indicate that the affinity and specificity motifs identified here in in vitro screens determine the functional properties of signal transduction molecules that contain these motifs.
Figure 7.
CD28KNI recruited Grb2 but lost the ability to bind PI3Kp85-C SH2. Jurkat T cells expressing either CD28wt or CD28KNI were activated with anti-CD28 mAbs or left unstimulated. CD28 was immunoprecipitated in the presence of soluble biotinylated Grb2-GST and PI3Kp85-C SH2-GST fusion proteins, and association of these proteins with CD28 was revealed by Western blot analysis.
Discussion
The evolutionary pressures that shape enzymatic and signal transduction processes involve selection on both affinity and specificity of interaction. In contrast, the library screening strategies that have been used to date primarily involve affinity-based selections. We report here an efficient strategy to rapidly identify ligands to biological targets that have been selected not only on the affinity but also on the specificity of this interaction. We document the value of specificity screening by the identification of a highly specific Grb2 binding motif that differs from the binding motif deduced by affinity selection. It has previously been noted that none of the naturally occurring Grb2 SH2 ligands fully conform to the optimal binding motif that was defined for this molecule. The fact that a subset of the physiological ligands for Grb2 SH2 does conform to the specificity motif defined here lends direct support to the validity of our strategy. We demonstrate that a positively charged residue at p+1 is compatible with Grb2 binding, but not with binding to a set of other SH2 domains. Furthermore, when the hybrid SH2 binding motif within the CD28 receptor is displaced by the Grb2 specificity motif, Grb2 signaling is maintained upon CD28 stimulation, whereas the ability to bind PI3Kp85-C SH2 is lost. This observation is consistent with sequence and structural analysis of SH2 domains. Accommodation of a lysine at p+1 in a phospho-peptide structure that binds Grb2 SH2 may be facilitated by interaction of the lysine ɛ-amino group with Gln-106 (βD-strand) within the binding pocket (37). In contrast, lysine side chains may be excluded at p+1 for ligands of the C-terminal and N-terminal SH2 domains of p85 because of charge repulsion by nearby lysine residues in both domains (40, 41). A similar effect of electrostatic or steric clashes has previously been proposed to explain the difference in fine specificity between the phospholipase Cγ1 C-terminal and the SHP-2 N-terminal SH2 domains (42).
Aside from the possible relevance of this technique for our understanding of in vivo protein interactions, this strategy should be of significant use for the development of lead components in drug development. The peptide motifs previously identified for Grb2 SH2 have been used as lead components for the development of small molecule Grb2 inhibitors by a number of groups. However, as shown here, these ligands display a substantial cross-reactivity with other SH2 domains and thereby lack the selectivity required for pharmacological agents. In contrast, the ability to directly identify ligands that are selective for a target protein (or in some cases display a desirable cross-reactivity) should form a potentially more appealing strategy for the identification of novel lead molecules. Finally, the ability to define ligand interaction patterns by flow cytometry as demonstrated here may form a useful tool to describe physical interactions between cellular components in proteomics.
Acknowledgments
We thank Tjasso Blokzijl for technical assistance, Meindert Lamers and Titia Sixma for help with structural analysis, Erik Noteboom and Anita Pfauth for expert technical assistance, and Mariette Oosterwegel for providing mCD28 cDNA. This research was supported by the Dutch Cancer Society (Grant NKI 97-1442), and A.C.W. was supported by the Sylvia and Charles Viertel Senior Medical Research Fellowship.
Abbreviations
- SH2
Src homology 2
- SH3
Src homology 3
- GST
glutathione S-transferase
- PE
phycoerythrin
- PI3Kp85
phosphatidylinositol 3-kinase-associated p85
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sudol M. Oncogene. 1998;17:1469–1474. doi: 10.1038/sj.onc.1202182. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D, Koch C A, Grey L, Ellis C, Moran M F, Pawson T. Science. 1990;250:979–982. doi: 10.1126/science.2173144. [DOI] [PubMed] [Google Scholar]
- 3.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 4.Songyang Z, Shoelson S E, McGlade J, Olivier P, Pawson T, Bustelo X R, Barbacid M, Sabe H, Hanafusa H, Yi T, et al. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oligino L, Lung F D, Sastry L, Bigelow J, Cao T, Curran M, Burke T R, Jr, Wang S, Krag D, Roller P P, et al. J Biol Chem. 1997;272:29046–29052. doi: 10.1074/jbc.272.46.29046. [DOI] [PubMed] [Google Scholar]
- 6.Shakespeare W, Yang M, Bohacek R, Cerasoli F, Stebbins K, Sundaramoorthi R, Azimioara M, Vu C, Pradeepan S, Metcalf C, et al. Proc Natl Acad Sci USA. 2000;97:9373–9378. doi: 10.1073/pnas.97.17.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton B E, Gold L, Zichi D A. Chem Biol. 1995;2:633–638. doi: 10.1016/1074-5521(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 8.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 9.Buday L, Downward J. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 10.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. Nature (London) 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 11.Xie Y, Pendergast A M, Hung M C. J Biol Chem. 1995;270:30717–30724. doi: 10.1074/jbc.270.51.30717. [DOI] [PubMed] [Google Scholar]
- 12.Tari A M, Hung M C, Li K, Lopez-Berestein G. Oncogene. 1999;18:1325–1332. doi: 10.1038/sj.onc.1202422. [DOI] [PubMed] [Google Scholar]
- 13.Gram H, Schmitz R, Zuber J F, Baumann G. Eur J Biochem. 1997;246:633–637. doi: 10.1111/j.1432-1033.1997.00633.x. [DOI] [PubMed] [Google Scholar]
- 14.Hart C P, Martin J E, Reed M A, Keval A A, Pustelnik M J, Northrop J P, Patel D V, Grove J R. Cell Signalling. 1999;11:453–464. doi: 10.1016/s0898-6568(99)00017-0. [DOI] [PubMed] [Google Scholar]
- 15.Furka A, Sebestyen F, Asgedom M, Dibo G. Int J Pept Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 16.Crameri A, Cwirla S, Stemmer W P. Nat Med. 1996;2:100–102. doi: 10.1038/nm0196-100. [DOI] [PubMed] [Google Scholar]
- 17.Kessels H W, van Den Boom M D, Spits H, Hooijberg E, Schumacher T N. Proc Natl Acad Sci USA. 2000;97:14578–14583. doi: 10.1073/pnas.97.26.14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meo T, Gramsch C, Inan R, Hollt V, Weber E, Herz A, Riethmuller G. Proc Natl Acad Sci USA. 1983;80:4084–4088. doi: 10.1073/pnas.80.13.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houghten R A, Pinilla C, Blondelle S E, Appel J R, Dooley C T, Cuervo J H. Nature (London) 1991;354:84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 20.Erb E, Janda K D, Brenner S. Proc Natl Acad Sci USA. 1994;91:11422–11426. doi: 10.1073/pnas.91.24.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cwirla S E, Peters E A, Barrett R W, Dower W J. Proc Natl Acad Sci USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLafferty M A, Kent R B, Ladner R C, Markland W. Gene. 1993;128:29–36. doi: 10.1016/0378-1119(93)90149-w. [DOI] [PubMed] [Google Scholar]
- 23.Muller K, Gombert F O, Manning U, Grossmuller F, Graff P, Zaegel H, Zuber J F, Freuler F, Tschopp C, Baumann G. J Biol Chem. 1996;271:16500–16505. [PubMed] [Google Scholar]
- 24.Marengere L E, Songyang Z, Gish G D, Schaller M D, Parsons J T, Stern M J, Cantley L C, Pawson T. Nature (London) 1994;369:502–505. doi: 10.1038/369502a0. [DOI] [PubMed] [Google Scholar]
- 25.Songyang Z, Gish G, Mbamalu G, Pawson T, Cantley L C. J Biol Chem. 1995;270:26029–26032. doi: 10.1074/jbc.270.44.26029. [DOI] [PubMed] [Google Scholar]
- 26.Fukazawa T, Reedquist K A, Panchamoorthy G, Soltoff S, Trub T, Druker B, Cantley L, Shoelson S E, Band H. J Biol Chem. 1995;270:20177–20182. doi: 10.1074/jbc.270.34.20177. [DOI] [PubMed] [Google Scholar]
- 27.Fixman E D, Fournier T M, Kamikura D M, Naujokas M A, Park M. J Biol Chem. 1996;271:13116–13122. doi: 10.1074/jbc.271.22.13116. [DOI] [PubMed] [Google Scholar]
- 28.Ward A C, Monkhouse J L, Hamilton J A, Csar X F. Biochim Biophys Acta. 1998;1448:70–76. doi: 10.1016/s0167-4889(98)00120-7. [DOI] [PubMed] [Google Scholar]
- 29.Jones N, Master Z, Jones J, Bouchard D, Gunji Y, Sasaki H, Daly R, Alitalo K, Dumont D J. J Biol Chem. 1999;274:30896–30905. doi: 10.1074/jbc.274.43.30896. [DOI] [PubMed] [Google Scholar]
- 30.van der Geer P, Hunter T. EMBO J. 1993;12:5161–5172. doi: 10.1002/j.1460-2075.1993.tb06211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mancini A, Niedenthal R, Joos H, Koch A, Trouliaris S, Niemann H, Tamura T. Oncogene. 1997;15:1565–1572. doi: 10.1038/sj.onc.1201518. [DOI] [PubMed] [Google Scholar]
- 32.Kim H H, Tharayil M, Rudd C E. J Biol Chem. 1998;273:296–301. doi: 10.1074/jbc.273.1.296. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Nishimura R, Kashishian A, Batzer A G, Kim W J, Cooper J A, Schlessinger J. Mol Cell Biol. 1994;14:509–517. doi: 10.1128/mcb.14.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett A M, Tang T L, Sugimoto S, Walsh C T, Neel B G. Proc Natl Acad Sci USA. 1994;91:7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel W, Ullrich A. Cell Growth Differ. 1996;7:1589–1597. [PubMed] [Google Scholar]
- 36.Hof P, Pluskey S, Dhe-Paganon S, Eck M J, Shoelson S E. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 37.Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverria C, Furet P, Caravatti G, Fretz H, Schoepfer J, Grutter M G. Nat Struct Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- 38.Gay B, Furet P, Garcia-Echeverria C, Rahuel J, Chene P, Fretz H, Schoepfer J, Caravatti G. Biochemistry. 1997;36:5712–5718. doi: 10.1021/bi962642y. [DOI] [PubMed] [Google Scholar]
- 39.Okkenhaug K, Wu L, Garza K M, La Rose J, Khoo W, Odermatt B, Mak T W, Ohashi P S, Rottapel R. Nat Immunol. 2001;2:325–332. doi: 10.1038/86327. [DOI] [PubMed] [Google Scholar]
- 40.Breeze A L, Kara B V, Barratt D G, Anderson M, Smith J C, Luke R W, Best J R, Cartlidge S A. EMBO J. 1996;15:3579–3589. [PMC free article] [PubMed] [Google Scholar]
- 41.Nolte R T, Eck M J, Schlessinger J, Shoelson S E, Harrison S C. Nat Struct Biol. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]
- 42.Kay L E, Muhandiram D R, Wolf G, Shoelson S E, Forman-Kay J D. Nat Struct Biol. 1998;5:156–163. doi: 10.1038/nsb0298-156. [DOI] [PubMed] [Google Scholar]