Abstract
The recent completion of the deletion of all of the nonessential genes in budding yeast has provided a powerful new way of determining those genes that affect the sensitivity of this organism to cytotoxic agents. We have used this system to test the hypothesis that genes whose transcription is increased after DNA damage are important for the survival to that damage. We used a pool of 4,627 diploid strains each with homozygous deletion of a nonessential gene to identify those genes that are important for the survival of yeast to four DNA-damaging agents: ionizing radiation, UV radiation, and exposure to cisplatin or to hydrogen peroxide. In addition we measured the transcriptional response of the wild-type parental strain to the same DNA-damaging agents. We found no relationship between the genes necessary for survival to the DNA-damaging agents and those genes whose transcription is increased after exposure. These data show that few, if any, of the genes involved in repairing the DNA lesions produced in this study, including double-strand breaks, pyrimidine dimers, single-strand breaks, base damage, and DNA cross-links, are induced in response to toxic doses of the agents that produce these lesions. This finding suggests that the enzymes necessary for the repair of these lesions are at sufficient levels within the cell. The data also suggest that the nature of the lesions produced by DNA-damaging agents cannot easily be deduced from gene expression profiling.
The development of DNA microarray methods for genome-wide analysis of gene expression provides a powerful way to determine the overall functional state of the cell. The technology is used in two general ways. In the first application an expression profile is obtained in unperturbed cells and tissues to obtain information on their biology, for example, to develop a “molecular taxonomy” of tumors to produce subclassifications of tumor types that cannot be obtained with traditional methods. This classification can lead to improved predications of patient outcome (1–4) and possibly predict sensitivity to anticancer agents (5). A comparison of gene expression profiles between primary tumors and metastases has also identified genes that may be involved in the etiology of metastasis (6, 7), and a comparison between normal and malignant tissues can identify possible diagnostic markers of cancer as well as potential therapeutic targets (8, 9).
The second general way in which gene expression profiling has been used is to examine changes in the transcriptional profile after a treatment or change in environmental conditions (10–15). Many of these transcriptional responses provide insight into the underlying biology; for example, to identify cell-cycle-regulated genes and genes regulated by the diauxic shift in Saccharomyces cerevisiae (10, 16), the genes that respond to serum in human fibroblasts (13), or in identifying the functions of unknown genes and specific targets of drug action in yeast (14). In studies of this kind it is often assumed that genes induced by a given stress or cytotoxic treatment are those needed for adaptation or protection of the cell against that treatment. If this assumption is correct, expression profiling after a damaging agent would be a powerful method for identifying the genes conferring resistance to that agent, and hence provide information on its mechanism. Recent publications have, in fact, suggested that several of the genes induced by DNA-damaging agents are involved in the repair of DNA damage and hence in the protection of the cell against such treatments (17–19). However, the assumption that genes whose expression increases in response to a particular cytotoxic agent are those that protect against the damage caused by the agent has not been formally tested. Here we use a pool of strains of budding yeast, S. cerevisiae, with deletion of all nonessential genes to directly test this hypothesis.
Deletion of the genes has been accomplished by an international consortium, the Saccharomyces Genome Deletion Project, that has replaced all of the ≈6,200 known open reading frames (ORFs) of yeast by using a PCR-mediated gene deletion strategy (20). In addition to a selectable marker, two molecular bar codes or “‘tags,” unique 20-base oligonucleotide sequences, are in the replacement cassette. These tags, after PCR amplification, can be detected by hybridization to the corresponding complementary sequence in a high-density oligonucleotide array, thus enabling the relative abundances of each tag, and hence the abundances of each deletion strain, to be determined (20). We have recently shown that this system can detect essentially all of the known nonessential genes involved in UV resistance as well as identifying some novel genes that when deleted cause sensitivity to UV damage (21).
In the present study we have compared the transcriptional response of budding yeast to four DNA-damaging agents with the sensitivity profiles to the same agents given under the same conditions. We define the sensitivity profiles as the repertoire of genes that when deleted cause sensitivity to that agent. Thus, by comparing the gene expression profile with that of the sensitivity profile for each of the cytotoxic agents, we can directly test the hypothesis that there is a relationship between changes in expression of individual genes and the impact of loss of that specific gene on the sensitivity of the cell to the particular agent. Our results demonstrate that there is little if any relationship between genes induced after DNA damage and the genes necessary for survival to the particular DNA-damaging agent. This finding suggests that the genes that protect against DNA-damaging agent cannot be inferred from the transcriptional response of cells to these agents.
Materials and Methods
Yeast Strains.
Genotypes of the parental yeast strain BY4743 and construction of the homozygous diploid deletion strains have been described previously (20). All of these completed strains can be obtained from Research Genetics (Huntsville, AL) or EUROSCARF (Frankfurt, Germany). In the present study, we used a pool of 4,627 strains representing homozygous deletion of the nonessential genes. For the transcriptional response to DNA-damaging agents we used the parental diploid strain BY4743 grown under conditions identical to those for the strains in the pool.
Treatment with DNA-Damaging Agents.
We performed clonogenic survival experiments with the parental strain, BY4743, to determine the exposure conditions for each agent to produce 20–50% cell killing. The exposure conditions for gene expression profiling with the parental strain and for sensitivity testing with the deletion pool were identical. In each case aliquots were grown in YPD (yeast extract/peptone/dextrose) medium at 30°C, and shaken at 300 rpm to mid-exponential phase (OD600 = 0.5–1.0). For the UV treatment, cells were pelleted, resuspended in ice-cold PBS, and dispersed in 150-mm Petri dishes to a depth of no more than 1 mm. Cells were immediately irradiated with 200 J/m2 UVC in a UV light box (Stratagene, Stratalinker, La Jolla, CA; 254 nm). Cells were then pelleted and resuspended in YPD and reincubated as above. For the ionizing radiation (IR) treatment, cells were irradiated with 200 Gy in YPD at room temperature by using a 137Cs source (Mark 1 Model 3 from J.L. Shepherd, San Fernando, CA; 33 Gy/min) and immediately returned to the orbital shaker at 30°C and 300 rpm. For the cisplatin (Sigma) and hydrogen peroxide (Sigma) treatments, cells were treated at 1 mM for 1 hr at 30°C, samples were taken immediately, and the remaining cells were pelleted, washed in cold PBS, resuspended in cold YPD, and reincubated as above. After 1 hr, samples were taken and the remaining cells were washed and resuspended in fresh YPD. For all of the gene expression experiments three cultures were mock treated in parallel with the treated cultures. The cells were collected at the end of the treatment period and pelleted at 8,000 × g for 3 min, and the pellets were snap frozen at −80°C.
For the experiments with the deletion pool the treated and control cultures were pelleted immediately after exposure and inoculated into prewarmed YPD medium at OD600 = 0.05 (106 cells per ml). Before reaching OD600 = 1.0, the cultures were diluted 1:20 into fresh YPD medium to maintain exponential growth. Cultures were harvested and genomic DNA was extracted 18 hr after treatment.
PCR Amplification, Microarray Hybridization, and Data Acquisition with Deletion Pool.
PCR amplification, microarray hybridization, and data acquisition were as described (21). Briefly, after isolation of genomic DNA from the treated and untreated pools, the isolated DNA was used as template in two PCRs that amplify the two tags from each strain in the pool by using biotinylated PCR primers complementary to common regions in the transplacement cassette. For both the treated and untreated pool, we combined the PCR products with oligonucleotides complementary to nontag regions of the PCR product, heat denatured the mixtures, and hybridized them to purpose-built oligonucleotide microarrays (DNA TAG3, Affymetrix, Santa Clara, CA) for 16 hr at 42°C. After staining with streptavidin-phycoerythrin (Molecular Probes), arrays were scanned at an emission wavelength of 560 nm by using an Affymetrix GeneChip Scanner (21). The hybridization intensities for each of the array elements were determined by using the Affymetrix GeneChip software.
Analysis of Deletion Pool Data.
For the analysis of strain prevalence in the pool, each strain was represented by four values of signal intensity (sense and antisense array elements for each of the two tags). These four values were averaged to give a single value for each strain present in the pool. To assess the degree of sensitivity to each of the DNA-damaging agents a ratio of treated/untreated signal for each strain was calculated. Those strains with lower signals in the treated compared with the untreated pool had a low ratio (sensitive strains), whereas those with similar signals from treated and untreated had ratios close to 1 (unaffected strains). These ratios were then ordered to produce a ranking, with the most sensitive strain having the lowest ratio. The final sensitivity ranking for each of the DNA-damaging agents was obtained by ordering the average ratio for the three replicate experiments for each agent.
Gene Expression Profiling.
An overnight culture of the parental strain, BY4743, was diluted to OD600 = 0.05 into fresh YPD at 30°C and 300 rpm in an orbital shaker for 5 hr before exposure to the DNA-damaging treatments. For the UV and IR treatments, samples were collected and cells were harvested at 30, 60, and 120 min after treatment. For all of the 1-hr exposure to the drugs, samples were taken at the end of the exposure, and further samples were taken 30 and 60 min later. Poly(A)+ RNA was isolated (Pharmacia Biotech) and converted into double-strand cDNA (Invitrogen), and complementary RNA biotinylated probes were prepared by in vitro transcription (Enzo Biochem) according to the Affymetrix Gene Expression Technical Manual. Probes were hybridized to whole yeast genome microarrays (YG-S98, Affymetrix), washed, and stained with the GeneChip Fluidics Station and scanned with the Affymetrix GeneChip Scanner according to manufacturer’s instructions. To minimize the problems of variations in the perfect match (PM) and mismatch (MM) signals for each of the 16 probes used for each gene on the Affymetrix array we used a recently developed statistical analysis (22) and associated “d-chip” software (http://www.biostat.harvard.edu/complab/dchip/). Only genes that were called present by the Affymetrix Microarray Suite Software and having at least a 2-fold greater gene expression ratio at the 90% confidence level were considered significant.
Results
Identification of the Deletion Strains Sensitive to the DNA-Damaging Agents.
We first performed pilot experiments with the diploid wild-type strain BY4743 to determine the dose giving between 50% and 80% cell survival in the wild-type strain. The agents used (and doses) were UV radiation (200 J/m2), IR (200 Gy), hydrogen peroxide (1 mM for 1 hr), and cisplatin (1 mM for 1 hr). We divided the pool of 4,627 deletion strains into two and treated one with one of the above agents with the other serving as the mock-treated control. We performed each experiment in triplicate and for each experiment determined the ratio of hybridization signal in the treated to that in the untreated sample. The strains were then ranked in ascending order of the mean value of this ratio (treated/untreated) to obtain a ranking of the strains in order of decreasing relative loss in the treated sample. We have shown previously for UV irradiation that this gives a semiquantitative ranking of relative sensitivities for cell killing, with the most sensitive strains having the lowest ratios (21). As previously reported (21), we found that the three replicate experiments gave very similar rankings for each of the agents, thus providing a high degree of confidence in the average ranking.
For the purpose of the comparison with gene expression profiling we chose to analyze the 100 genes whose deletion produced the greatest sensitivity to each DNA-damaging agent. We chose 100 because this would be expected to include essentially all of the hypersensitive and moderately sensitive strains with a minimum number of false positives. For example, with UV irradiation the top 100 ranked strains identified 29 of a possible 32 strains previously reported to be UV sensitive in addition to several deletion strains not previously known to be UV sensitive (21). However, most of the highly sensitive strains were in the top 50 (i.e., ranking 1 to 50). For example, with UV irradiation 22 known repair genes were in the top 50 with 2 in the second 50. For IR 11 strains with deletions in DNA repair genes were ranked in the top 50 strains with 2 in the second 50.
Although the mechanism of cell killing by each of the agents used is different, damage to DNA is a common feature. Consistent with this is the fact there was considerable overlap between deletion strains that were sensitive to each agent. For example, there was approximately 50% overlap of strains in the top 100 for sensitivity for IR, UV, and cisplatin, with somewhat less overlap between these three agents and hydrogen peroxide (≈35%). The number of strains with overlapping and unique sensitivities to each agent is shown in Table 1. Table 2 shows the 20 strains that were in the top 100 for all four of the DNA-damaging agents. The genes deleted in these strains are involved in postreplication repair (RAD5, HPR5, RAD18), homologous recombination (RAD59, MUS81, SAE2, MMS4, RAD57, RAD54, RAD55), nucleotide excision repair (RAD1), DNA-damage checkpoints (RAD9, MEC3, RAD17), and miscellaneous (RPL20A, YBR099C, THR1, NPL6, YLR426W, YIM1). Of those in the miscellaneous category, YBR099C is unlikely to encode a protein involved in DNA-damage repair, as the ORF overlaps that of MMS4 on the opposite strand, so its deletion would also disrupt the Mms4 protein. The absence of some genes in this list (such as RAD6 in postreplication repair and RAD52 in homologous recombination) can be explained by the low abundance of the relevant deletion strains in the control cultures (21).
Table 1.
Overlap of deletion strains in the top 100 in sensitivity to the DNA-damaging agents
| Agent | IR | UV | Cisplatin | H2O2 | Unique | All |
|---|---|---|---|---|---|---|
| IR | 100 | 48 | 45 | 35 | 32 | 20 |
| UV | 48 | 100 | 47 | 36 | 26 | 20 |
| Cisplatin | 46 | 48 | 100 | 35 | 30 | 20 |
| H2O2 | 35 | 36 | 35 | 100 | 48 | 20 |
The number of deletion strains in common in the top 100 most sensitive for each of four DNA-damaging agents. The identities of the 20 genes in the deletion strains that are common in the top 100 for each of the four agents (column headed “All”) are shown in Table 2.
Table 2.
Genes producing sensitivity to all the DNA-damaging agents
| Gene | Protein function | Sensitivity ranking
|
|||
|---|---|---|---|---|---|
| IR | UV | Cis | H2O2 | ||
| RAD5 | DNA helicase involved in postreplication repair | 1 | 6 | 7 | 10 |
| RAD9 | Involved in cell cycle checkpoints after DNA damage | 3 | 15 | 31 | 31 |
| MEC3 | DNA damage checkpoint; in complex with Rad17p and Ddc1p | 4 | 17 | 84 | 40 |
| RAD59 | RAD52 epistasis group; involved in dsb repair | 5 | 13 | 15 | 27 |
| MUS81 | DNA repair; interacts with Rad54p | 6 | 8 | 13 | 30 |
| SAE2 | Initiation of meiotic recombination and DNA repair | 7 | 19 | 8 | 11 |
| RAD57 | RAD52 epistasis group; involved in dsb repair; forms dimer with Rad55p | 8 | 18 | 28 | 12 |
| YBR099C | Hypothetical ORF on opposite strand to MMS4 | 10 | 12 | 16 | 18 |
| MMS4 | Repairs alkylating agent damage | 11 | 10 | 9 | 8 |
| THR1 | Homoserine kinase; involved in threonine biosynthesis | 14 | 14 | 33 | 37 |
| RPL20A | Ribosomal protein L20A (L18A) | 15 | 35 | 39 | 6 |
| NPL6 | Nuclear protein targeting | 18 | 63 | 96 | 33 |
| RAD55 | RAD52 epistasis group; interacts with Rad51p and Rad57p | 20 | 26 | 30 | 19 |
| RAD54 | RAD52 epistasis group; interacts with Rad51p and Mus81p | 22 | 20 | 54 | 14 |
| RAD17 | DNA damage checkpoint required for G2 arrest | 29 | 37 | 95 | 50 |
| YLR426W | Member of short chain alcohol dehydrogenase/ribitol dehydrogenase family | 30 | 87 | 62 | 100 |
| YIM1 | Mitochondrial inner membrane protease | 37 | 95 | 79 | 86 |
| RAD18 | Postreplication repair; partner of Rad6p | 39 | 21 | 35 | 9 |
| HPR5 | DNA helicase involved in DNA repair | 42 | 22 | 21 | 4 |
| RAD1 | DNA endonuclease involved in nucleotide excision repair | 81 | 5 | 3 | 58 |
The identities of the 20 genes whose homozygous deletion produces sensitivity to all four of the DNA-damaging agents are shown along with the sensitivity rankings to each of the agents. Cis, cisplatin; Rad17p, for example, indicates the protein encoded by RAD17; dsb, double-strand break.
Analysis of Gene Induction After Exposure to the DNA-Damaging Agents.
We used the identical doses and treatment conditions as used with the deletion pool to define the transcriptional response of wild-type cells to the same DNA-damaging agents. For UV and IR the exposure time was short (0.5 and 6 min, respectively) and we took samples at 30 min, 1 hr, 2 hr, and 4 hr after the exposure. The other two conditions involved drug exposures for 1 hr and we took samples for mRNA analysis immediately, 30 min, 1 hr, and 2 hr after the end of the 1-hr exposure period. For all of the conditions we obtained three replicate control samples, collected immediately after the end of the mock exposure to each of the DNA-damaging agents.
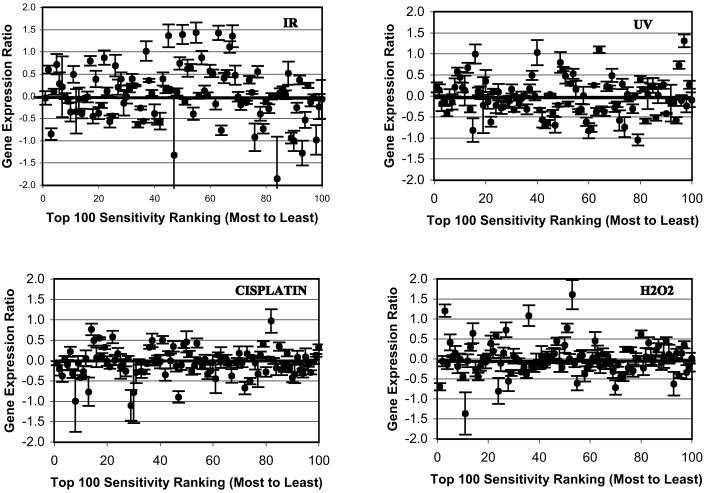
To address the question of whether the genes affecting sensitivity to the DNA-damaging agents are up-regulated in response to DNA damage we calculated the fold induction for each gene at each of the time points after treatment. Fig. 1 shows a plot of the fold induction for each of the 100 genes whose deletion produced the greatest sensitivity to the DNA-damaging agents examined. The ranking is from 1 to 100 with the lowest-ranked strains having the greatest sensitivity in the competitive hybridization assay. For clarity we have averaged the fold induction in the wild-type strain over the first three time points after exposure to the agents. This period encompassed the largest changes in gene expression produced by the agents. A visual examination of Fig. 1 suggests that there is no generalized increase in expression over the averaged time period of those genes whose products conferred resistance to the DNA-damaging agents. To ensure that we were not losing real changes in gene expression at single time points, we also fitted the best fitting trend of gene expression for the genes 1 to 100 for each of the four time points after exposure to each agent. There was no trend in gene expression as a function of sensitivity ranking for any of the time points (data not shown). Table 3 shows the log2 of the mean value of the ratio of gene expression in the treated to control samples for all 100 genes for each of the time points after exposure. In no case is there a significant increase in gene expression over this group of 100 genes. Thus, although expression level of some of the genes is increased, the increase is balanced by a decrease in expression in an equivalent number of genes. This analysis shows that there is no generalized increase in expression of genes that are involved in the resistance of S. cerevisiae to these DNA-damaging agents.
Figure 1.
The relationship between gene expression and sensitivity to the four DNA-damaging agents. In each panel the 100 most sensitive strains with homozygous deletion of different genes are ranked from 1 to 100 from most to least sensitive strains. For each gene in these 100 most sensitive deletion strains the ratio of gene expression compared with controls is averaged for the first three time points after exposure of the wild-type strain to the DNA-damaging agent. The y axis is plotted as log2 of gene expression ratio, which gives equal distance to a doubling and halving of the gene expression in the treated compared with control samples. Each gene is shown ± 1 standard error of the expression ratio based upon the three different time points for that gene.
Table 3.
Mean gene expression ratios (log2) for the 100 most sensitive strains
| Time after exposure, hr | IR | UV | Cisplatin | H2O2 |
|---|---|---|---|---|
| 0 | — | — | 0.05 (0.06) | 0.08 (0.12) |
| 0.5 | 0.09 (0.16) | 0.03 (0.10) | −0.07 (0.09) | 0.02 (0.11) |
| 1 | −0.02 (0.19) | −0.11 (0.10) | −0.13 (0.13) | 0.04 (0.09) |
| 2 | 0.09 (0.9) | 0.04 (0.10) | −0.08 (0.09) | −0.17 (0.08) |
| 4 | 0.01 (0.13) | −0.11 (0.10) | — | — |
The 95% confidence interval is given in parentheses.
With the above analysis, however, it might be possible to miss an increase in expression of a small number of genes by averaging expression levels over the 100 genes. We therefore examined all genes induced by a factor of 2 or more by each of the DNA-damaging agents at each time point after exposure and determined how many of the these genes conferred resistance to the DNA-damaging agents. Any increase over what would be expected by chance from the total number of induced genes would suggest that at least some of the genes necessary for survival were transcriptionally activated by DNA damage. As can be seen from Table 4, there is no suggestion that this is the case: At no time point after treatment are the numbers of “resistance” genes more represented in the list of induced genes compared with that expected by chance.
Table 4.
Total number of genes induced greater than 2-fold by the DNA-damaging agents and total number of the 100 most effective “resistance” genes that are induced at each time period after exposure
| Time after exposure, min | IR
|
UV
|
Cisplatin
|
H2O2
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. >2× | No. in top 100 >2× | Expected by chance | No. >2× | No. in top 100 >2× | Expected by chance | No. >2× | No. in top 100 >2× | Expected by chance | No. >2× | No. in top 100 >2× | Expected by chance | |
| 0 | — | — | — | — | — | — | 53 | 0 | 0.85 | 423 | 3 | 6.80 |
| 30 | 689 | 12 | 11.07 | 320 | 5 | 5.14 | 181 | 1 | 2.91 | 281 | 5 | 4.51 |
| 60 | 432 | 8 | 6.94 | 183 | 1 | 2.94 | 325 | 2 | 5.22 | 141 | 3 | 2.27 |
| 120 | 156 | 2 | 2.51 | 351 | 5 | 5.64 | 149 | 0 | 2.39 | 94 | 0 | 1.51 |
| 240 | 467 | 7 | 7.50 | 247 | 2 | 3.94 | — | — | — | — | — | — |
The number of genes induced by more than 2-fold by each of the DNA-damaging agents at each time point after treatment is shown both for the total number of genes and for the number of genes within the top 100 whose deletion causes the greatest sensitivity. The columns headed “Expected by chance” show the number of genes induced by more than 2-fold in the top 100 sensitive strains that would be expected by chance if there were no relationship between the genes induced and those whose deletion produces sensitivity to the agents. It is calculated as 100 × N/6224, where N = number of genes induced at that time period.
Discussion
In the present study we asked the question of whether the genes important for survival of S. cerevisiae to DNA-damaging agents are the ones whose transcription is increased after exposure to these agents. The recent completion by the Saccharomyces Deletion Project of the systematic deletion of each ORF in budding yeast has enabled this question to be addressed in a comprehensive manner. For all nonessential genes a systematic comparison can be made of sensitivity and gene expression by using the same strains under identical conditions of exposure. We chose four different agents that damage DNA in different ways and that require different enzymes to repair the damage. For IR the principal toxic lesion is the DNA double-strand break, for UV it is a pyrimidine dimer, for hydrogen peroxide it is DNA single-strand breaks and base damage, and for cisplatin it is interstrand and/or intrastrand DNA cross-links (23).
Our data show no evidence of a relationship between the genes whose expression is increased after these different DNA-damaging agents and the genes involved in protecting against cytotoxicity to the same agents. We demonstrate this in two ways. First, we found no overall increase in expression levels in the top 25, 50, or 100 genes whose deletion produces sensitivity to the agents used. Second, we found that although the expression levels of some genes involved in resistance to the DNA-damaging agents (such as RAD51 and RAD54) were increased after DNA damage, the number of such genes was no more than was expected by chance, given the large number of genes with increased expression levels.
Although this conclusion is contrary to the widespread view that at least some of the genes induced after DNA damage are involved in protecting the cell against the damage, ours is not the first evidence against this assumption. For example, Jelinski et al. (12), who studied the response of S. cerevisiae to four alkylating agents, tert-butyl hydroperoxide, and IR, found a remarkably different transcriptional response among these DNA-damaging agents, with only 21 genes (12 induced and 9 repressed, of which none were DNA-repair genes) responding to all of the agents. Similarly, Gasch et al. (24) found few changes in the transcription of genes involved in DNA-damage repair in S. cerevisiae exposed to methyl methanesulfonate (MMS) or IR, although they did identify one cluster of 9 of approximately 500 induced genes that they considered a specific signature of DNA damage. These included two genes involved in homologous recombination (RAD51 and RAD54) as well as the ribonucleotide reductase subunit genes RNR2 and RNR4. We also found in the present study that all 9 of these genes were induced by more than factor of 2 by all four of the DNA-damaging agents.
Two reasons have been proposed for the small proportion of genes known to be involved in protecting against DNA damage that are induced by DNA-damaging agents. The first is that so-called DNA-damaging agents also damage many other cellular macromolecules, which in turn could induce expression of protein chaperone and proteasome genes (as has been observed for MMS treatment (12, 15). The other (and somewhat overlapping) explanation is that cytotoxic treatments produce a general stress response that is independent of DNA damage (15). Either of these explanations would be consistent with the small proportion of genes specifically required for protection against DNA damage in the general response to the agents. However, although DNA-damaging agents undoubtedly cause considerable damage to other cellular macromolecules and elicit a generalized stress response, the possibility that they also induce the expression of genes required to protect the cells from the insult is not supported by our data, as we were able to examine the expression of all nonessential genes that were important for the survival to the different DNA-damaging agents used. The fact that we found no increased gene expression (relative to the population as a whole) in the small subset of the 100 genes most important for survival after exposure to each agent strongly suggests that there are few, if any, genes necessary for surviving DNA damage that are induced at least by an acute exposure to these agents.
Despite these conclusions, the finding that DNA-damaging agents induce the expression of genes involved in DNA repair (18, 19, 24) must be considered. However, as so many transcripts are changed after environmental stress or DNA damage, there is high probability that genes involved in DNA damage would be part of this response without necessarily contributing to cellular protection. For example, with some 150 genes having a categorized cellular role of “DNA repair” (https://www.incyte.com/proteome/YPDsearch-long.html), an average of 5 “DNA repair” genes would be expected by chance to be in every 200 induced genes. However, direct evidence that genes involved in DNA repair can be induced by DNA damage without contributing to the response to the damage is provided by earlier work showing that when the promoter regions of RAD2 and RAD54 responsible for increased gene expression were deleted so that no induction occurred after UV or IR exposure, respectively, there was no change in the sensitivity of the mutated strains to UV or to IR (25, 26). Other evidence that wild-type levels of at least some repair proteins are sufficient to provide full resistance to DNA damage is provided by the lack of effect of overexpression of RAD52 mRNA on sensitivity of yeast to MMS (27). The apparently nonspecific nature of the DNA-damage response has also been reported for cell cycle-regulated genes, with the induction of most cell cycle genes in cells arrested in S phase after MMS treatment similar to that in cells arrested in G2/M after IR exposure (24).
The present data imply that despite the extensive changes in gene expression in response to the different DNA-damaging agents, few if any of these changes were necessary to protect the viability of the cell against these agents. This implication suggests that endogenous levels of the various proteins involved in protecting against DNA damage are at sufficient levels to provide full resistance to the lesions produced by the agents used. This is perhaps not surprising. With the exception of UV irradiation, it is unlikely that the richness of the highly conserved DNA-repair pathways in prokaryotes and eukaryotes evolved to cope with the precise lesions produced by the DNA-damaging agents used in the present study. However, in recent years the crucial involvement of recombination to repair broken DNA during its replication as well as to maintain genomic stability has become apparent (28). Thus, in addition to their widely recognized role in the proper segregation and shuffling of genetic material during meiosis, the enzymes of recombination are now seen to play a vital role in repairing damage similar to that introduced by IR that occurs normally during DNA synthesis as a result of stalled replication forks or other processes that break DNA (29–31). As the generation of reactive oxygen species, including hydrogen peroxide, is also a universal feature of aerobic metabolism (32), it is also reasonable that the enzymes necessary for the repair of oxidative DNA damage are at sufficient levels to protect the cell. Thus, the requirement for the integrity of DNA, its need to break and rejoin during normal cellular processes, and its vulnerability to damage during normal replication, oxidative metabolism, and environmental insults all contribute to the need for sufficient levels of endogenous DNA-repair enzymes. This is probably the reason that most, if not all, of the enzymes protecting against DNA damage do not need to be induced after damage by exogenous agents.
Not addressed by the present study is the question of whether expression profiling can provide information on the mechanism of action of agents (e.g., drugs) that have non-DNA targets. One such approach is to compare the gene expression profile after treatment with a large number of expression profiles of single-gene mutants, and deduce the target from the similarity of the expression profile with that of a known gene deletion (14, 33). Another relevant question is whether any of the genes whose expression was changed by the DNA-damaging agents are involved in repairing DNA damage other than that leading to cell death (for example damage leading to mutations or genomic instability). However, a search of the genes involved in mismatch repair and genes whose mutations lead to chromosomal instability (34) failed to support this possibility—these genes were not induced after the exposures given in the present study.
In summary, the present data show that few, if any, of the genes involved in repairing the various potentially lethal DNA lesions produced in this study, including double-strand breaks, pyrimidine dimers, single-strand breaks, base damage, and DNA crosslinks, are induced in response to exposure to the agents that produce these lesions. Although some genes involved in repairing these lesions are induced by DNA damage, their number is no more than can be accounted for by chance, assuming that gene induction is nonspecific. This finding raises serious questions about any conclusions deduced from gene expression profiling as to the nature of DNA lesions in cells or to the genes involved in their repair.
Acknowledgments
This work was supported by National Institutes of Health Grants R01 CA15201 and P01 CA67166 (to J.M.B.) and HG00205 (to R.W.D.), a grant from Aventis (to R.W.D.), and Australian National Health and Medical Research Council Fellowship 997034 (to G.W.B.).
Abbreviations
- IR
ionizing radiation
- MMS
methyl methanesulfonate
References
- 1.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Nature (London) 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee A, Richards W G, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shipp M A, Ross K N, Tamayo P, Weng A P, Kutok J L, Aguiar R C, Gaasenbeek M, Angelo M, Reich M, Pinkus G S, et al. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 5.Kihara C, Tsunoda T, Tanaka T, Yamana H, Furukawa Y, Ono K, Kitahara O, Zembutsu H, Yanagawa R, Hirata K, et al. Cancer Res. 2001;61:6474–6479. [PubMed] [Google Scholar]
- 6.Clark E A, Golub T R, Lander E S, Hynes R O. Nature (London) 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 7.Saha S, Bardelli A, Buckhaults P, Velculescu V E, Rago C, St Croix B, Romans K E, Choti M A, Lengauer C, Kinzler K W, Vogelstein B. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 8.Welsh J B, Sapinoso L M, Su A I, Kern S G, Wang-Rodriguez J, Moskaluk C A, Frierson H F, Jr, Hampton G M. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 9.Tackels-Horne D, Goodman M D, Williams A J, Wilson D J, Eskandari T, Vogt L M, Boland J F, Scherf U, Vockley J G. Cancer. 2001;92:395–405. doi: 10.1002/1097-0142(20010715)92:2<395::aid-cncr1335>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 10.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 11.Jelinsky S A, Samson L D. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelinsky S A, Estep P, Church G M, Samson L D. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 14.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 15.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckardt-Schupp F, Klaus C. Biochimie. 1999;81:161–171. doi: 10.1016/s0300-9084(99)80049-2. [DOI] [PubMed] [Google Scholar]
- 18.Schaus S E, Cavalieri D, Myers A G. Proc Natl Acad Sci USA. 2001;98:11075–11080. doi: 10.1073/pnas.191340698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 21.Birrell G W, Giaever G, Chu A M, Davis R W, Brown J M. Proc Natl Acad Sci USA. 2001;98:12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Wong W H. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickoloff J A, Hoekstra M F, editors. DNA Damage and Repair. Totowa, NJ: Humana; 1998 and 2001. [Google Scholar]
- 24.Gasch A P, Huang M, Metzner S, Botstein D, Elledge S J, Brown P O. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siede W, Robinson G W, Kalainov D, Malley T, Friedberg E C. Mol Microbiol. 1989;3:1697–1707. doi: 10.1111/j.1365-2958.1989.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 26.Cole G M, Mortimer R K. Mol Cell Biol. 1989;9:3314–3322. doi: 10.1128/mcb.9.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dornfeld K J, Livingston D M. Mol Cell Biol. 1991;11:2013–2017. doi: 10.1128/mcb.11.4.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores-Rozas H, Kolodner R D. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisby M, Rothstein R, Mortensen U H. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton M D, Walker G C. Proc Natl Acad Sci USA. 2001;98:8342–8349. doi: 10.1073/pnas.111036998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel B, Flores M J, Viguera E, Grompone G, Seigneur M, Bidnenko V. Proc Natl Acad Sci USA. 2001;98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henle E S, Linn S. J Biol Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 33.Marton M J, DeRisi J L, Bennett H A, Iyer V R, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, et al. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Kolodner R D. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]