Abstract
At slightly acidic pH, the association of two d(5mCCTCACTCC) strands results in the formation of an i-motif dimer. Using NMR methods, we investigated the structure of [d(5mCCTCACTCC)]2, the internal motion of the base pairs stacked in the i-motif core, the dimer formation and dissociation kinetics versus pH. The excellent resolution of the 1H and 31P spectra provided the determination of dihedral angles, which together with a large set of distance restraints, improve substantially the definition of the sugar-phosphate backbone by comparison with previous NMR studies of i-motif structures. [d(5mCCTCACTCC)]2 is built by intercalation of two symmetrical hairpins held together by six symmetrical C•C+ pairs and by pair T7•T7. The hairpin loops that are formed by a single residue, A5, cross the narrow grooves on the same side of the i-motif core. The base pair intercalation order is C9•C9+/5mC1•5mC1+/C8•C8+/C2•C2+/T7.T7/C6•C6+/C4•C4+. The T3 bases are flipped out in the wide grooves. The core of the structure includes four long-lived pairs whose lifetimes at 15°C range from 100 s (C8•C8+) to 0.18 s (T7•T7). The formation rate and the lifetime of [d(5mCCTCACTCC)]2 were measured between pH 6.8 and 4.8. The dimer formation rate is three to four magnitude orders slower than that of a B-DNA duplex. It depends on pH, as it must occur for a bimolecular process involving non cooperative association of neutral and protonated residues. In the range of pH investigated, the dimer lifetime, 500 s at 0°C, pH 6.8, varies approximately as 10−pH.
INTRODUCTION
In slightly acidic solution, oligonucleotides containing cytidine stretches can adopt a four-stranded structure, the i-motif, built by two parallel duplexes intercalated into each other in a head to tail orientation. The strands of each duplex are held together by hemiprotonated C•C+ pairs (1). Tight stacking of the intercalated pairs generates a rigid core of long-lived base pairs. In most of the i-motif structures reported, the core is formed of intercalated C•C+ pairs but T•T pairs can be also incorporated in the i-motif core as shown for the tetramers of d(5mCCTCC) (2), d(5mCCTCTCC) and d(5mCCTCCCTCC) (3).
In the course of a systematic research of oligonucleotides containing non-C residues that can associate into i-motif structures, we found that d(5mCCTCACTCC) and d(5mCCTCICTCC) form stable hemiprotonated structures whose NMR spectra exhibit an exceptionally good resolution. In contrast, the poorly resolved spectrum of d(5mCCCCACCCC) shows the formation of several intercalated structures that could not be resolved. A 5mC residue, providing a marker readily identifiable, was incorporated at the 5′ end of all the oligonucleotides investigated.
The titration of the multimer of d(5mCCTCACTCC) shows the formation of a dimer. The high definition structure (root mean square deviation, RMSD = 0.66 Å) computed using 278 distance restraints derived from NOESY experiments and 17 dihedral restraints from E COSY and 1H-31P COSY experiments shows two symmetry related hairpins associated into an i-motif structure.
The structures of at least 15 i-motif monomers, dimers or tetramers have been solved by X-ray or NMR methods in the last 12 years, but little is known about the i-motif formation–dissociation kinetics (4). The spectra of d(5mCCTCACTCC) and [d(5mCCTCACTCC)]2 exhibit several well-resolved NMR lines allowing the measure of the proportion of each species. This provided favorable conditions for a kinetic study of the equilibrium between the monomer and the i-motif dimer. kon, the dimerization rate of d(5mCCTCACTCC) and Kdis, the dimer dissociation constant, were measured between pH 6.8 and 4.8 at 0 and 15°C. The dimer dissociation kinetics were measured versus pH on NMR spectra collected at interval right after dilution of concentrated fully dimeric samples.
MATERIALS AND METHODS
Oligonucleotide preparation and NMR samples
The oligonucleotides were synthesized on a 15 µM scale, purified by chromatography on a DEAE column and dialyzed against water. The strand concentration was determined from the absorbance measured at neutral pH using the A260 value of 68 000 M−1 cm−1. The strand concentration was 5–10 mM in the structural studies, 1–3 mM in proton exchange experiments and 50 µM to 0.5 mM in the kinetics studies of the mononer–dimer equilibrium.
The samples were dissolved either in H2O/2H2O (9/1, v/v) or in 99.98% 2H2O with 1 mM ethylene diamine tetra-acetic acid and 0.2 mM dimethyl silapentane sulphonate (DSS) for 1H and 13C chemical shift references. The 31P spectra were scaled on trimethyl phosphate (3.45 p.p.m. downfield of H3PO4 85%). In the structural studies, the sample pH was adjusted close to the cytidine pK (pKN3 = 4.3) with NaOH and HCl solutions. In the kinetics experiments, the samples were buffered by 20 mM potassium phosphate and 0.5 mM sodium acetate. The chemical shift of the acetate methyl protons provided a marker for pH determination according to: pH = 4.63 − log(1.909 − δacetate)/(δacetate − 2.076) at 0°C.
Imino proton exchange
The proton exchange methodology has been extensively described (5). Imino proton exchange from a base pair requires disruption of the pair. The imino proton of C•C+ pairs exchanges with water at each opening event, so that the exchange time is equal to the base pair lifetime (6). The H-bonded imino proton of T and G residues is transferred to water from the open base pair via a proton acceptor (e.g. phosphate) acting as a catalyst. The base pair lifetime is obtained in that case as the extrapolation of the imino proton exchange time at infinite proton acceptor concentration. Exchange times longer than minutes were obtained from the deuteration rate of a protonated sample diluted into 2H2O. Values in the range of 10−2 to seconds were determined by magnetization transfer from water. Exchange times in the range of milliseconds were identified with the imino proton longitudinal relaxation times.
Association and dissociation kinetics of the dimer of d(5mCCTCACTCC)
The monomer–dimer equilibrium, kon [monomer]2 = koff [dimer], is the balance of a bimolecular reaction that depends on the collision rate and effectiveness with which each collision results in strand association and on the dimer dissociation rate.
In a solution where the oligonucleotide is initially in the monomeric form, the dimer concentration versus time, [D]t, depends on the oligonucleotide concentration, [s], the dimer fraction at equilibrium, f, and the dimerization rate kon:
| 1 |
where
| 2 |
The dimer dissociation constant may be expressed as a function of the oligonucleotide concentration and of the dimer fraction at equilibrium by:
| 3 |
The dimer lifetime
| 4 |
The dimer and monomer concentrations were determined from the area or from the peak height of several markers. The C imino and amino protons, the T imino protons, T7 methyl protons, A(H8), A(H2) and T7(H6) may be used to measure the dimer concentration. The monomer concentration was measured using the adenine aromatic protons. Two dimer markers allowing a better accuracy were selected, in particular for the determination of the shortest reaction rates or for experiments requiring extreme sample dilution. The methyl proton peak of T7(CH3) was selected for its larger intensity and T3(H3) for its short relaxation time (30–50 ms) allowing fast accumulation rate without signal saturation. In the range of pH investigated, the association rate varies approximately as the product: [s] 10−pH. It could be measured between pH 6.8 and 4.8. Measurements at lower pH would require sample dilution inappropriate to NMR measurements.
To measure kon, the oligonucleotide solution was heated at 100°C in the NMR tube, rapidly cooled during ∼15 s by immersion in a beaker containing water at the temperature of the experiment (0 or 15°C) and inserted into the NMR probe at controlled temperature. To determine koff, a small volume of a 5 mM fully dimeric solution was rapidly diluted by a factor of 10–100 into the NMR tube. The NMR tube was maintained during ∼15 s in a water bath at controlled temperature for temperature equilibration and inserted into the NMR probe. The magnet homogeneity was always preset with a dummy sample. The dead time to start the kinetics measurements was ∼1 min. The number of FID in each block of the series of spectra recorded to follow the kinetics was adapted to the reaction rate and to the sample concentration. The recovery delay was optimized according to the relaxation time of the observed proton. The sample temperature, controlled on the first accumulated spectrum by the proton frequency of the DSS methyl proton, was always found within 2°C of the preset temperature.
NMR methods
The NMR experiments were performed on a 500 MHz Varian Inova spectrometer equipped with a penta probe.
In H2O solution, the Jump and return (JR) detection sequence was used for water suppression (7) with maximum sensitivity at 13.5 p.p.m. The NOESY experiments were performed with (256 × 2048) complex points, a spectral width of 12 KHz in both dimensions and a relaxation delay of 1 s.
The NOESY, TOCSY and ECOSY experiments in 2H2O solution were acquired with (256 × 2048) complex points, spectral widths of 4 kHz in both dimensions and a relaxation delay of 3 s. The 1H2HO signal was saturated with a 2 s low power pulse. The mixing time was randomly varied by ±5% in order to reduce the spurious contribution of zero-quantum coherence signals at short mixing time (8). The TOCSY experiments used 10 MLEV-17 repetitions (total time 15 ms) (9). The 1H-31P TOCSY (10) and 1H-31P COSY (11) experiments were collected with (120 × 2048) complex points and spectral widths of 1.6 and 0.6 kHz in the 1H and 31P dimensions, respectively. The HSQC experiments in natural abundance were performed with (130 × 2048) complex points, spectral widths of 4 and 7 kHz in the 1H and 13C dimension, respectively, and a relaxation delay of 3 s.
The 2D data were processed on INDIGO workstation (Silicon Graphics, Inc.) using the Felix 97.2 software (Biosym). Apodization in both dimensions included a 1 Hz exponential broadening, a sine-bell with a 45–60° phase shift in 1H experiments or 80° in heteronuclear experiments and a Felix skew factor of 1. In H2O solution, the residual water signal was reduced using the time domain convolution function of the Felix software and the spectra were multiplied by ad hoc 1/sin function in the t2 dimension in order to correct the frequency response to JR detection.
Distance restraint and molecular dynamics
The inter-proton distance restraints were obtained from the build-up of NOE cross peaks measured at 5°C with mixing times of 30, 50, 70, 90 and 250 ms in 2H2O and of 50, 70, 90 and 150 ms in H2O. The cross-peak volumes were scaled by reference to the H5-H6 cross peaks (2.45 Å) or to the intra-residue methyl-H6 cross peaks (2.9 Å) in the case of cross peaks involving a methyl group. The intra-residue distances were restrained to the measured values with upper and lower bounds of 10–20%, depending on the spectral resolution. The inter-residue distance restraints were sorted into three categories with upper and lower limits of 1.8–2.9, 1.8–3.7 and 2.9–4.7 Å. In the case of cross peaks involving a pair of geminal protons and a third partner (e.g. H2′/H2″-H1′), or two pairs of geminal protons (amino-H2′/H2″ or H2′/H2″-H5′/H5″), we used only the strongest cross peak as a restraint. To avoid distance misevaluation due to spin diffusion, the distance(s) derived from the weakest cross peak(s) was (were) considered as lower bounds. This procedure accounts for the relatively large number of repulsive restraints used in the structural calculation (Table 1). The structures were calculated using the simulated annealing method of the X-PLOR 3.851 program (12) as reported previously (3). The inter-proton distances shorter than 4.7 Å were systematically searched on the computed conformers using the MOLMOL 2.4 software (13). Distances incompatible with the NOESY spectra were excluded in further calculations by repulsive constraints of 4.2 Å. This procedure was used only for well-resolved non-exchangeable protons. The computed conformers were minimized by 300 Powell cycles, sorted according to their energy and aligned on the C1′-N1 vectors of the paired residues. The 10 structures of lowest energy were selected for structural analysis. The pairwise RMSD the geometry parameters and SDs were computed using X-PLOR and homemade softwares. The structures were visualized using MOLMOL 2.4.
Table 1.
Distance and dihedral restraints, violations and deviations from the ideal geometry of [d(5mCCTCACTCC)]2
| Intra-residue restraintsa | 103 | ||
| Inter residue restraints exchangeable protonsb | 45 | ||
| Inter residue restraints, non-exchangeable protons | 40 | ||
| Base-pairing restraintsc | 20 | ||
| Repulsive restraintsd | 90 | ||
| Number of restrained dihedral anglese | 17 | ||
| Violations of input restraintsf | Number | RMSD | |
| NOE violations >0.2 Å | 4.2 ± 1.3 | 0.067 ± 0.05 | |
| Dihedral violations >6° | 3.2 ± 0.7 | 4.4 ± 1.4 | |
| Deviations from ideal geometry | |||
| Angular deviation deviations >6° | 6.4 ± 0.7 | 1.7 ± 0.5 | |
| Deviations on bonds length >0.05Å | 0 | 0.008 ± 0.001 | |
| Improper deviation >6° | 0 | 0.9 ± 0.05 | |
| Number of van der Waal contacts | 0 | ||
| Pairwise RMSD in structural computation (Å) (f) | Bases | Sugar | P group |
| 0.38 | 0.41 | 1.1 | |
aThe same intra-residue distance restraints were imposed between each pair of symmetry-related residue.
bThe same inter residue distance restraints were imposed to two for each strand in order to enforce their equivalence.
cThree for each C•C+ pairs and two for pair T7•T7.
dDistance restrains defined by a lower bound (cf. Material and Method section).
eThe Pseudorotation angle of each residue and the ɛ angle of eight residues.
fComputed for the 10 selected conformers.
RESULTS
Exchangeable proton spectra and multimer stoichiometry
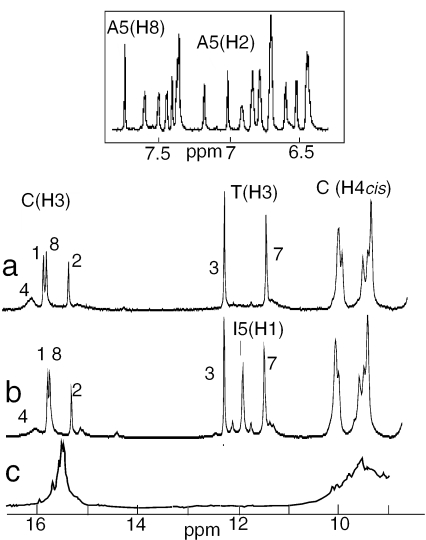
The proton spectrum of the multimer of d(5mCCTCACTCC) at acidic pH (Figure 1a) shows the imino proton peaks (15–16 p.p.m.) and two amino proton groups (∼9.5 and 8.4 p.p.m.) characteristic of hemiprotonated C•C+ pairs. The observation of a single peak for each thymidine imino proton (11.3 and 12.1 p.p.m.) is a first indication of the strand equivalence within the multimer. The 2H2O spectrum of the aromatic-H1′ region displayed in the inset of Figure 1 shows the excellent spectral resolution. The proton spectra of the multimer of d(5mCCTCICTCC) (Figure 1b) exhibits the extra I(H1) proton peak (10.9 p.p.m.). Otherwise, both spectra are quite similar, indicating the formation of comparable structures. The spectrum of d(CCCCACCCC) (Figure 1c) shows also the formation of hemiprotonated C•C+ pairs but the poor spectral resolution indicates multiple strand arrangements that could not be resolved.
Figure 1.
Exchangeable proton region of the multimers of (a) d(5mCCTCACTCC); (b) d(5mCCTCICTCC)]2; and (c) d(5mCCCCACCCC). The imino protons are labeled by the residue number. The well-resolved aromatic-H1′ proton region of the multimer of d(5mCCTCATCTCC) in 2H2O solution is shown in inset. T = 5°C, pH 4.3.
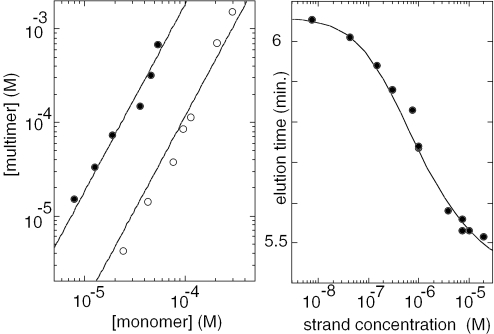
The multimer stoichiometry was determined by NMR titration and gel filtration chromatography. The NMR titration was performed at pH 5.6 in order to reduce the i-motif stability so as to shift the monomer and dimer concentrations at equilibrium in a range of values accessible to NMR detection. The observation of distinct NMR lines for the equivalent protons of the monomer and multimer species shows that the association–dissociation kinetics is slow on the NMR time scale. The NMR spectra of d(5mCCTCACTCC) solutions (25 µM to 2 mM) show that the multimer concentration increases as the square of the monomer concentration (Figure 2, left) and thus reveals the formation of a dimer. The dimer dissociation constants corresponding to the fits are 5.5 × 10−6 and 8.7 × 10−5 M−1 at 0 and 25°C, respectively.
Figure 2.
Determination of the multimer stoichiometric. Left panel: NMR titration at equilibrium. pH 5.6, 0°C (closed circles) and 25°C (open circles). The multimer concentration that increases as the power of two of the monomer concentration reveals dimer formation. Right panel: determination of the stoichiometry by gel filtration chromatography, pH 4.5, at room temperature. The monomer–multimer inter-conversion time is shorter than the elution times. A single peak is eluted at a position depending on the strand concentration. The fit of the peak position versus the strand concentration of the eluted solution was computed for a dimer–monomer equilibrium with a dissociation constant 5 × 10−7 M−1.
The chromatograms of d(5mCCTCACTCC) solutions injected on a gel filtration column at 22°C, pH 4.5, show a single peak whose elution time decreases with the strand concentration. This indicates that the monomer–dimer inter-conversion time is shorter that the elution time (5–6 min). The elution times, telu, plotted in the right panel of Figure 2 versus the eluted oligonucleotide concentration, [s], were fitted according to expression: telu = (1 − f) tmono + ftdimer, where tmono and tdimer are the elution times of the monomeric and dimeric species. The dimer fraction, f, is related to the dissociation constant by Kdis = [s](1 − f)2/f. The fit displayed was obtained with the dissociation constant: Kdis = 5 × 10−7 M−1.
Spectral identifications
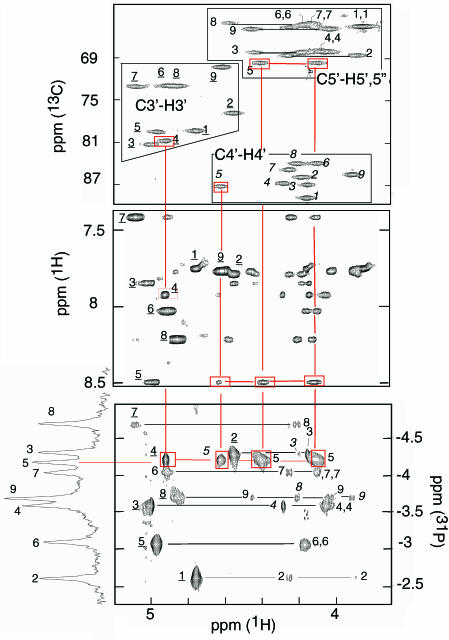
The observation of nine spin systems shows the strand equivalence within the dimer. The non-exchangeable protons of each spin system were identified using TOCSY and NOESY experiments at short mixing times (30 and 50 ms) according to standard procedures (14). The H4′ and H5′/H5″ protons were distinguished from each other by their connectivity to 13C4′ (82–92 p.p.m.) or 13C5′ (68–63 p.p.m.) by an HSQC experiment in natural abundance (Figure 3). The 3′ and 5′ end residues were identified on the 1H-31P TOCSY spectrum displayed in Figure 4 by their lack of 31P-H3′ or 31P-H5′/H5″ connectivities. Starting from the end residues, the nine spin systems were sequentially connected by the (H3′)n−1-(31P)n-(H4′/H5′/H5″)n cross peaks.
Figure 3.
Proton assignments and sequential through-bond identifications in [d(5mCCTCATCTCC)]2. The cross peaks are labeled by the residue number (underlined for H3′ italic for H4′). Upper panel: identification of the H3′, H4′, H5/H5″ protons on the HSQC spectrum in natural abundance. Central panel: Aromatic-H3′/H4′/H5′/H5″ region of the NOESY spectrum (mixing time 250 ms). Lower panel: 1H-31P TOCSY experiment providing sequential identification through (H3′)n+1-(31P)n-(H4′-H5′/H5″)n cross peaks. The red lines connect the C4(H3′)-A5(31P), A5(31P)-A5(H4′/H5′/H5″) cross peaks to the corresponding cross-peaks of the HSQC and NOESY spectra. Solution condition: T = 5°C, pH 4.4, 2H2O solution, strand concentration 6 mM.
Figure 4.
Selected regions of the NOESY (mixing time 50 ms) and 1H-31P COSY spectra of [d(5mCCTCACTCC)]2. The H3′ and some H5′ protons are labeled by the residue number, black and red, respectively. Top panel: H2′/H2″-H3′ intra-residue cross peaks and sequential (H2′/H2″)n-(H5′)n+1 cross-peaks between: (α) T7(H2″)-C8(H5′); (β) C8(H2″)-C9(H5′); (χ) C2(H2″)-C3(H5′). With the exception of C8(H2″)-C9(H5″) (data not shown) the corresponding H2′/H2″-H5″ cross peaks are absent. Central Panel: The strong intensity of the intra-residue H3′-H6/H8 cross peaks indicates the N-conformation of C6, T7, C8 and C9. Lower panel: The 1H-31P COSY spectrum shows systematic weak H3′-31P COSY cross peaks for the sugar in the N conformational range. Solution condition: T = 5°C, pH 4.4, 2HO solution, strand concentration 6 mM.
The exchangeable protons of pairs 5mC1•5mC1+, C2•C2+ and C8•C8+ were identified by the intra-residue imino/amino H5/H6 NOESY cross peaks. Selective saturation at −5°C of the broad 16.1 p.p.m. peak shows a noe to C4 (H4cis) that assigns this peak to the imino proton of pair C4•C4+. The imino protons of pairs C1•C1+ and C6•C6+ are not observed owing to fast exchange with water. N3 protonation shifts the amino protons of non-paired cytidines by 1.7 and 2 p.p.m. (15). As expected for hemiprotonated cytidines, the amino proton chemical shifts of [d(5mCCTCACTCC)]2 are about midway between those of neutral and protonated cytidines and they are unchanged between pH 4.3 and 6.8. The thymidine imino protons were identified by the intra-residue NOESY cross peaks observed at long mixing time with their own methyl groups. In the dimer of d(5mCCTCITCC), I(H1) was assigned by its noe to I(H2).
Dimer structure
i-Motif dimer can be formed by association of two hairpins, either in head-to-tail or in head-to-head orientation (16). The cytidine imino protons of [d(5mCCTCATCC)]2 are noe-connected to a single set of amino proton peaks. This indicates that the C•C+ pairs are symmetrical and, since the paired cytidines of i-motif belong to parallel strands, this implies that the two hairpin loops are on the same side of the i-motif core.
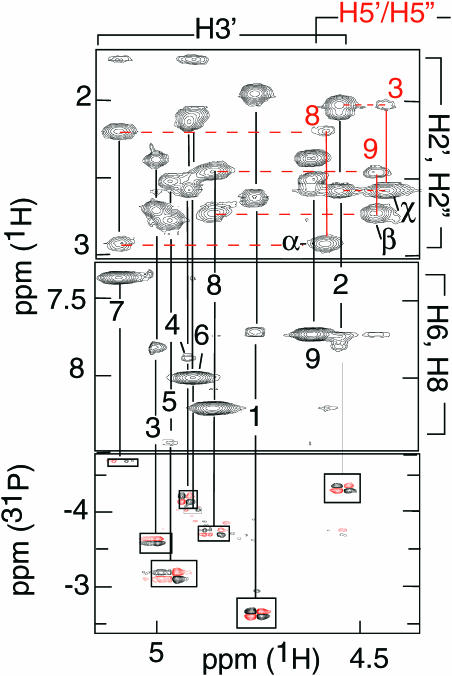
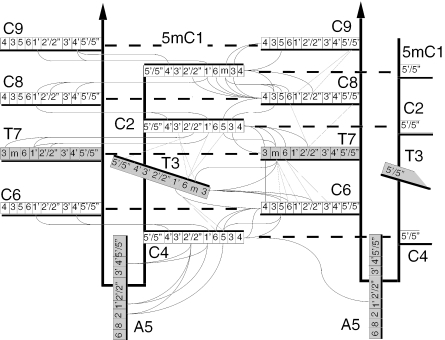
The C•C+ pairs stacked in i-motif structures are connected by well-characterized NOESY cross-peaks (3). The base stacking order, C9/5mC1/C8/C2/T7/C6/C4, was determined by the H1′-H1′ and amino/imino cross peaks connecting C9/5mC1/C8/C2/T7 and C6 to C4 (Figure 5). The reciprocal amino-H2′/H2″ cross peaks between C1•C1+ and C8•C8+ and between C2•C2+ and T7•T7 (Figure 5) are characteristic of pairs stacked with contacting faces oriented in the 3′ direction (17). The NOESY cross peaks between T7 (H3, H6 and methyl protons) and C6 are indicative of stacking interactions (Figure 5). The NOESY cross peaks connecting T3 to C2, T7, C6 and C4 and the fast exchange of T3(H3) suggest that T3 is flipped out of the i-motif core (Figure 5).
Figure 5.
The short inter-residue distances (<4.7 Å) used to compute the structure of [d(5mCCTCATCC)]2. The faces of the bases oriented in the 5′ direction are labeled with a black heavy line. The non-cytidine residues are shaded gray. A dashed heavy line connects the base-paired residues.
A5 forms a mini-loop between the base-paired 5mC1-C2-T3-C4 and C6-T7-C8-C9 segments of each hairpin. The A5 loops may cross either the narrow or the wide grooves. The adjacent C4•C4+ and C6•C6+ pairs are stacked by the faces oriented in the 5′ direction. At this step, the P–P inter-strand distances across the i-motif narrow and wide grooves are typically 9 ± 1 and 14.5 ± 1 Å. It is impossible that a single residue could span the wide groove without severe distortion of the adjacent C•C+ pairs. The presence of an imino proton on pair C4•C4+ (16.1 p.p.m.; Figure 1) and the chemical shifts of C4 and C6 amino protons show that both residues form regular hemiprotonated pairs. This implies therefore that A5 crosses the i-motif narrow groove.
The glycosidic angles were constrained by the aromatic-H1′ and -H3′ intra-residue distances. The sugar puckers were characterized by the aromatic-H3′, H2″-H4′ and H1′-H4′ distances and by the coupling constants derived from the H1′-H2″/H2″ cross peaks of E. COSY experiments. The ɛ angles were obtained from the 3J 1H3′-31P coupling constants measured on a 1H-31P COSY experiment (Figure 4). The weak intra residue H6-H5′/H5″ cross peaks observed for residues 1, 3, 6, 7, 8 and 9 correspond to distances >4.2 Å. The H6-H5′/H5″ distances depend on γ and χ dihedral angles, and to a lesser extend on the sugar pucker. Considering the glycosidic orientations and sugar puckers of these residues, one may compute that H5′-H6 and H5″-H6 distances >4.2 Å imply that the γ angles are in the narrow conformational range: 20° > γ > 100°.
The sugar backbone was also constraint by the short distances derived from the strong sequential (H2′/H2″)n–(H5′/H5″)n+1 cross peaks observed at the steps between C2-T3, T7-C8 and C8-C9 (Figure 4). For non-ambiguous identification of these cross peaks, the H5′ and H5″ protons of T3, C8 and C9 were identified stereo-specifically. In a nucleoside with γ angle in the range of 20–100°, H3′ is close to H5′ (3.8–4.1 Å) and far from H5″ (4.5–8 Å). H5′ and H5″ were thus distinguished from each other by the intensities of their cross peaks with H3′.
Proton exchange and base pair lifetimes
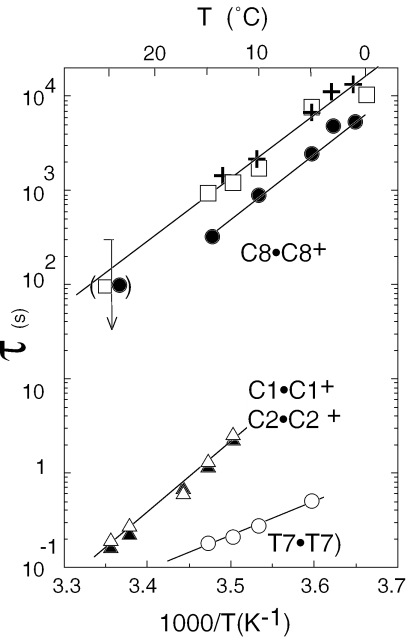
C imino proton exchange is limited by the opening rate of the hemiprotonated pairs (6). The fast exchange rates of the imino protons of the outer pairs, C9•C9+ and C4•C4+ and of C6•C6+, the pair stacked to the sequentially adjacent T7•T7 pair, indicate lifetimes <1 ms at 0°C. The lifetime of C8•C8+ measured by H/D substitution followed in real time is ∼2 h at 0°C. The lifetimes of C1•C1+ and C2•C2+, the pairs stacked on each side of C8•C8+, are ∼100 times shorter (Figure 6).
Figure 6.
Base pair lifetimes, amino proton exchange times and dimer lifetime versus temperature at pH 5.6. Lifetimes of pairs C8•C8+ (closed circles); C1•C1+ (closed triangles); C2•C2+ (open triangles) and T7•T7 (open circles). Exchange time of the H-bonded amino proton of pair 5mC1•5mC1+ (open square). The exchange time of the poorly resolved H-bonded amino protons of pairs C8•C8+ and C2•C2+ are comparable. Dimer lifetime (crosses).
The rate of catalysis by phosphate of T3(H3) is comparable with that of the free thymidine. When the phosphate concentration increases, the exchange time of T7(H3) tends towards a limit value corresponding to a base pair lifetime of 0.18 s at 15°C. The six external cytidine amino protons and the H-bonded amino protons of C4, C6 and C9 exchange in <3 min at 0°C, pH 5.6. The exchange times of the H-bonded amino protons of pairs 1, 2 and 8 are extremely long: ∼5 h at 0°C for the well-resolved amino proton of 5mC1. The proton exchange rates measured on the dimer of d(5mCCTCACTCC) and of d(5mCCTCICTCC) are similar. The rate of exchange catalysis by phosphate of I5(H1) that is only two times slower than that of the inosine monomer shows that I5 is not H-bonded in the dimer.
Association kinetics, dimer lifetime and dissociation constant
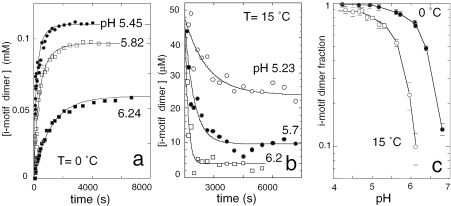
The dimer concentration measured as a function of the time in d(5mCCTCACTCC) solutions initially monomeric is displayed in Figure 7a. As it may be expected for a bimolecular process involving protonated residues, the formation rate of [d(5mCCTCACTCC)]2 increases linearly with the strand concentration (data not shown) and depends on pH. The time constants for dimer formation, Δ, were obtained from the fits displayed in Figure 7a according to Equation 1.
Figure 7.
Association–dissociation kinetics of [d(5mCCTCACTCC)]2 and dimer fraction at equilibrium as a function of pH. (a) Dimerization kinetics of d(5mCCTCACTCC). The oligonucleotide, 0.125 mM, was initially monomeric. The dimer concentration, measured on NMR spectra collected as a function of the time, tends towards a pH dependent equilibrium. The fits are computed according to Equation 1 with f and kon values of 0.48 and 3.4 M−1 s−1 at pH 6.24 (closed square); 0.75 and 20.7 M−1 s−1 at pH 5.82 (open square); 0.88 and 57.8 M−1 s−1 at pH 5.45 (closed circle). (b) Dissociation kinetics of [d(5mCCTCACTCC)]2. The measurements started right after dilution to 45 µM of a concentrated fully dimeric solution. The dimer lifetimes, 1130, 480 and 158 s at pH 5.23, 5.7 and 6.2, respectively, were obtained from the exponential fits. (c) Fraction of i-motif dimer at equilibrium versus pH. Closed circles: T = 0°C, the strand concentration is 0.33 mM. Open circles: T = 15°C, strand concentration 0.1 mM.
The dimer fraction at equilibrium, f, was measured for each sample used in the kinetics measurements (Figure 7c). The rate constants, kon, determined from Δ and f values according to Equation 2, are plotted versus pH in Figure 8a. The rate constant for association of neutral and protonated C-rich strands is expected to vary with pH as F(pH), the product of the fractions of protonated and neutral cytidine: F(pH) = 10(pKN3 − pH)/(1 + 10(pKN3 − pH))2 where pKN3 is the cytidine pK. According to this model, kon versus pH was fitted with expression:
| 5 |
The fits displayed in Figure 8a yield values of 380 ± 80 and 569 ± 150 M−1 s−1 at 0 and 15°C, respectively. The dissociation constant computed from the dimer and monomer concentrations at equilibrium (Equation 3) varies approximately as 10−2pH (Figure 8b). The dimer lifetime, toff, derived from the dissociation constants and kon values (Equation 4) is displayed in Figure 8c. The large error bars on toff values are the propagation of the error on the dissociation constants measured in the range of pH where the monomer concentration in much smaller as that of dimer.
Figure 8.
Kinetics and dissociation constants of [d(5mCCTCACTCC)]2 as a function of pH. (a) Bimolecular formation rate of [d(5mCCTCACTCC)]2 versus pH. The fit are computed according to Equation 5 with values of 380 ± 80 M−1 s−1 at 0°C (closed circle) and 569 ± 150 M−1 s−1 at 15°C (open circle). (b) Dissociation constant of the i-motif dimer versus pH at 0°C (closed circle) and 15°C (open circle). The dissociation constants were obtained from the monomer and dimer concentrations at equilibrium (Equation 3). The lines with a slope of 2 show the 10 2pH variation of Kdis. Indicative error bars are displayed for some data points. (c) Dimer lifetime at 0°C (filled circle) and 15°C (open circle), derived from the kon values displayed in a and from the dissociation constant Kdis of b.
The dimer dissociation kinetics were also measured on spectra recorded as a function of the time, right after dilution of a fully dimeric sample (Figure 7b). The toff values measured at pH 5.6 versus temperature are displayed in Figure 6. The activation energy derived from the plot of toff versus 1/T is 76.7 kJ/mol and toff values measured versus pH are displayed in Figure 8c. It is noteworthy that the measured lifetimes are in good agreement with those computed from Kdis and kon values.
DISCUSSION
Dimer structure and internal motions
The structure of [d(5mCCTCACTCC)]2 displayed in Figure 9 was computed with the distance and dihedral restraints listed in Table 1. The good resolution of the NMR spectrum provides a high definition structure that exhibits characteristics common to i-motif structures. The structural parameters derived from the analysis of 10 selected conformers are displayed in Table 2. The restraints violations and the deviations from ideal geometry of the computed structures are listed in Table 1.
Figure 9.
Structure of the i-motif dimer of d(5mCCTCATCTCC). (a) Schematic representation. The hemiprotonated structure is formed of two symmetry related hairpins connected together by six C•C+ pair and by the T7•T7 pair. The base of T3 is flipped out in the wide grove. The 5mC1-C2-C3-C4 and C6-T7-C8-T9 segments of each hairpin line the i-motif narrow grooves. A single residue, A5, loops across the narrow groove. (b) View normal to the narrow groove of the lowest-energy structure. The background hairpin is drawn in thin lines. (c) View normal to the wide groove of the 10 selected conformers. (d) The stacking arrangement in the tetramer of d(5mCCTCC) is shown for comparison. [d(5mCCTCC)]4 is formed by two non-equivalent duplexes. Simultaneous opening of T3 and reclosing of T3 results in duplex inter conversion (2). This motion is not observed in [d(5mCCTCACTCC)]2 due to the non-equivalence of the thymidines.
Table 2.
Backbone dihedral angles (α−ζ), glycosidic angles (χ), pseudo-rotation angles (P), helical twist (h) in the i-motif dimer of [d(5mCCTCACTCC)]4
| Residue | α (ϒ) | β (ϒ) | γ (ϒ) | δ (ϒ) | ɛ (ϒ) | ζ (ϒ) | χ (ϒ) | Π (ϒ) | η twist (°) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 79 ± 3 | 140 ± 8 | 252 ± 5.6 | 305 ± 33 | 224 ± 6 | 103 ± 4 | b | ||
| 2 | a | a | 187 ± 22 | 81.9 ± 18 | 248 ± 2 | 308 ± 3 | 195 ± 2 | 56 ± 3 | 16 ± 3 |
| 3 | 279 ± 2 | 217 ± 3 | 53 ± 3 | 153 ± 2 | 297 ± 4 | 88.1 ± 6 | 258 ± 3 | 163 ± 4 | |
| 4 | 190 ± 17 | 176 ± 8 | 103 ± 18 | 153 ± 4 | 253 ± 4 | 239 ± 24 | 225 ± 6 | 125 ± 45 | |
| 5 | 297 ± 26 | 80 ± 15 | 165 ± 12 | 129 ± 20 | 266 ± 25 | 133 ± 35 | 261 ± 9 | 124 ± 7 | |
| 6 | a | a | 83 ± 7 | 105 ± 29 | 226 ± 9 | 263 ± 3 | 233 ± 5 | 35 ± 4 | |
| 7 | 269 ± 10 | 105 ± 10 | 103 ± 7 | 121 ± 4 | 52 ± 9 | 137 ± 32 | 206 ± 2 | 32 ± 7 | −4.1 ± 4 |
| 8 | a | 128 ± 17 | 30 ± 4 | 99 ± 4 | 135 ± 2 | 269 ± 40 | 236 ± 4 | 46 ± 2 | 41 ± 4 |
| 9 | a | a | 43.6 ± 23 | 113 ± 11 | 258 ± 3 | 28 ± 3 | 25 ± 2 |
The parameters and SDs were measured from 10 selected conformers. The angle average values are given when at least 7 out of the 10 measured values are clustered within a sector of 60°.
aThis criterion convergence is not reached.
bThe helical twist is the angle between the projection of the C1′-N1′ vectors of sequentially adjacent bases in a plane perpendicular to the helix axis. It is not displayed for non-stacked bases.
The i-motif core
The dimer is built by two symmetrical hairpins including six C•C+ pairs and a T•T pair. The T3 bases are symmetrically tilted in the wide grooves. T3 unstacking leaves an interval between C2•C2+ and C4•C4+ allowing intercalation of the sequentially adjacent T7•T7 and C6•C6+ pairs. The stacking interval between the C•C+ pairs is 3.1 ± 0.1 Å. It is larger between pairs T7•T7 and C6•C6+: 3.9 ± 0.1 Å. The helical twist between the paired cytidines, 16° at step 1–2 and 25° at step 8–9 are typical of values observed in i-motif structures. As observed previously in the related structures of [d(5mCCTCC)]4 (2) and of [d(5mCCTCTCC)]4 (3), the orientation of the bases of the T•T pair results in a reduction of the helical twist at the 5′ C-T step and in larger twist at the T-C3′ step (Table 2). The groove widths were defined as the average of the 25 inter-strand distances connecting the five C1′, C2′, C3′, C4′ and O4′ atoms of contacting sugars across the narrow and wide grooves. The narrow groove width is 5.2 ± 0.3 Å at the steps where the bases are stacked by the faces oriented in the 3′ direction. As it is always observed in i-motif structures, it is slightly broader, 6.3 ± 0.5 Å, at the steps where the bases are stacked by the faces oriented in the 5′ direction. It is much broader, 9.2 ± 0.2 Å, at the steps between T3/T7 and T3/C6. The wide groove width is 11.4 ± 1 Å on the average.
The A5 loop
Simulations enforcing A5 looping across the wide grooves result in disruption of pair C4•C4+ and in inter proton distances inconsistent with the NMR spectra. The sugar of A5 is positioned by the distance restraints derived from five noe cross peaks with the base and sugar protons of C4 and the A5 base is oriented by the A(H2)-C4 (H5 and H1′) cross peaks in a direction nearly perpendicular to the adjacent C4•C4 plane (Figure 5). The absence of any inter-residue noe connectivities to A5 (H8) and amino protons is consistent with orientation of the C8-N6 edge of A5 outside of the i-motif core. The modest overlap of the A5 bases and the distance between the base planes, ∼5 Å, rule out stacking interactions. I5 in [d(5mCCTCICTCC)]2 and A5 in [d(5mCCTCICTCC)]2 are connected to pair C4•C4+ by similar noe cross peaks. This indicates comparable orientations of the purine bases in each structure.
Several dimeric and monomeric i-motif structures have been described. In all the cases the loops crossing the narrow grooves (underlined residues in the sequences below) contain either two residues, as in the cases of the dimer of d(5mCCTCTCC) (2) and of the monomeric i-motif of d(CCTTTCCTTTACCTTTCC) (18), three residues in the case of [d(CCCCTGTCCCC)]2 (19) and of the monomeric i-motif of d(CCCTAACCCTAACCCTAACCC) (20) or four residue as shown for the dimer of d(TCCCGTTTCCA) (21). The present study shows that a single residue can span the i-motif narrow groove without notable distortion of the adjacent C•C+ pairs. In the extremely stable d(GCGAAGC) hairpin, a mini loop containing a single adenine, A4, has been described (22,23), but in this example, A4 is stacked to the adjacent G3 residue which forms a non-standard G4•A5 pair.
Sugar–phosphate backbone
As observed previously in other i-motif structures (20), the 31P spectrum of [d(5mCCTCACTCC)]2 exhibits a good spectral dispersion (2.07 p.p.m.; Figure 3). The 31P spectrum of [d(5mCCTCICTCC)]2 is nearly super imposable, except for the 31P at each side of the purine residue (Supplementary Table S1). It has been proposed that the 31P chemical shifts of B-DNA duplexes are correlated with the ɛ dihedral angles (24). It may be noticed that the 31P chemical shifts of [d(5mCCTCACTCC)]2 and of the i-motif structure of the human telomeric repeats (20) show a very poor correlation with the ɛ angle values derived from the 3J 1H3′-31P coupling constant.
The BI and BII conformations of the phosphate backbone are characterized by the (ɛ−ζ) difference (25). The (ɛ−ζ) values measured for the non-paired T3 and A5 residues, 209 ± 9° and 133 ± 59°, respectively, indicate that the phosphor atom at their 3′ side (P4 and P6) adopt the uncommon BII conformation whereas the (ɛ−ζ) values measured for the other residues correspond to BI conformation, as this is generally observed in i-motif structures. The observation that all the residues with strong (weak) H6/H8- H3′ cross peaks exhibit weak (strong) H3′-31P cross peaks (Figure 4) suggests some correlation between the sugar puckers and ɛ dihedral angles.
The NOESY cross peaks corresponding to short sequential (H2″)n–(H5′)n+1 distances of 2.4–3 Å observed at steps 2–3, 7–8 and 8–9 of [d(5mCCTCACTCC)]2 were not observed in previous NMR studies of i-motif structures, owing possibly to the poor resolution of the corresponding regions of the NOESY spectra. Examination of i-motif structures from NMR or X-ray studies indicates that short (H2″)n–(H5′)n+1 distances are not systematic features. For example, the measure of the 12 × 4 sequential distances between H2′/H2″ and H5′/H5″ protons of the crystal structure of [d(CCCC)]4 [(26), PDB accession no. 190D] shows only four distances <3 Å, all between H2″ and H5′ protons. Furthermore, several H5′/H5″-H2′/H2″ distances are also very short at several steps of the crystal structure of the d(CGCGAATTCGCG)2 duplex [(27), PDB accession no. 1BNA). Consequently, we conclude that short H2″-H5′ distances are not characteristic of i-motif structure but the consequence of a combination of six ((ν2,ɛ,ζ)n and (α,β,γ)n+1) dihedral angles.
Comparison with related structures
Figure 9 shows that the structural motif of the core of [d(5mCCTCACTCC)]2 is closely related to that of the i-motif of [d(5mCCTCC)]4 (2). This tetramer is built by intercalation of two symmetrical non-equivalent duplexes. The stacking order is C5-5mC1-C4-C2-(T3)-T3-C2-C4-5mC1-C5. In the duplex whose residues are underlined, the thymidines (T3) are looped out. In the other duplex T3 forms a T3•T3 pair. The exchange cross peaks observed between the homologous protons of each duplex show that the simultaneous closing of the (T3) residues and opening of the T3•T3 pair exchange the duplex structures at a rate close to 1 s at 0°C. Figure 9 shows that equivalent inter-conversion process in [d(5mCCTCACTCC)]2 would involve opening of pair T7•T7 and simultaneous pairing of the open T3 residues. The absence of a detectable proportion of a dimer species with open T7 thymidines and paired T3 is a consequence of the non-equivalence of the T residues in the dimer. A [d(5mCCTCACTCC)]2 structure with paired T3 and open T7 bases may be modeled without noticeable van der Waal's contact and the comparison of the observed and simulated structures give no indications about their free energy difference.
Internal motions in [d(5mCCTCACTCC)]2
The dimer core includes three long-lived hemiprotonated pairs: C1•C1+, C8•C8+ and C2•C2+. Despite the structural similarities of the dimer and [d(5mCCTCC)]4 structures (Figure 9), the comparison of their base pair opening kinetics shows that in the dimer, the lifetime of pairs C1•C1+, C8•C8+ and C2•C2+ is ∼10-fold that of their counterparts in the tetramer.
The dimer lifetime and the exchange times of the H-bonded amino proton of the three long-lived C•C+ pairs are comparable (Figure 6). This strongly suggests that amino proton exchange requires dimer dissociation. The huge difference between the exchange times of H-bonded and external amino protons has been noticed in previous studies of i-motif structures (6). In [d(5mCCTCACTCC)]2 rotation of the amino group around the C4-N4 bond seems entirely hindered for residues C1, C8 and C2. This is most probably a consequence of the tight stacking of these residues. The comparison of the exchange time of the H-bonded amino protons with that of the free cytidine residue, 0.15 s at 0°C, pH 5.6, (28) provides an upper bound value of 10−5 for the dissociation constants of pairs 1.8 and 2.
Kinetics of the monomer–dimer equilibrium
The salient feature that emerges from the kinetics study is the slowness of the rates of dimer formation and dissociation. This observation is consistent with the spectacular hysteresis observed in the melting and renaturation profiles of i-motif structures (4). The formation and dissociation kinetics of [d(5mCCTCACTCC)]2 shows striking differences with that of Watson–Crick duplexes (29,30). The association rate of complementary oligonucleotides into a Watson–Crick duplex ranges between 5 × 105 and 107 M−1 s−1. It is independent of pH (∼7) and weakly depends on the oligonucleotide length and temperature. The lifetime at 20°C of Watson–Crick duplexes containing a number of base pairs comparable with [d(5mCCTCACTCC)]2 is in the range of 0.15–1 s.
Between pH 4.8 and 6.8, the association rate, kon, of two d(5mCCTCACTCC) strands into an i-motif dimer vary as 10−pH (Figure 8a). The origin of this pH dependence is trivial. It reflects the variation of the concentration of the protonated C+ partner. It may be predicted that for pH values smaller than the cytidine pKN3, kon should decrease for a symmetrical reason: the rarefaction of the neutral C partner. Nevertheless, once taken into account the effect of pH on the concentration of the interacting partners, we find that the pH independent rate, (Equation 5), is slower by three to four magnitude orders than the association rate of two complementary strands into a Watson–Crick duplex. The variation of with temperature corresponds to small activation energy of 13 ± 22 kJ/mole. Oligonucleotide association in Watson–Crick duplexes is described by a model involving the formation of a correct nucleus of a few base pairs, followed by rapid ‘zipping’ into the fully paired duplex (31). The formation of an i-motif structure must involve additional steps. By analogy with this ‘nucleation-zipping’ model, one may imagine a plausible scenario including: (i) the formation a hemiprotonated nucleus; (ii) adequate intercalation during the nucleus elongation of a third strand with appropriate orientation; and (iii) association, in parallel orientation with this stand, of a fourth strand that finally locks the strand assembly into a long-lived structure. Only the initial (monomer) and final (i-motif) states are populated to measurable extents. Strand mispairing at any step of this pathway should result in unproductive dissociation of the short-lived intermediate species. It may be therefore proposed that this is the multiplicity of the steps required to achieve the formation of a stable structure that accounts for the slow formation kinetics of the i-motif structure.
[d(5mCCTCACTCC)]2 formation requires the protonation of six cytidines. Hence, a fully cooperative dimerization process would occur at a rate depending on the power of six of the proton concentration. The measured rate that increases as the proton concentration shows clearly that dimerization does not entail the cooperative formation of six C•C+ pairs and suggests that elongation of the partially hemiprotonated structure occurs step by step via successive formation of hemiprotonated C•C+ pairs.
The lifetime of [d(5mCCTCACTCC)]2 is in the range of hours around pH 4.8 and it decreases approximately as 10−pH when the pH is raised (Figure 8a). However, the comparison of spectra at pH 4.8 and 6.8, give no indication for a structural change accounting for the reduction of the dimer lifetime.
The half protonation pH, pH1/2, of hemiprotonated pairs depends on the cytidine pKN3 and on the base pair dissociation constant: pH1/2 ≈ pKN3 − log(Kdis) (32). Hence, with a lower bound estimation of 10−5 for their dissociation constants (cf. above), pairs 1.8 and 2 should be stable up to pH 9.3.
The two outer C•C+ pairs and C6•C6+ have extremely short lifetimes and presumably a modest stability. It is probably that at high pH, transient un-protonation increases the open-state lifetime of these pairs and results in destabilization of the inner pair by end fraying. If the disruption of the outer pairs strongly reduces the dimer lifetime, then a small proportion of open pair, undetectable on the dimer spectrum, may efficiently contribute to the reduction of the i-motif lifetime. According to this interpretation, it may be conjectured that the i-motif stability at high pH is strongly dependent on the outer pair stability.
In the monomolecular i-motif structures of d(CCTTTCCTTTACCTTTCC) (18) and of d(CCCTAACCCTAACCCTAACCC) (20), the outer pairs are to some extent protected by the loops that cross the grooves and there lifetimes are at least 10-fold that of the outer pairs of [d(5mCCTCACTCC)]. This may be in relation with the better stability of the folded structures at high pH and with the observation that the lifetime of the folded d(CCCTAACCCTAACCCTAACCC) structure is the same at pH 5 and 6 (33). Structures designed with C-rich oligonucleotides terminated with complementary residues allowing the formation of an i-motif core protected at each end by parallel duplex stacked on the outer C•C+ pairs may provide interesting models to test the effect of end fraying on the stability of i-motif structure.
At last, it may be noticed that the pH dependence of kon and koff results in a reduction of the dimer stability by a factor of 100 when the pH is raised by one unit. This clearly shows that a stabilizing environment, in particular for the outer hemiprotonated pairs, is a sine qua non requirement for the existence of i-motif in the cell environment and for a biological function of i-motif structures.
Acknowledgments
M.C. thanks ICSN for financial support. The coordinates of the [d(5mCCTCACTCC)]2 conformer of lowest energy and the restraints used in the molecular dynamics have been submitted to the protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973, USA (acquisition number: 2AWV and RCSB034409). Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gehring K., Leroy J.-L., Guéron M. A tetrameric DNA structure with protonated cytidine–cytidine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 2.Nonin S., Leroy J.-L. Structure and conversion kinetics of a bi-stable DNA i-motif: broken symmetry in the DNA [d(5mCCTCC)]4 tetramer. J. Mol. Biol. 1996;261:399–414. doi: 10.1006/jmbi.1996.0472. [DOI] [PubMed] [Google Scholar]
- 3.Leroy J.-L. T·T pair intercalation and duplex interconversion within i-motif tetramers. J. Mol. Biol. 2003;333:125–139. doi: 10.1016/s0022-2836(03)00945-8. [DOI] [PubMed] [Google Scholar]
- 4.Mergny J.-L., Lacroix L. Kinetics and thermodynamics of i-DNA formation: phosphodiester versus modified oligodeoxynucleotides. Nucleic Acids Res. 1998;26:4797–803. doi: 10.1093/nar/26.21.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guéron M., Leroy J.-L. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 6.Leroy J.-L., Gehring K., Kettani A., Guéron M. Acid multimers of oligo-cytidine strands: stoichiometry, base-pair characterization and proton exchange properties. Biochemistry. 1993;32:6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- 7.Plateau P., Guéron M. Exchangeable proton NMR without base-line distortion, using strong pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 8.Macura S., Wüthrich K., Ernst R.R. The relevance of J cross-peaks in two-dimensional NOE experiments of macromolecules. J. Magn. Reson. 1992;47:351–357. [Google Scholar]
- 9.Bax A., Davis D.G. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 10.Kellogg G.W. Proton-detected hetero-TOCSY experiments with application to nucleic acids. J. Magn. Reson. 1992;98:176–182. [Google Scholar]
- 11.Sklenar V., Bax A. Measurements of 1H-31P NMR coupling constant in double stranded DNA fragments. J. Am. Chem. Soc. 1987;109:7225–7226. [Google Scholar]
- 12.Brünger A.T. X-PLOR Version 3, A System for X-Ray Crystallography and NMR. New Haven, CT: Yale University; 1990. [Google Scholar]
- 13.Koradi R., Billeter M., Wütrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 14.Phan A.T., Guéron M., Leroy J.-L. Methods for the investigation of unusual DNA motifs. Methods Enzymol. 2001;261:383–441. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 15.Razka M. Mononucleotides in aqueous solution: Proton magnetic resonance studies of amino groups. Biochemistry. 1974;13:4616–4622. doi: 10.1021/bi00719a023. [DOI] [PubMed] [Google Scholar]
- 16.Nonin S., Phan A.T., Leroy J.-L. Solution structure and base pair opening kinetics of the i-motif dimer of d(5mCCTTTACC): a noncanonical structure with possible roles in chromosome stability. Structure. 1997;5:1231–1246. doi: 10.1016/s0969-2126(97)00273-6. [DOI] [PubMed] [Google Scholar]
- 17.Leroy J.-L., Guéron M. Solution structure of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connection between amino protons and sugar protons. Structure. 1995;3:101–120. doi: 10.1016/s0969-2126(01)00138-1. [DOI] [PubMed] [Google Scholar]
- 18.Han X., Leroy J.-L., Guéron M. An intramolecular i-motif: the solution structure and base-pair opening kinetics of d(5mCCTTTCCTTTACCTTTCC) J. Mol. Biol. 1998;278:949–965. doi: 10.1006/jmbi.1998.1740. [DOI] [PubMed] [Google Scholar]
- 19.Catasti P., Chen X., Deaven L.L., Moyzis R.K., Bradbury E.M., Gupta G. Cystosine-rich strands of the insulin minisatellite adopt hairpins with intercalated cytosine+.cytosine pairs. J Mol Biol. 1997;272:369–382. doi: 10.1006/jmbi.1997.1248. [DOI] [PubMed] [Google Scholar]
- 20.Phan A.T., Guéron M., Leroy J.-L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 21.Gallego J., Chou S.H., Reid B.R. Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer. J. Mol. Biol. 1997;273:840–856. doi: 10.1006/jmbi.1997.1361. [DOI] [PubMed] [Google Scholar]
- 22.Hirao I., Kawai G., Yoshizawa S., Nishimura Y., Ishido Y., Watanabe K., Miura K. Structural features and properties of an extraordinarily stable hairpin-turn structure of d(GCGAAGC) Nucleic Acids Res. 1994;22:576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padrta P., Stefl R., Kralik L., Zidek L., Sklenar V. Refinement of d(GCGAAGC) hairpin structure using one- and two-bond residual dipolar couplings. J. Biomol. NMR. 2002;24:1–14. doi: 10.1023/a:1020632900961. [DOI] [PubMed] [Google Scholar]
- 24.Roongta V.A., Jones C.R., Gorenstein D.G. Effect of distortions in the deoxyribose phosphate backbone conformation of duplex oligodeoxyribonucleotide dodecamers containing GT, GG, GA, AC, and GU base-pair mismatches on 31P NMR spectra. Biochemistry. 1990;29:5245–5258. doi: 10.1021/bi00474a005. [DOI] [PubMed] [Google Scholar]
- 25.Hartmann B., Piazzola D., Lavery R. BI-BII transitions in B-DNA. Nucleic Acids Res. 1993;21:561–568. doi: 10.1093/nar/21.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L., Cai L., Zhang X., Rich A. Crystal structure of a four-stranded intercalated DNA: d(C4) Biochemistry. 1994;33:13540–13546. doi: 10.1021/bi00250a005. [DOI] [PubMed] [Google Scholar]
- 27.Drew H.R., Wing R.M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R.E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kettani A., Leroy J.-L., Guéron M. Amino proton exchange process in mononucleosides. J. Am. Chem. Soc. 1997;119:1108–1115. [Google Scholar]
- 29.Riesner D., Römer R. In: Physico-Chemical Properties of Nucleic Acid. Duchesne J., editor. Vol. 2. London: AcademicPress; 1973. [Google Scholar]
- 30.Howorka S., Movileanu L., Braha O., Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc. Natl Acad. Sci. USA. 2001;98:12996–13001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantor C.R., Schimmel P.R. Biophysical Chemistry Part III. The Behavior of Biological Macromolecules. New York: Freeman; 1980. Kinetics of confirmational change; pp. 1211–1239. [Google Scholar]
- 32.Leroy J.-L., Nonin S., Han X., Phan A.T., Guéron M. Switching and looping in i-motif structures. In: Sarma R.H., Sarma M.H., editors. Structure and Motion and Expression. of biological macromolecules. Schenectady, NY: Adenine Press; 1998. pp. 49–62. [Google Scholar]
- 33.Phan A.T., Mergny J-L. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]