Abstract
A method providing absolute transcript concentrations from spotted microarray intensity data is presented. Number of transcripts per µg total RNA, mRNA or per cell, are obtained for each gene, enabling comparisons of transcript levels within and between tissues. The method is based on Bayesian statistical modelling incorporating available information about the experiment from target preparation to image analysis, leading to realistically large confidence intervals for estimated concentrations. The method was validated in experiments using transcripts at known concentrations, showing accuracy and reproducibility of estimated concentrations, which were also in excellent agreement with results from quantitative real-time PCR. We determined the concentration for 10 157 genes in cervix cancers and a pool of cancer cell lines and found values in the range of 105–1010 transcripts per µg total RNA. The precision of our estimates was sufficiently high to detect significant concentration differences between two tumours and between different genes within the same tumour, comparisons that are not possible with standard intensity ratios. Our method can be used to explore the regulation of pathways and to develop individualized therapies, based on absolute transcript concentrations. It can be applied broadly, facilitating the construction of the transcriptome, continuously updating it by integrating future data.
INTRODUCTION
Recent developments in molecular techniques, such as serial analysis of gene expression (SAGE), massive parallel signature sequencing (MPSS) and microarray technology, have opened for genome-wide exploration of the transcriptome (1–3). Such data increase our understanding of complex biological processes and diseases and are becoming useful in the design of molecular therapies (4). SAGE and MPSS provide quantitative and comparable measures of the transcript abundance, whose universality allows for integration into future studies. The complexitity of SAGE and MPSS has, however, limited their utility (5). Efficient production of spotted glass-slide arrays has made the microarray technology to a widespread technique that is more suitable for high-throughput analysis. The technique has provided valuable information on the relative transcript levels in tissues, but differences in experimental protocols and normalization methods make direct comparison of datasets between microarray studies very difficult (6). Improved methods to extract useful information from such data that lead to absolute rather than relative transcript concentrations would be of high value (6–8), facilitating the building up of an universal transcript database. This is the goal of several public data repositories, including, for example, the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/projects/geo/) and SAGEmap (http://sagemap.wr.usgs.gov/index.asp).
Extraction of absolute transcript levels from spotted microarray data is complicated owing to significant experimental variation and noise originating in the production and hybridization processes (7–9). The use of probes with different length and base composition, leading to differences in hybridization efficiency between probes, makes assessment of absolute levels difficult. Most analyses are based on intensity ratios between two biological samples, hybridized together in a single experiment. Normalization of the ratios reduces the influence of systematic effects, though absolute levels are lost as well as possibly important biological information (10–12). Analysis based on intensities per se rather than ratios opens for calculating accurate transcript levels.
We have developed a model based on a new principle that enables estimation of absolute transcript levels on a genome-wide scale by extended exploitation of microarray data. Once the concentrations have been estimated, new analyses are possible, including within sample comparison, merging of datasets with a design lacking connectivity or based on amplified and non-amplified starting materials, cross-platform and cross-species comparisons and more general meta-analyses. The technique was thoroughly validated on datasets with known mRNA concentrations. Moreover, we estimated the transcript concentrations of 10 157 genes and expressed sequence tags (ESTs) in 12 cervix cancers and a pool of 10 human cancer cell lines, and found values consistent with quantitative real-time PCR (qRT-PCR) data and with previously publised data (13). We generated new views into the transcriptome, by comparing transcript abundance between genes or groups of genes within a population. The model follows the different steps of the microarray experiment, incorporating information associated with array, cDNA synthesis, hybridization and scanning characteristics. We computed the joint posterior distributions of the absolute transcript levels of all genes, describing dependencies between genes, both within and between individual samples. Uncertainties from sample preparation to imaging were coherently propagated in a global statistical approach, leading to realistically large confidence intervals around estimated concentrations.
Few methods quantifying transcript concentrations from spotted microarray data have been developed so far. The approach proposed by Dudley et al. (14) requires hybridization of each sample with a reference of known concentration. Other methods rely on calibration of each array with additional techniques (15). The present method is the first quantifying absolute transcript levels from spotted microarray data without the need for calibration of each sample or gene individually. There are a few quantitative methods based on in situ synthesized arrays (16,17) and, notably, (18) which takes an empirical Bayesian approach, but the data produced from them are scarce, probably because of a limited access to such arrays. The possibility to directly use the spotted microarray technology for the estimation of absolute transcript concentrations opens for a more comprehensive generation of transcript databases. Results reported here were based on spotted cDNA microarrays, which feature particularly large experimental variation. Our technique can also be directly applied to spotted oligoarrays and can handle experiments based on amplified as well as non-amplified material.
MATERIALS AND METHODS
Principles
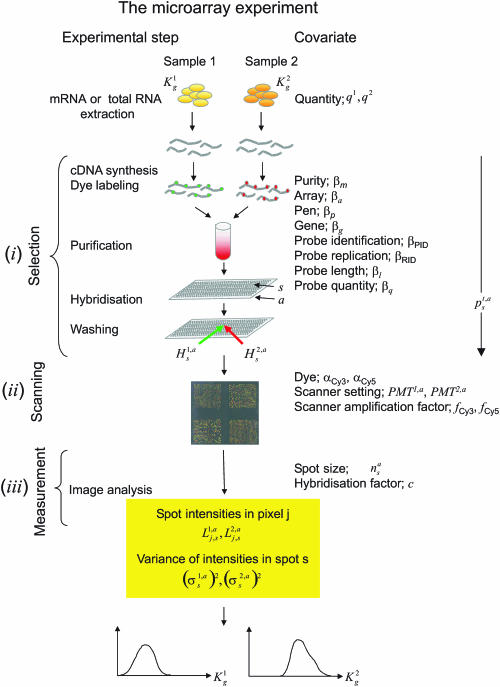
The idea is to follow conceptually the mRNA molecules through the microarray experiment, from cDNA synthesis to hybridization and subsequent washing (Figure 1). We modelled mathematically the process as a simple selection, where each molecule had a certain probability of being kept in the experiment. This probability depended on known experimental covariates, like mRNA purity, array, pen, gene and probe identification, replication, length and quantity. We treated scanning and image analysis as an integral part of the experiment and used associated covariates, such as dye, scanner setting and spot size. Also characteristics specific for the scanner and hybridization technique were included: the scanner amplification factor was needed to account for differences in the intensity response among scanner types; the hybridization factor identified the absolute scale of the estimates. Both factors were determined in two off-line calibration experiments.
Figure 1.
Illustration of the microarray experiment. The various steps of the experiment and the corresponding covariates used in the model are listed with their symbols. The model consists of three levels: (i) selection, (ii) scanning and (iii) measurement. In (i), and mRNA molecules for gene g present in sample 1 and 2 undergo a selection process. Each molecule succeeds or fails in each of the experimental steps: cDNA synthesis, dye labelling, purification, hybridization and washing. Success for each molecule is modelled as a Bernoulli coin toss. The success probability depends on properties of the molecule and of the experiment (covariates). Molecules of the same gene can have different covariates, e.g. if they hybridize on different spots with different probes. If probe is in excess, molecules can be modelled as independent variables and the number of remaining molecules after each step is binomially distributed. The probability of successfully passing through the entire experiment is the product of the probabilities of surviving each individual step. Nested binomial variables are binomial and the final number of molecules ready for being scanned is binomial with two parameters: the unknown original number of transcripts per gene in each sample and the selection probability, modelled as in Equation 1. Level (ii) describes the translation of the bound molecules remaining after washing (, on array a, spot s, for sample t = 1, 2) into fluorescence intensities, as in Equation 2. Measurement error (iii) of pixel-wise intensities (on array a, pixel j on spot s for sample t = 1, 2) is assumed to be normally distributed as in Equation 3. This model allows to obtain estimates of absolute concentrations and together with their posterior marginal probability density, as sketched at the bottom.
Basic data are the average fluorescence intensities, background corrected or not, and standard deviations of each spot on the microarray slide. Intensities should be within the linear range of the scanner, and saturated intensities should either be excluded or corrected (19). No transformation nor normalization are done. Non-connected datasets are allowed as long as the design includes at least one loop, like a self–self or dye–swap hybridization. About 50 genes must be spotted at least in duplicates, their number being independent of the number of genes in the analysis, but related to identifiability of probe and pen effects. Our method succeeds in obtaining absolute concentrations because it makes explicit use of probe and spot related covariates like probe length and quantity, to describe probe-dependent hybridization efficiency. By means of duplicate spotting, we have many transcripts with more than one probe, and the effect of probe-dependent covariates can be estimated and further incorporated into the estimates of concentrations for genes spotted only once. Experiments with amplified material are handled like those with non-amplified ones, but estimates are transformed back to original scale.
The model is simple and natural. We performed Bayesian estimation of its parameters and calculated the posterior joint distribution of all absolute transcript concentrations using Markov Chain Monte Carlo (MCMC; http://www.statslab.cam.ac.uk/~mcmc/) (20). This distribution reflects biological variation of and dependencies between the numbers of transcripts. Based on this distribution we estimated the number of transcripts for each gene in each sample together with their uncertainty, described by 95% credibility intervals. The values were given in terms of number of transcripts per µg of total RNA, mRNA or per cell, depending on the experimental protocol.
Covariates
The steps of the microarray experiment were modelled as a binomial selection process, using covariates associated with cDNA synthesis, dye labelling, purification, hybridization and washing (Figure 1 and Supplementary Data 1). The corresponding covariates were sample purity, array, pen, gene, probe replication (RID), probe identification (PID), probe length and probe quantity. Replicated genes had both PID and RID effects, where PID accounted for different probes and RID for replications of equal probes. The number of base pairs in the probe sequence was used as probe length. Probe quantity was the mean spot fluorescence intensity in a test slide of each printing series that was stained for single-stranded DNA by use of SYBR green II (Molecular Probes). Probe quantity was included, although probe in excess was assumed, since hybridization efficiency of high density probes may be reduced (21). About 50 genes must be spotted at least in duplicates to be able to estimate the effect of the probe and spot related covariates. Estimates are more precise if the duplicated genes are chosen to span over the range of probe length and quantity (not of the concentrations). Additional probe-related covariates may improve estimates further.
Covariates associated with scanning were dye, photo multiplier tube (PMT) voltage and the scanner amplification factor. The dye covariate represented the dye effect in both labelling and scanning. The amplification factor was a measure of the increase in intensity per unit of increase in PMT voltage. The factor was determined once for each dye and scanner as the slope in log-linear plots of intensity versus PMT voltage (19). A covariate associated with image analysis was the hybridization factor, used to scale the estimated values to the true number of transcripts. It was determined with weighted linear regression of estimates versus true values in a dataset based on samples with known transcripts concentrations. Such control samples in general show a more efficient cDNA synthesis and dye labelling than biological samples, owing to the high purity of these molecules. The samples are therefore not useful for calibration of intensities. However, after cDNA synthesis and labelling, the binding behaviour to the array slides of cDNA synthesized from the control samples resembles that of the cDNA from biological samples. This step is not dependent on the quality of the applied mRNAs, justifying the use of control samples for validation and determination of the hybridization factor. Under ordinary stable experimental settings it is sufficient to determine this factor once for each hybridization machine.
Statistical methods
The known quantity of material for sample t on array a is denoted as qt,a, e.g. the weight of mRNA after amplification or of total RNA, as in our study on cervix cancer. For each gene g, let denote the unknown number of transcripts per weight unit present in sample t (Figure 1). Let be the measured intensity for sample t in pixel j in spot s on array a. The non-linear model that relates these data to the number of transcripts consists of three layers: (i) a model for the selection process, describing the proportion of target molecules (from the original ) that have survived the several steps of the experiment until washing of the hybridized slides; (ii) a model for the scanning process of the hybridized slides; and (iii) a model for measurement and residual errors.
In (i), the molecules undergo a series of processes from cDNA synthesis to hybridization and washing (Figure 1). Let be the number of pixels in spot s on array a and the total number of pixels in all spots related to gene g on array a. After successful cDNA synthesis, labelling and purification, a proportion of the molecules candidates to reach the correct spots for hybridization. Here c is the hybridization factor per pixel. Each of these molecules has a success probability to hybridize and to remain fixed after subsequent washing, independently of other molecules. This independency corresponds to the usual probe in excess assumption. As discussed in Supplementary Data 1, also accounts for successful cDNA synthesis, dye labelling and purification and it depends on biological and experimental conditions described by covariates. Let be the unknown number of molecules in sample t that succeeds in hybridizing on spot s on array a, and resists subsequent washing, thus being available for imaging. Then we obtain the simple model
where g is the gene spotted in spot s on array a and
| 1 |
The β's represent effects of the various covariates for spot s on array a (βa array, βp pen, βg gene, βPID probe identification and βRID probe replication), [probe length] is the number of base pairs of the probe in spot s, [probe quantity] is the SYBR green intensity, [purityt] is the purity of sample t and exp(β0) is the global baseline selection probability. When non-connected datasets are analysed jointly, an effect βe is required for each connected subset. Identifiability of all parameters is assured (Supplementary Data 2).
In (ii), the expected scanned intensity on spot s, array a, is modelled as
| 2 |
where PMTt,a is the PMT-voltage used during scanning of sample t on array a, fCy3 or fCy5 are the known scanner amplification factors, while αCy3 and αCy5 are unknown chemical and optical dye effects.
In (iii), we assume for the pixel-wise intensity measurement ,
| 3 |
where is a normally distributed error term with mean zero and a spot varying variance . By conditional independence of the pixel-wise intensities, only the spot-wise mean intensity is required in computations. is estimated directly from the intensities as their sample variance in each spot.
In the statistical analysis of several arrays and samples, many of the unknown parameters are shared, like array, dye, pen, gene and probe related effects; all data involving sample t contribute information on the unknowns . To assure statistical identifiability, some genes must be spotted at least in duplicate. The number of required replicated genes is independent on the total number of spotted genes, since replicates are used to estimate the common parameters. The whole dataset must include at least one self–self array or a dye–swap or a longer loop, necessary to identify the relative dye effect αCy3/αCy5. Beyond this, we do not require a connected design. To facilitate estimation, the model is reparametrized, so that the baseline β0, βe, βg, βm and αCy5 are estimated only on the basis of the variances in the binomials. Data relative to non-duplicated genes and samples hybridized only once are not used to estimate variances (Supplementary Data 2). MCMC was implemented to compute the joint and marginal posterior distributions of all unknowns of interest (Supplementary Data 3). The joint distribution describes dependencies between variables, e.g. between 's for various genes and samples. A priori nothing is assumed on the number of transcripts. The model naturally introduces dependency through shared experimental factors, so that the quantities are dependent. Observed dependencies in the data are then attributed backwards in part to this experimental dynamics. The residual unexplained dependence is summarized by the posterior joint distribution of the 's. Estimates of parameters are marginal posterior modes with 95% symmetric credibility intervals.
Microarray experiments
Microarray slides produced at the Microarray Facility at Health Enterprise Rikshospitalet-Radiumhospitalet were used. The slides contained 18 432 spots printed with 32 pens. The probes were human cDNAs of known genes and ESTs, selected from the Research Genetics 40K I.M.A.G.E. clone selection (Invitrogen). Probe lenth ranged from 525 to >2000 bp. A total of 17 DNA control probes (Lucidea Universal ScoreCard, Amersham Biosciences) were printed in equal amounts on six subarrays. These control spots were used for validation of our method and for determination of the hybridization factor optimal for the experiments on cancers and cell lines. Samples from 12 cervix cancers (FIGO stages 2b–4a) and a pool of 10 cancer cell lines, originating from mammary gland and cervix adenocarcinoma, liver hepatoblastoma, testis embryonal carcinoma, glioblastoma, melanoma, liposarcoma, lymphoma, B-lymphocyte myeloma and T lymphoblast leukaemia, were analysed. Total RNA was isolated from the tumours by use of Trizol reagent (Life Technologies), whereas total RNA from the cell lines was commercially available (Stratagene). Labelled cDNA was synthesized from 20 µg total RNA using Superscript II transcriptase (Life technologies) and Fairplay Microarray Labeling kit (Stratagene) in the presence of either Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia). Each tumour sample was co-hybridized with the cell line sample in a dye–swap design, yielding totally 24 microarrays analysed jointly. Two control samples, each containing 17 different mRNA sequences pre-mixed at specific concentrations, were included in the hybridization mixture for analysis of the control spots (Lucidea Universal ScoreCard, Amersham Biosciences). A total of 0.5 µl of each control sample was used, corresponding to a number of transcripts in the range of 5.8 × 105–5.8 × 109. RNA purity was optimal and equal for all samples and was therefore currently not used in our model. The slides were imaged at a resolution of 10 µm using an Agilent G2565BA scanner (Agilent Technologies). The laser power and the PMT voltage were 100%. Saturated spot intensities were corrected as described previously (19). Spot and background intensities were quantified using the GenePix 4.1 image analysis software (Axon Instruments). Bad spots, regions with high unspecific binding of dye and weak spots that were not automatically detected by the software were filtered out and excluded from the further analysis. The background signal was very low for control spots, and hence no background correction of intensities was necessary. For other spots, we performed analysis for intensities both background corrected (background subtraction) and not. A detailed description of materials is given in Supplementary Data 4.
Quantitative real-time PCR
The estimated transcript concentrations of eight genes that covered the whole concentration range were compared with data obtained by use of qRT-PCR. The TaqMan PCR system (Applied Biosystems) with a 7500 Sequence Detector (Perkin-Elmer) was used. cDNA was synthesized from 2 µg of the total RNA used in the microarray experiments by use of Superscript II transcriptase (Life Technologies). Pre-designed, gene-specific TaqMan probe and primer sets (Applied Biosystems), consisting of a specific fluorogenic probe and a pair of oligonucleotides, were used to run standard qPCRs for CDK4, CTNNB1, HK2, MYC, CSTA, PPT2, CCND1 and PDK2 (Supplementary Data 5). We employed 1 ng cDNA for all but the low abundant genes CCDN1 and PDK2, 10 ng cDNA were used, to increase the signal. The reactions were carried out in triplicate in a 25 µl reaction volume and a 96-well plate format. The transcript concentration of each gene was calculated using the standard curve method and presented relative to the expression of TBP, which served as an internal, endogenous control (22).
RESULTS
Validation of the methodology
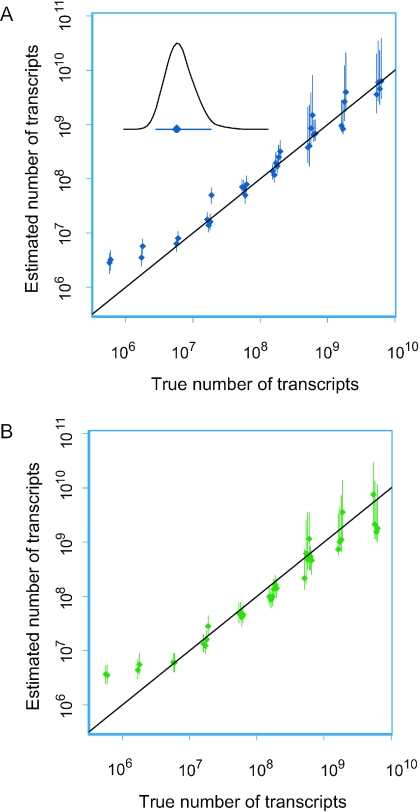
Two different dye–swap experiments using samples with known transcript concentrations bound to the control spots of our arrays were used for validation of our method. The spot intensities covered the whole detection range, from near background values to saturation. There was a highly linear relationship between estimates and true numbers of transcripts in double logarithmic plots (Figure 2). Scaling of the data in the first experiment (Figure 2A) led to a hybridization factor c of 4.31 × 10−10 and estimates in good agreement with the true numbers of transcripts. The lowest numbers (<107) were, however, overestimated, possibly because low intensity spots had more noise (14,16,17). The uncertainty of our estimates increased for the most abundant molecules with numbers >109.
Figure 2.
Validation of the methodology to estimate absolute numbers of transcripts. Control samples with 17 genes of known mRNA concentrations were used, each printed on six spots with six different pens. The data in (A) and (B) are based on two different dye–swap experiments and show estimated numbers of transcripts (y-axis) and true ones (x-axis). Positions on the x-axis are slightly shifted to facilitate visualization. Diagonal lines are shown; the fit is good when the line passes through the credibility interval. The inset in (A) shows the posterior probability density of the number of transcripts for a gene with estimated 5.8 × 107 mRNA molecules (mode) and its 95% credibility interval. The data in (A) were used to determine the hybridization factor, which was found to be 4.31 × 10−10. Analysis of the data in (B), using the hybridization factor from (A) showed a strong concordance between the two estimates, although the numbers of transcripts were slightly underestimated.
We used the hybridization factor c = 4.31 × 10−10 determined from experiment in Figure 2A to scale the data in the second experiment (Figure 2B). This also resulted in values consistent with the true ones, showing good reproducibility in our estimates (Figure 2B). There was a systematic underestimation of log10 concentrations by 0.1, which was small enough not to influence the estimated values significantly, since log10 concentrations were in the range 6–10. The optimal hybridization factor determined from the data of Figure 2B was 3.45 × 10−10 and hence only slightly different from the other one. For further comparison, the hybridization factor based on the control samples included in the cancer data was found to be 5.94 × 10−10, in agreement with the findings of the validation experiments. Estimates of the transcript concentrations obtained using either of the three hybridization factors were not significantly different.
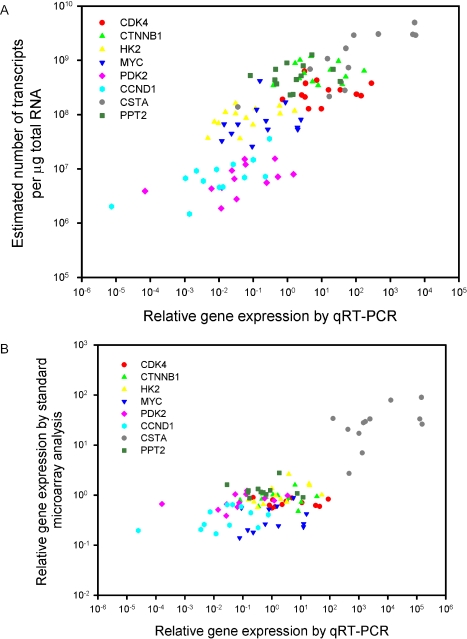
We performed a further validation of our method by comparing the transcript concentrations of eight genes in cancer cell lines and primary tumours with corresponding data achieved with qRT–PCR. The transcript concentrations of 10 157 genes and ESTs in 12 cervical tumours and in a pool of 10 cancer cell lines were determined. Eight genes covering the whole concentration range were selected for qRT–PCR. We found a clear and strong correspondence between the qRT–PCR data and the transcript concentrations determined with our method (Figure 3A). The best agreement occurred at intermediate and high concentrations, reflecting the increased uncertainties of both methods in quantification of low abundant transcripts. There was also positive correlation between estimated concentration and PCR data for some individual genes, despite a limited within-gene variability and few data points. While Figure 3A is based on not-background corrected data, Supplementary Figure 1 relates to background subtracted intensities. The difference is minimal, since background was not particularly high in the involved spots. The validation clearly showed that our technique was reproducible and stable, and estimates were reliable.
Figure 3.
qRT-PCR validation of the methodology to estimate absolute numbers of transcripts. Transcript concentration (number of transcripts per µg total RNA) of 10 157 genes and ESTs in 12 cervical tumours and a pool of 10 cancer cell lines was determined with our technique. Our estimated concentrations and the standard log-ratio data of eight genes covering the whole concentration range were plotted against the corresponding data achieved with qRT-PCR in (A) and (B), respectively. TBP was used as endogenous control of the qRT-PCR data. The data in (A) show a strong correlation between our estimates and the qRT-PCR data (r = 0.79, P < 0.0001, Pearson product moment correlation for all points). There was also positive correlation, sometimes significant, for some individual genes, despite a limited concentration range (r = 0.81, P = 0.0007 for CSTA; r = 0.71, P = 0.007 for CCND1; r = 0.50, P = 0.09 for HK2; r = 0.48, P = 0.1 for PDK2). The correlation remained if each estimated concentration was first normalized dividing it with the corresponding estimated concentration of TBP in the same sample and then plotted against the qRT–PCR data, since the estimate concentrations of this gene differed little among the tumours (P < 0.0001, data not shown). In (B), the cell line data served as a reference sample. The log-ratios in (B) were calculated relative to the intensities measured for the cell lines. All intensities were first normalized, using standard lowess normalization. There was also a significant correlation between these data and the qRT-PCR data (r = 0.77, p < 0.0001) which seemed mainly to be due to the highly expressed CSTA. Analysis of individual genes showed a significant correlation only for HK2 (r = 0.58, P = 0.05). Many genes with highly different concentration estimates (A) had similar standard log-ratio expressions (B). The concentrations can be compared directly between tumours and genes, whereas the log-ratios can be compared only between tumours for the same gene since their values depend on the transcript level of the reference sample.
Standard log-ratio expressions based on normalized intensities of tumour and cell line samples also showed a significant correlation to qRT–PCR data (Figure 3B), although the correlation was not as good as for the absolute concentration estimates when individual genes were considered. Many genes had approximately the same log-ratio although their absolute transcript concentrations differed considerably (Figure 3A), demonstrating the additional information that is achieved in absolute measures. We hypothesize that such information may increase our knowledge of transcript dosage and pathway regulation in a way that cannot be achieved by means of standard log-ratio expressions. The log-ratio expressions were also significantly correlated to the concentration estimates (data not shown). However, while concentrations can be compared directly between tumours and genes, this is not the case for log-ratio expressions, which can be compared only between tumours.
Transcript concentrations in cancer cell lines and cervix tumours
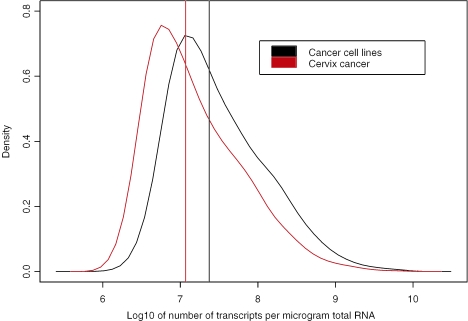
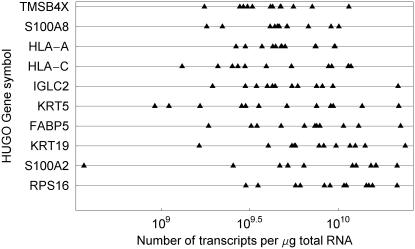
To test our model we analysed further the transcript concentration of 10 157 genes and ESTs determined for 12 cervical tumours and a pool of 10 cancer cell lines. We listed estimated concentrations for each gene and for each tumour, equipped with its 95% confidence interval in a table available at http://www.nr.no/pages/samba/area_emr_smbi_transcount. Similarly for the cancer cell lines, we reported estimated concentrations and confidence intervals for each gene in a second table, which also includes the mean concentrations of the cervix tumours. This is also available at the same web site. Concentrations are reported based on intensities both background corrected and not. Background corrected concentrations are systematically slightly smaller than not-background corrected ones, the difference being minimal and of interest only for low concentrations <3 × 106. The transcript distributions were skewed, with a heavy tail towards higher concentrations for both the cell lines and tumours (Figure 4). The concentrations ranged from 6.9 × 105 to 1.1 × 1010 transcripts per µg total RNA for the cell lines and from 9.9 × 105 to 9.1 × 109 for the tumour, here using averages over the 12 estimates. Limitation in the sensitivity of the microarray technology leads to lack of data for many genes at low expression. We therefore expect the existence of transcripts at lower concentrations than 105 as well. The cell lines had a slightly higher median concentration (2.36 × 107) than the tumours (1.17 × 107). The genes with the highest mean transcript concentration in the tumours are listed in Table 1, and include genes known to be involved in growth (TMSB4X, FABP5, TRAM2 and CPA4), immune response (HLA-A, S100A8, HLA-C, S100A10 and IGLC2), metabolism (RPS16 and CPA4), cell communication (LAMR1 and ITGA3) and signal transduction (LAMR1 and ITGA3).
Figure 4.
Posterior probability density of transcript concentrations (number of transcripts per µg total RNA) for cancer cell lines (black) and cervix cancer (red). The data of 10 157 genes and ESTs were included, and the calculations were based on a pool of 10 cell lines and 12 cervix tumours. The median value of each distribution is shown as a vertical line and was slightly higher for the cell lines than for cervix cancer. Both distributions were skewed towards higher values, and less abundant transcripts were more frequent than high abundant ones.
Table 1.
Genes of high transcript concentration in cervix cancer
| HUGO Gene symbol | Cervix tumour mean (×109) | Lower limit conf. int. (×109) | Upper limit conf. int. (×109) | Probability in top 20 | Reporter ID number | Gene ontology information |
|---|---|---|---|---|---|---|
| Genes of high transcript concentration in cervix cancer | ||||||
| TMSB4X | 9.05 | 7.71 | 9.33 | 1 | 868368 | Growth |
| HLA-A | 7.83 | 5.54 | 8.16 | 1 | 853906 | Immune response |
| S100A8 | 7.02 | 4.61 | 7.43 | 1 | 562729 | Immune response |
| HLA-C | 6.76 | 4.90 | 7.07 | 1 | 810142 | Immune response |
| KRT5 | 5.68 | 3.56 | 5.97 | 1 | 592540 | Unknown |
| KRT19 | 5.22 | 3.12 | 5.68 | 1 | 810131 | Unknown |
| S100A2 | 5.02 | 2.47 | 5.57 | 0.98 | 810813 | Unknown |
| FABP5 | 4.85 | 2.47 | 5.73 | 0.99 | 281039 | Growth |
| IGLC2 | 4.45 | 3.98 | 4.63 | 1 | 66560 | Immune response |
| RPS16 | 4.27 | 3.40 | 4.44 | 1 | 853151 | Metabolism |
| 701677 | 3.62 | 1.74 | 4.36 | 0.89 | Unknown | |
| RPS27A | 3.47 | 2.50 | 3.67 | 0.99 | 877827 | Unknown |
| IGHG1 | 3.38 | 2.04 | 3.60 | 0.94 | 289337 | Unknown |
| S100A10 | 3.11 | 1.92 | 3.28 | 0.89 | 756595 | Unknown |
| 753862 | 2.97 | 1.92 | 3.15 | 0.88 | Immune response | |
| LAMR1 | 2.91 | 1.97 | 3.14 | 0.89 | 884644 | Cell communication |
| Signal transduction | ||||||
| ITGA3 | 2.89 | 1.57 | 3.54 | 0.76 | 755402 | Cell communication |
| Signal transduction | ||||||
| 207029 | 2.83 | 1.97 | 3.02 | 0.86 | Unknown | |
| TRAM2 | 2.73 | 1.56 | 4.00 | 0.77 | 207550 | Growth |
| CPA4 | 2.36 | 1.22 | 3.05 | 0.48 | 359285 | Growth |
| Metabolism | ||||||
Mean transcript concentration (number of transcripts per µg total RNA) based on 12 tumours, lower and upper limit of the 95% confidence interval and the probability to be among the 20 genes with highest transcript concentration are listed. Background was very low and intensities were hence not corrected for this. Genes or ESTs without symbol are indicated by their probe identification number.
Transcript concentrations in the cell lines were estimated precisely, since the sample was hybridized 24 times. Much more data were available her than for the 12 tumour samples, each hybridized only twice. Reported 95% credibility intervals for each gene in the cell lines represent the precision achievable with our technique, given this level of replication of the experiment. Credibility intervals for each of the 12 cervix tumours depict the uncertainty of our estimates based on a single dye–swap. The accuracy of these estimated concentrations depends on the number of spots available for each gene. The standard deviation of the log-concentration for each of the 10157 genes and ESTs in the 12 cervix tumours ranged between 3.6 and 9.82. The corresponding coefficient of variation ranged between 0.53 and 1.059. There were only 217 genes with a coefficient of variation >1. This shows that our confidence intervals have a good precision.
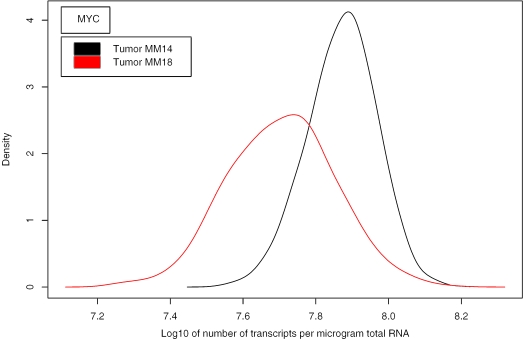
Biological variability between the 12 cervix tumours is described by the spread of the estimated concentrations. These could differ with a factor 10–100 between tumours (Figure 5), consistent with a large heterogeneity in the molecular composition even among tumours of the same histological type (23). In Supplementary Figure 3A we present low abundant transcripts. There was comparable heterogeneity on log-scale for genes with low and high concentration. Background corrected data are displayed in Supplementary Figures 2 and 3B. The high precision in our estimates enabled us to identify significant differences in the transcript concentration of many genes between individual tumours that were consistent with differences in qRT-PCR data, as demonstrated for the MYC gene in Figure 6. The biological heterogeneity was also reflected by differences in the median concentration of all transcripts, ranging from 6.00 × 106 to 2.31 × 107 transcripts per µg total RNA in the 12 tumours (data not shown).
Figure 5.
Transcript concentration (number of transcripts per µg total RNA) for 10 genes in cervix cancer with highest estimated mean concentration. Each point represents the estimated value of a single tumour, showing large differences in transcript concentration among the tumours. The within gene range (max–min) varies from 10 (TMSB4X; C4A) to 100 (S100A2; MAG).
Figure 6.
Posterior probability density of the transcript concentration (number of transcripts per µg total RNA) for the oncogene MYC in two different cervix tumours. The mode of this density is the estimated concentration as listed at http://www.nr.no/pages/samba/area_emr_smbi_transcount. There was a significant difference in the concentration between the tumours (P < 0.001, Kolmogorov–Smirnov test). The qRT-PCR data (relative to TBP) were 0.24 for MM14 and 0.023 for MM18, in agreement with our estimates.
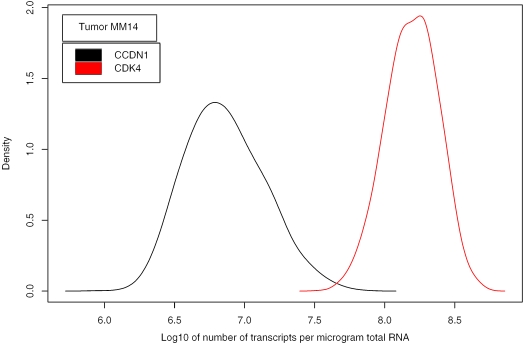
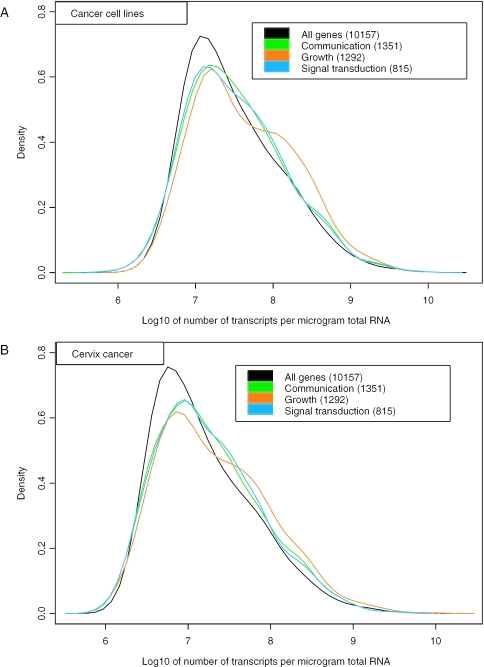
Our method allows us to compare the transcript concentration of different genes within a single sample. The accuracy was high enough for detection of significant differences within individual tumours, differences that were consistent with qRT-PCR data. This is exemplified in Figure 7, showing the transcript distribution of the cell cycle control genes CCND1 and CDK4, both involved in regulation of the G1 phase of the cell cycle. Similar analyses were also performed on groups of genes for the cell lines and tumour population. The genes involved in communication, growth and signal transduction were selected from gene ontology databases using the GoMiner software (24), and the transcript distributions were compared among the ontology groups (Figure 8). The distributions were skewed towards higher values for all groups of genes and only minor differences in the form and median values were observed. The skewed form and broad concentration range are therefore general characteristics that are valid also for relatively large subgroups of genes involved in a specific biological process. We found, however, that the relative frequency of highly concentrated transcripts was larger in the selected groups than in the entire set of genes.
Figure 7.
Posterior probability density of the transcript concentration (number of transcripts per µg total RNA) for the cell cycle control genes CCND1 and CDK4 in a cervix tumour. Both genes are involved in the G1 phase of the cell cycle. The mode of this density is the estimated concentration as listed at http://www.nr.no/pages/samba/area_emr_smbi_transcount. There was a significant difference in the concentration between the genes (P < 0.001, Kolmogorov-Smirnov test). The qRT-PCR data (relative to TBP) were 0.013 for CCND1 and 4.12 for CDK4, in agreement with our estimates.
Figure 8.
Posterior probability densities of transcript concentrations (number of transcripts per µg total RNA) of genes known to be involved in communication (green), growth (orange) and signal transduction (blue) for cancer cell lines (A) and cervix cancer (B). The calculations were based on a pool of 10 cancer cell lines and 12 cervix tumours. The number of genes in each functional group is indicated. Some genes were shared by the groups. The distribution of all 10 157 genes and ESTs is also shown (black). All distributions were skewed to the right and had similar median values.
DISCUSSION
We have developed a method for estimating precisely the transcript concentration of individual genes directly from spotted microarray intensity data. The method allows to compare concentrations of different transcripts within and between single tumours, which opens for new insight into transcript dosage and pathway regulation. In contrast, standard ratio estimation only allows comparison of the same transcript between two tumour samples.
The method is generally applicable, since the information about each microarray experiment required to achieve satisfactorily accurate estimates is easily available, though currently rarely made public. We encourage experimenters to make all data describing the experimental procedure available, submitting both channels separately, scanning parameters and measures of probe quantity to public repositories for microarray data like Arrayexpress (http://www.ebi.ac.uk/arrayexpress/), GEO and Cibex (http://cibex.nig.ac.jp/index.jsp). Since spotted microarrays are a widespread technology, our method will then facilitate the introduction of novel approaches to the study of the transcriptome.
Our method is based on four main ideas: we incorporate an extended number of covariates compared with other models (7); we treat unequal number of replicates per gene; we use the binomial process, which better depicts experimental dynamics and allows for estimation of the critical parameters β0, βg and αCy3/αCy5; and we avoid normalization and imputation of missing values and build a bottom-to-top coherent stochastic model, fully propagating uncertainty. These elements were crucial for achieving reliable estimates of transcript concentrations. In datasets based on known transcript concentrations we demonstrated a high accuracy of our estimates, especially at intermediate concentrations. Our results were better than in Dudley et al. (14), which reported a significant discrepancy between estimated and true concentrations both at intermediate and lower levels. The accuracy was in fact comparable with that achieved from methods based on in situ synthesized arrays (16,17), despite this technology uses standardized manufacturing and hybridization, so that probe specific biases are highly reproducible and predictable (25). Moreover, in datasets based on cervix tumours and cancer cell lines we found concentrations ranging from 105 to 1010 transcripts per µg total RNA. Assuming an RNA content of ∼1 µg per 105 cells (26), this corresponds to a range of 1–105 transcripts per cell. Previously published data of transcript numbers in humans are scarce. Zhang et al. (27) reported numbers ranging from 1 to 5300 per cell in human cancers, as determined by SAGE. However, higher numbers, up to 30 000 transcripts per cell, have been reported for genes in the mouse liver (13,28), making the number of 105, estimated for the highly abundant genes in our study, plausible. The skewed form of the transcript distributions observed in our work is also in agreement with earlier reports (27), and may be caused by underestimation at low concentrations, since the corresponding weak spots on the array are more frequently excluded than the bright ones. Comparison of our estimates and standard log-ratios with qRT-PCR data for a limited number of genes suggested that our estimates were more reliable than the ratios in reflecting the transcript levels. Discrepancies between microarray and PCR data have been reported with worries in previous studies (29). Our approach overcomes some of these difficulties, extracting information from microarray data that are more compatible with PCR results. Note that standard qRT-PCR itself does not provide accurate absolute expression levels (30). The accuracy of our method was high enough to detect significant differences in the transcript concentration within individual tumours as well as between tumours, differences that were consistent with qRT-PCR data. Our estimates therefore reliably revealed true transcript concentrations with satisfactory precision.
There are limitations of our methodology. Cross-hybridization and unspecific binding are not taken into account, and possible splice-variants for some of the genes or degree of homology between probe sequence and RefSeq sequence have not been considered. Currently, no analysis tools for microarray data are addressing these aspects. Other covariates could easily be included in our model when available, such as target length and labelling efficiency, probably leading to higher accuracy in the estimates. Of importance is also the slow convergence of the currently implemented MCMC algorithm. Results can require up to a few days of computation time. Our software may require some ad hoc implementation, specific to new datasets and covariates.
A major advantage of our model is that it can be directly applied on one or multi-colour experiments and on data from spotted oligoarrays, using base composition of the probes as covariates rather than the probe length. Moreover, the hierarchical structure enables integration of biological information about the samples, such as patient survival data, and known interactions between genes, in a coherent Bayesian setting. If the mRNA weight is not available, and significant variability in the proportion of mRNA in total RNA is suspected, or if the hybridization factor is not available, it is possible to scale each sample so that the sum of estimated transcripts are equal. Direct comparison of such scaled concentrations is still possible between and within samples, but the interpretation as absolute concentrations is lost.
Our method possesses several further beneficial features. No normalization and imputation of missing values is needed. Our model performs automatically unsupervised normalization, very similarly to ANOVA based methods (11), since the main factors are present in Equation 1; we incorporate explicitly more sources of variability, including scanning. Current normalization methods are often platform-dependent and based on hypotheses on the gene expressions difficult to test. Misuse of normalization is rather common in practice (31). The need for balanced designs, also for linear mixed effect models (11), often leads to discarding genes or requires imputation of missing values. Current methods for imputation fail if the missing mechanism is not at random or if the level of missing exceeds 20% (32). Our method does not impute missing values but can directly handle unbalanced datasets.
Another characteristic of our method is that few constraints are imposed on the experimental design. The reference design is common because it allows in-house re-utilization of results. However, it requires stable reference samples, it leads to low statistical power, while the reference is uselessly measured many times (33). Our method allows for re-utilization of results without the need of a reference. Thus it opens for new possibilities of meta-analyses (34). Such analyses are currently built on top of statistical tests to detect differential expressions (35,36). Since the result of these tests may depend on experimental protocol and microarray platform, bias may lead to wrong conclusions. With our method, data from different studies can be combined at the basic level of transcript concentrations, regardless of whether studies use amplified or non-amplified starting material, cDNA or oligonucleotide platforms. By making more experimental information public available, such as spot intensities and standard deviations of each channel, scanner settings and measures of probe quantity and sequence length, all microarray data can be re-used in new investigations, leading to a better exploitation of the data and more precise results. In particular, our method may contribute to new insight into the regulation of pathways and be useful in the development of improved therapeutic strategies in which knowledge of the absolute concentrations is directly utilized.
SUPPLEMENTARY DATA
Supplementary data is available at NAR online.
Supplementary Material
Acknowledgments
We thank L. Holden, E. Hovig, M. Langaas, O. Myklebost, T. Speed, T. Stokke and B. Ylstra for useful discussions, V. Nygard for help with the software interface. Financial support was provided by The Norwegian Research Council (FUGE Bioinformatics; StAR; GeneStat), The Norwegian Microarray Consortium, Health Enterprise Rikshospitalet-Radiumhospitalet, The Norwegian Cancer Society and The Dutch BSIK/BRICKS Consortium.
Conflict of interest statement. None declared.
REFERENCES
- 1.Velculescu D.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnason D., Luo S., McCurdy S., Foy M., Ewan M. Gene expression analysis by massive parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 3.Brown P., Botstein D. Exploring the new world of the genome with DNA microarrays. Nature Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 4.Lash A.E., Tolstoshev C.M., Wagner L., Schuler G.D., Strausberg R.L., Riggins G.J., Altschul S.F. SAGEmap: a public gene expression resource. Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polyak K.P., Riggins G.J. Gene discovery using the serial analysis of gene expression technique: implications for cancer research. J. Clin. Oncol. 2001;19:2948–2958. doi: 10.1200/JCO.2001.19.11.2948. [DOI] [PubMed] [Google Scholar]
- 6.Holloway A., vanLaar R., Tothill R., Bowtell D. Options available—from start to finish—for obtaining data from DNA microarrays II. Nature Genet. 2002;32:481–489. doi: 10.1038/ng1030. [DOI] [PubMed] [Google Scholar]
- 7.Butte A. The use and analysis of microarray data. Nature Rev. Drug Discov. 2002;1:951–960. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- 8.Slonim D. From patterns to pathways: gene expression data analysis comes to age. Nature Genet. 2002;32:502–508. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- 9.Churchill G. Fundamentals of experimental design for cDNA microarrays. Nature Genet. 2002;32:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- 10.Quackenbush J. Microarray data normalisation and transformation. Nature Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 11.Kerr M., Martin M., Churchill G. Analysis of variance for gene expression microarray data. J. Comput. Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- 12.Newton M., Kendziorsky C., Richmond C.e. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J. Comput Biol. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- 13.Barth R., Gross K., Gremke L., Hastie N. Developmentally regulated mRNAs in mouse liver. Proc. Natl Acad. Sci. USA. 1982;79:500–5004. doi: 10.1073/pnas.79.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudley A., Aach J., Steffen M.A., Church G.M. Measuring absolute expression with microarrays with calibrated reference sample and an extended signal intensity range. Proc. Natl Acad. Sci. USA. 2002;99:7554–7559. doi: 10.1073/pnas.112683499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Townsend J., Hartl D. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3:research 0071.1–0071.16. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Held G., Grinstein G., Tu Y. Modeling of DNA microarray data by using physical properties of hybridization. Proc. Natl Acad. Sci. USA. 2003;100:7575–7580. doi: 10.1073/pnas.0832500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hekstra D., Taussig A., Magnasco M., Naef F. Absolute mRNA concentrations from sequence-specific calibration of oligonucleotide arrays. Nucleic Acids Res. 2003;31:1962–1968. doi: 10.1093/nar/gkg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dror R., Murnick J., Rinaldi N., Marinescu V., Rifkin R., Young R. Bayesian estimation of transcript levels using a general model of array measurement noise. J. Comput. Biol. 2003;10:433–452. doi: 10.1089/10665270360688110. [DOI] [PubMed] [Google Scholar]
- 19.Lyng H., Badiee A., Svendsrud D.H., Hovig E., Myklebost O., Stokke T. Profound influence of non-linearity in microarray scanners on gene expression ratios: analysis and procedure for correction. BMC Genomics. 2004;5:10. doi: 10.1186/1471-2164-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaumont M., Rannala B. The Bayesian revolution in genetics. Nature Rev. Genet. 2004;5:251–261. doi: 10.1038/nrg1318. [DOI] [PubMed] [Google Scholar]
- 21.Peterson A., Heaton R., Georgiadis R. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mocellin S., Rossi C., Pilati P., Nitti D., Marincola F. Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol. Med. 2003;9:185–195. doi: 10.1016/s1471-4914(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 23.Wong Y., Selvanayagam Z., Wei N., Porter J., Vittal R., Hu R., Lin Y., Liao J., Shih J., Cheung T., et al. Expression genomics of cervical cancer: molecular classification and prediction of radiotherapy response by DNA microarray. Clin. Cancer Res. 2003;9:5486–5492. [PubMed] [Google Scholar]
- 24.Zeeberg B., Feng W., Wang G., Wang M., Fojo A.T., Sunshine M., Narasimhan S., Kane D., Reinhold W., Lababidi S., et al. Gominer: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C., Wong W. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. Vol 1. Canada: John Wiley and Sons, Inc.; 2000. [Google Scholar]
- 27.Zhang L., Zhou W., Velculescu V.E., Kern S.E., Hruban R.H., Hamilton S.R., Vogelstein B., Kinzler K.W. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 28.Hastie N., Held W., Toole J. Multiple genes coding for the androgen-regulated major urinary proteins of the mouse. Cell. 1979;17:449–457. doi: 10.1016/0092-8674(79)90171-5. [DOI] [PubMed] [Google Scholar]
- 29.Etienne W., Meyer M., Peppers J., Meyer R. Comparison of mRNA gene expression by RT–PCR and DNA microarray. Biotechniques. 2004;36:618–626. doi: 10.2144/04364ST02. [DOI] [PubMed] [Google Scholar]
- 30.Shih S., Robinson G., Perruzzi C., Calvo A., Desai K., Green J., Ali I., Smith L., Senger D. Molecular profiling of angiogenesis markers. Am. J. Pathol. 2002;161:35–41. doi: 10.1016/S0002-9440(10)64154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T. Normalisation for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 33.Townsend J. Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics. 2003;4:41. doi: 10.1186/1471-2164-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreau Y., Aerts S., De Moor B., De Stooper B., Dabrowski M. Comparison and meta-analysis of microarray data: from the bench to the computer desk. Trends Genet. 2003;19:570–577. doi: 10.1016/j.tig.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes D.R., Barrette T.R., Rubin M.A., Ghosh D., Chinnaiyan A.M. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway disregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 36.Choi J.K., Yu U., Kim S., Yoo O.J. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19(Suppl. 1):i84–i90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.