Abstract
The complex gene expression responses of Anopheles gambiae to microbial and malaria challenges, injury, and oxidative stress (in the mosquito and/or a cultured cell line) were surveyed by using cDNA microarrays constructed from an EST-clone collection. The expression profiles were broadly subdivided into induced and down-regulated gene clusters. Gram+ and Gram− bacteria and microbial elicitors up-regulated a diverse set of genes, many belonging to the immunity class, and the response to malaria partially overlapped with this response. Oxidative stress activated a distinctive set of genes, mainly implicated in oxidoreductive processes. Injury up- and down-regulated gene clusters also were distinctive, prominently implicating glycolysis-related genes and citric acid cycle/oxidative phosphorylation/redox-mitochondrial functions, respectively. Cross-comparison of in vivo and in vitro responses indicated the existence of tightly coregulated gene groups that may correspond to gene pathways.
Current molecular, cellular, and genomic methods are now applied to the mosquito Anopheles gambiae because of its importance as the major vector for human malaria in sub-Saharan Africa. An important focus of such studies is the immune defense system of the mosquito, which may be implicated in the response to invasion by the Plasmodium parasite and, therefore, in the success or failure of malaria transmission.
As illustrated by the fruit fly Drosophila, insects possess a sophisticated innate immune system controlled by signal-transduction pathways that resemble, in some respects, the corresponding immune pathways of mammals (1). Drosophila immunity has served as a model and greatly facilitated corresponding studies in the mosquito. However, until recently, progress was slowed by cloning individual immune components one at a time. Genomic-scale investigation was initiated by the isolation and characterization of nearly 6,000 mosquito expressed sequence tags (ESTs) from subtracted and normalized cDNA libraries (2). These libraries, prepared from cultured hemocyte-like cell lines, identified numerous new putative components of the immune response. A fuller catalog can be expected soon from the annotation of the mosquito's whole-genome sequence (http://www.ncbi.nlm.nih.gov:80/cgi-bin/Entrez/map_search?chr=agambiae.inf). Of course, candidates identified by sequencing require experimental validation.
Here, we use first-generation cDNA microarrays for high-throughput expression profiling of mosquito defense reactions. We determine the molecular composition of the immune response mounted by a hemocyte-like cell line when challenged by bacteria or elicitors derived from microbial surfaces. As a control, we define the response of the cell line to oxidative stress after exposure to hydrogen peroxide. In addition, we compare and contrast the responses of adult mosquitoes when challenged by injection of bacteria or by a control sterile injury. In a pilot study, the effects of malaria infection are also examined.
Materials and Methods
Construction of Anopheles Microarrays.
A two-step PCR amplification of EST clone inserts used amino-modified T3 and T7 primers and bacterial cultures as primary templates. Ethanol-precipitated PCR products were resuspended in ArrayIt Microspotting solution (Telechem International, Sunnyvale, CA) and spotted on aminosilane-coated glass slides with the Omnigrid microarray spotter (GeneMachines, San Carlos, CA). The slides then were incubated at 60°C for 3 h and 100°C for 10 min to cross-link the DNA.
Probe Preparation and Microarray Hybridizations.
cmRNA was synthesized with the Ambion MEGAscript T7 RNA synthesis kit (Ambion, Austin, TX) from double-stranded cDNA primed with an oligo d(T)-T7 promoter sequence and complementary cDNA probes were synthesized and labeled with Cy-3-dUTP and Cy-5-dUTP fluorescent nucleotide analogs, in a random primed first-strand reverse-transcription reaction. After removal of unincorporated dNTPs with a Qiagen PCR purification kit (Qiagen, Chatsworth, CA), the probes were combined, lyophilized, and resuspended in hybridization buffer containing 50% formamide, 6× SSC, 0.5% SDS, 5 × Denhardt's reagent, and 0.5 mg/ml poly(A) DNA. Arrays were prehybridized in 6 × SSC, 0.5% SDS, and 1% (vol/vol) BSA at 42°C for 90 min, hybridized overnight at 42°C in humidified hybridization chambers, washed twice in 0.1 × SSC, 0.1% SDS (30 min), twice in 0.1 × SSC (15 min), rinsed with de-ionized H2O, and dried.
Microarray Data Analysis.
Intensities of hybridized probes were estimated with a GenePix 4000b (AXON Instruments, Foster City, CA) semiconfocal microarray scanner and software. The analyzed genes had median signal/background >3, median background <500, and median signal/noise >3, or showed exceptionally high intensity in one sample. The hybridization ratio for each probe was calculated by using the estimated ratio of medians after normalization of the total array intensity. The estimated fold regulation value of each clone was expressed as the log-2 value of the normalized ratio of medians. To ensure reproducible regulation profiles, genes were only selected for cluster analysis if regulated above a threshold cutoff, set as 3-fold in at least three experiments for Figs. 1 and 2, and 2-fold in two experiments in Fig. 3, respectively. Clustering analysis and graphic presentations were performed as described (3, 4) by using the CLUSTER and TREEVIEW software (available at http://rana.stanford.edu/software/). Expression patterns of known immunity genes matched previously published data, and for 12 selected genes in cell-line experiments were verified by semiquantitative PCR.
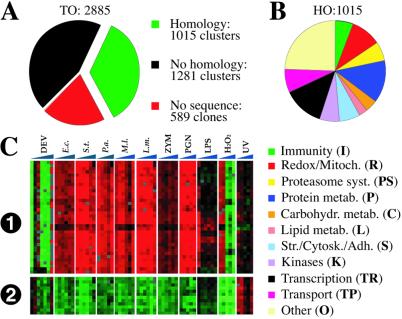
Figure 1.
Microarray methods. (A) Clone and clone cluster composition as described in the text. (B) Allocation of the HO genes to functional classes according to previous work (9) and manual database searches. (C). Reproducibility of un-averaged expression data (i) of a gene of complex splicing pattern and unknown function represented by 17 clones in the microarray and (ii) of a ribosomal protein gene represented by six clones. Two replicate spots (consecutive rows) on the array are shown for each clone. Normally, the 34 and 12 rows, respectively, would be averaged. Vertical sectors and columns represent profiles from seven developmental stages (DEV) and six time-course studies in cells challenged as in Fig. 2 or by UV irradiation. Note that, consistently, gene 1 is underrepresented in all developmental stages except the third (2nd instar larvae) and last (adult); it is overexpressed in all bacterial and PGN experiments but only in the first two time points of H2O2 challenge. Gene 2 is consistently down-regulated in all bacterial, PGN, and H2O2 experiments but is up-regulated by UV challenge.
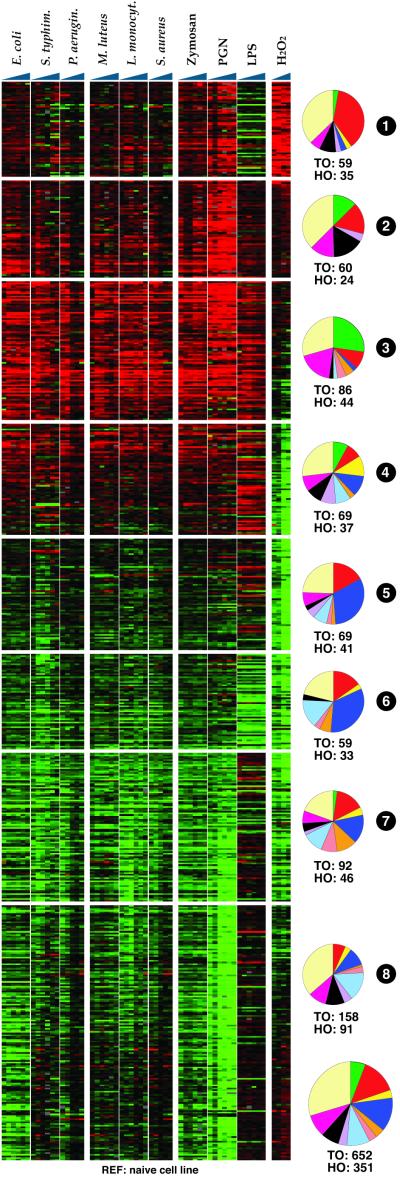
Figure 2.
Differential expression profiles of immune-competent cells challenged by the indicated bacteria, purified elicitors (Zymosan, PGN, LPS), or H2O2 and assayed in six time-course experiments (1, 4, 8, 12, 18, and 24 h after challenge). Only 652 TO and 351 TO genes showing at least 3-fold regulation in at least three experimental time points are included in the analysis. The reference probe was from unchallenged (naïve) cells. The SOM-sorted cluster matrix was divided into eight clusters based on transitions between distinguishable expression profiles. Pie charts show composition of each cluster and the total regulated set (Inset according to functional classes; see Fig. 1B). The complete gene content of each cluster is provided in Table 2 and Fig. 4).
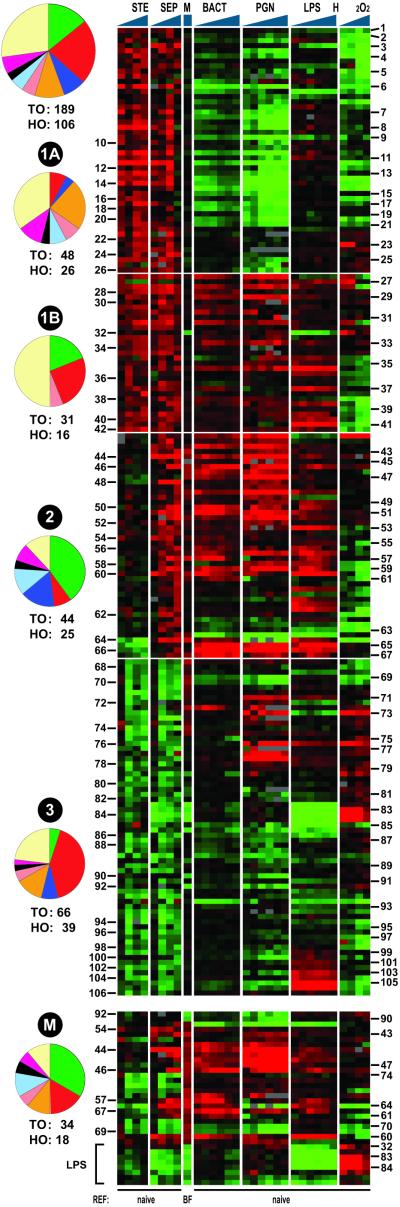
Figure 3.
In vivo responses to sterile (STE) and septic (SEP) injury of adult mosquitoes and cross-correlation with in vitro data from the cell line. The expression profiles (1, 6, 12, and 24 h) of genes showing at least 2-fold regulation in at least two of the eight STE and SEP experiments, or at least 2-fold regulation after malaria (M) challenge, were SOM-sorted into clusters 1, 2, and 3. The respective profiles from the cell-line challenge experiments (see Fig. 2) then were added after averaging the responses to six different bacteria (BACT). The combined expression profiles were again SOM-sorted, resulting in the subdivision of cluster 1 into 1A and 1B. Limited manual repositioning of genes within the clusters helped to highlight the patterns, including the existence of subclusters. The profiles of genes that were regulated after malaria challenge were used to construct the malaria-specific cluster M, which was analyzed by hierarchical clustering (Bottom). Numbers identify all 106 regulated HO genes, whose predicted functions are presented in Table 1. Brackets (Bottom) indicate a gene group that is strongly down-regulated by malaria and LPS (see also cluster 3). The complete gene content is provided in Table 3 and Fig. 5.
Cell Culture and Mosquito Challenge.
The A. gambiae cell line 4a-3B and adult female mosquitoes were maintained and challenged or infected as described (2, 5–7). Heat-killed bacteria were applied to the cell line at OD = 0.1, which ensured strong transcriptional regulation and probably over-stimulation (8). Peptidoglycan (PGN, 77140, Fluka) and lipopolysaccharide (LPS, L-2880, Sigma) were applied at 10 μg/ml, Zymosan (Sigma, Z-4250) was at 100 μg/ml, and the H2O2 concentration was 20 mM.
Results and Discussion
Microarray Analysis.
The cDNA microarrays were doubly spotted with PCR-amplified inserts of 3840 EST clones (2), of which 3,251 had been sequenced and assembled into 2,296 clone clusters, and 589 had not been sequenced (Fig. 1A). The combined total of sequenced clusters and nonsequenced clones is the maximal number of candidate genes [total of sequenced clusters and nonsequenced clones (TO), 2,885]. Of special interest were 1,015 gene clusters showing homology to genes of other organisms (HO) clusters (Fig. 1B) sufficient to assign all but 80 of these to functional classes. Differential gene expression profiles in response to a variety of challenges proved highly consistent between duplicate spots and clones of the same gene (Fig. 1C) and were averaged. They were displayed in red (overexpression) or green (underexpression), relative to the indicated reference sample. In time course studies (Figs. 2 and 3), data from successive experiments appeared as colored horizontal lines. We used both the self-organizing maps (SOMs) and hierarchical clustering methods (3, 4) to cluster genes with similar expression profiles. The SOMs were subdivided manually into clusters, according to notable transition in patterns of gene expression. In each study, the composition of differentially expressed clusters, in terms of functional classes, were displayed in accompanying pie charts. We monitored the 10 largest classes by consistent colors in the pie charts (see Fig. 1B). The full composition of the differentially expressed clusters was recorded in Table 1 and supporting Tables 2–4 (which are published on the PNAS web site, www.pnas.org).
Table 1.
Clusters of genes/functional classes
| Gene | Gene ontology | Cluster |
|---|---|---|
| I: Immunity | ||
| 31 | DOPA decarbox. | 1B |
| 33 | chitin-bind. | 1B |
| 38 | CD-36 | 1B |
| 44 | PGRP-LB | 2, M |
| 46 | GNBP | 2, M |
| 47 | CED-6-like | 2, M |
| 50 | FBN lectin | 2 |
| 53 | Cactus | 2 |
| 57 | aTEPIV | 2, M |
| 59 | serine pro. | 2 |
| 61 | serine pro. 14D | 2, M |
| 64 | FBN lectin | 2, M |
| 67 | LRR/Toll-like | 2, M |
| 78 | lectin | 3 |
| 106 | serpin | 3 |
| R: Redox/mit. | ||
| 17 | aldehyde deh. | 1A |
| 22 | mit. rib. prot. | 1A |
| 27 | cyt. c oxid. I (mit) | 1B |
| 28 | ferredoxin red. | 1B |
| 35 | cyt. P450 | 1B |
| 36 | oxidored. | 1B |
| 52 | cyt. P450 CYP-9 | 2 |
| 54 | ferredoxin red. | 2, M |
| 71 | mit. siderofl. | 3 |
| 73 | cyt. c oxid. | 3 |
| 74 | mit. ph. carrier | 3, M |
| 76 | NADH deh. 1 | 3 |
| 79 | NADH deh. 18 kDa | 3 |
| 84 | cyt. P450 | 3, M |
| 91 | Fe-S oxidored. | 3 |
| 94 | cyt. c oxid. | 3 |
| 95 | cyt. c red. | 3 |
| 96 | mit. ATP-synth. | 3 |
| 97 | NADH deh. B12 | 3 |
| 98 | NADH deh. 1a | 3 |
| 99 | NADH deh. 24 kDa | 3 |
| 102 | Fe-S oxidored. | 3 |
| 103 | mit. fum. hyd. | 3 |
| 104 | cyt. c oxid. Vib | 3 |
| P: Protein metab. | ||
| 25 | N-acetyl. Ph.-inos. | 1A |
| 51 | 4-nitrophenylph. | 2 |
| 55 | cystath. beta-synth. | 2 |
| 58 | peptidylpropylisom. | 2 |
| 63 | translocon assoc. | 2 |
| 100 | ribonucleoprot. | 3 |
| 101 | ribosomal prot. | 3 |
| 105 | glut. tRNA synth. | 3 |
| TP: Transport | ||
| 8 | mRNA recept./transp. | 1A |
| 9 | importin | 1A |
| 16 | innex.-domain transp. | 1A |
| 48 | dopam. transp. | 2 |
| 49 | vac. protein sort. | 2 |
| 83 | integ. membr. transp. | 3, M |
| C: Carboh. metab. | ||
| 3 | lactate deh. | 1A |
| 6 | alpha-glucosid. | 1A |
| 7 | Pepck | 1A |
| 10 | uridine kinase | 1A |
| 14 | trehalase | 1A |
| 18 | alpha-glucosid. | 1A |
| 68 | dihydrolip. Succ.tran. | 3 |
| 69 | isocitr. deh. | 3, M |
| 72 | GDP-D-man. Deh. | 3 |
| 86 | glyceral.-3-P deh. | 3 |
| 92 | 2-ketoglut. dehydr. | 3, M |
| L: Lipid metab. | ||
| 2 | fatty-acid CoA lig. | 1A |
| 19 | proteolipid hydr. | 1A |
| 29 | fatty-acid CoA lig. | 1B |
| 90 | ceramidase | 3, M |
| S: Str/Cytosk/Adh | ||
| 4 | tubulin-beta | 1A |
| 23 | actin binding | 1A |
| 43 | tubulin-beta | 2, M |
| 56 | cytoskeletal | 2 |
| 66 | cell surf. recept. | 2 |
| TR: Transcr/Nucl | ||
| 20 | transcr. fact. | 1A |
| 60 | DNA polym. | 2, M |
| 81 | histone-like tr. fact. | 3 |
| O: Others | ||
| 1 | beta-adaptin | 1A |
| 5 | CG12096 | 1A |
| 11 | CG5642 | 1A |
| 12 | CG11261 | 1A |
| 13 | CG5642 | 1A |
| 15 | aden. hom. cyst. | 1A |
| 21 | CG3634 | 1A |
| 24 | Ca channel | 1A |
| 26 | CG10347 | 1A |
| 30 | CG13004 | 1B |
| 32 | CG13319 | 1B, M |
| 34 | menin | 1B |
| 37 | cAMP phosphodiest. | 1B |
| 39 | CG4710 | 1B |
| 40 | CG14074 | 1B |
| 41 | G-coupled prot. | 1B |
| 42 | v-ATPase | 1B |
| 45 | transferase | 2 |
| 62 | chaperone | 2 |
| 65 | cats-up-like | 2 |
| 70 | dsRNA bind. RNase | 3, M |
| 75 | ATPase | 3 |
| 77 | CG10433 | 3 |
| 80 | CG14450 | 3 |
| 82 | Kisir/MSF-1 hom. | 3 |
| 85 | Hsp70-like | 3 |
| 88 | CG4785 | 3 |
| 89 | hypoxia-induced | 3 |
| 93 | acid phosphat. | 3 |
Cell Responses to Microbial Challenge and Oxidative Stress.
We have used an immune-competent, hemocyte-like cell line, 4a-3B (5), to determine gene expression responses to microbial challenge in a simplified in vitro system. The cells were challenged with six different Gram− and Gram+ bacteria species (heat killed to avoid differences in growth dynamics) at concentrations that would ensure strong transcriptional regulation (8). Three purified elicitors of microbial origin also were used. As a control, cells were challenged with H2O2, revealing the response to strong oxidative stress. The responses were studied in parallel time-course experiments, each with up to six time-points (1–24 h) which, together with the redundant sequence representation in the microarrays, ensured the robustness of the expression profiles.
Fig. 2 summarizes differential profiles based on 211,200 data points, from 652 TO and 351 HO genes that scored as differentially regulated relative to naïve (control) cells (see also Table 2 and Fig. 4, which is published as supporting information on the PNAS web site). The SOM software arranged the genes in a continuum of global profiles, which were easily subdivided into eight expression clusters (1–8, as labeled in Fig. 2) by considering notable transitions. Collectively, the expression clusters represented the 10 major functional gene classes in frequencies comparable to the microarray as a whole (compare Figs. 1B and 2 Inset). However, different clusters were strongly enriched in one or more functional gene classes.
Cluster 1 is distinguished by consistent and strong induction upon H2O2 treatment, and by numerous genes (13/35 HO) that are predicted to function in oxidation/reduction reactions or be expressed in mitochondria (redox/mitochondrial class). One of these genes encodes thioredoxin reductase, the key enzyme that regulates redox balance in insects (9). Conversely, cluster 3 shows strong induction of a different set of genes upon challenge by both bacteria and microbial elicitors, but not by H2O2. This set is strongly enriched in genes that belong to the immunity functional class (12/44) and depleted of genes belonging to several other functional classes, notably the redox/mitochondrial class (4/44). In short, the induction of cluster 1 can be considered a signature of the cellular response to oxidative stress, whereas induction of cluster 3 represents the distinctive signature of a core immune response to microbial challenge.
Additional up-regulation responses to microbial challenge are reflected in clusters 2 and 4. These clusters add 6 immunity genes to the 12 genes present in cluster 3, and thus, together encompass 18 of the 20 immunity genes present in Fig. 3. These three immunity clusters are enriched in additional functional classes. Together they include 14 of 31 transport genes; clusters 2 and 4 encompass 7/24 transcription/nuclear genes; cluster 4 alone includes 3/12 kinase genes and 4/11 proteasome genes. Although insects are known to detect and respond to different classes of microbes by using distinct signal transduction pathways (10), no subclusters differentiating the responses to Gram+ and Gram− bacteria were defined at the tested bacterial concentrations. This finding is likely due to over-stimulation, overlap of responses to various bacterial components exposed by heat-killing, or subsequent effects such as phagocytosis and digestion of debris.
Four other clusters, 5–8, are largely down-regulated by both microbial and oxidative challenge. Genes encoding ribosomal proteins and other components of protein metabolism are especially numerous (40 genes, as compared with 6 in clusters 1–4), possibly indicating incidence of apoptosis. The structural/cytoskeletal/adhesion class is also strongly represented in the down-regulated expression clusters (27 genes, as compared with 4 in clusters 1–4), suggesting that major changes in cell shape and adhesiveness may occur upon microbial challenge. Finally, carbohydrate and lipid metabolism genes are overrepresented in the down-regulated cluster 7 (9 genes, as compared with 5 in clusters 1–4).
In contrast to the uniformity of responses to different bacteria (and Zymosan, extract of yeast cell walls), two purified microbial elicitors, PGN and LPS, result in distinct responses. In response to LPS only, the immunity clusters 3 and 4 show strong, reasonably coherent gene activation, and only cluster 6 (which is especially rich in ribosomal proteins) is coherently down-regulated. Four other clusters (1, 5, 7, and 8) show noncoherent gene-specific effects of LPS, either positive or negative for different genes within the same expression cluster. The nature of the atypical LPS response is explored further below. In contrast, PGN induces strong coherent responses in most clusters. Its activation response is robust in clusters 1, 2, and 3, indicating an unusual concomitance of the oxidative stress and immune responses. Thus, PGN binding may be responsible for an oxidative burst which occurs in innate immunity and is induced by attachment of bacteria to immune-competent cells (11). In most other respects, the PGN profiles resemble the corresponding up or down responses to bacteria. The PGN-recognition protein gene PGRP-LB in cluster 2 is strongly up-regulated (over 130-fold after PGN challenge for 8 h), suggesting that this gene may indeed encode the bona fide mosquito receptor of PGN.
Adult Mosquito Responses to Sterile and Septic Injury and Malaria Infection.
To investigate the in vivo immune response against bacteria, one group of mosquitoes were pricked with a needle dipped in a thick suspension of live E. coli and Micrococcus luteus (septic injury), and another group were pricked with a sterile needle (sterile injury). For these experiments, we used the melanotically encapsulating malaria-refractory strain L3–5, in which robust immune responses have been reported (6, 12), and as reference, naïve uninjured mosquitoes sampled at 0 h. The experimental groups were sampled at 1, 6, 12, and 24 h, leading to the detection of a broad range of genes and suggestive evidence of complex temporal kinetics. The responses were weaker than in the cell lines, suggesting that only a minority of the mosquito's cells are implicated in these in vivo responses and are partially masked by nonresponding tissues (13).
Nevertheless, the in vivo experiments document clear responses involving 106 HO genes which are numbered in Fig. 3 and listed in Table 1 (see also Fig. 5, which is published as supporting information on the PNAS web site). Nearly half belong to only three functional classes: 24 redox/mitochondrial gene, 15 immunity genes, and 11 carbohydrate metabolism genes. The predominant responses observed in these experiments are seen in both sterile and septic injury and, therefore, can be ascribed to injury/wound healing. They include injury-induced up-regulation of 42 HO genes relative to naïve mosquitoes (Cluster 1) and injury-induced down-regulation of a second set of 39 genes (Cluster 3). Superimposed on these shared injury responses, only septic treatment activates a distinctive cluster 2, which represents the reaction to bacterial infection.
Cross-Comparison and Detailed Analysis of in Vitro and in Vivo Responses.
These in vivo responses were compared with the in vitro responses of the cell line to bacteria. For this cross-comparison, the response profiles of cultured cells to the six different species of bacteria were first averaged. The already clustered in vivo expression profiles of the 189 TO/106 HO genes then were combined with the respective profiles of the same genes from the cell-line study (Fig. 3, Fig. 5, Table 3). The three clusters then were subjected individually to SOM reanalysis. This cross-comparison clearly subdivided the in vivo cluster 1 into two, 1A and 1B, resulting in a total of four with remarkably distinct composition (Fig. 3 and Table 1). Importantly, the comparison also highlighted the existence of some specific features within the new clusters, as discussed below.
Cluster 1A.
This cluster is rapidly activated by injury but down-regulated in vitro in response to bacteria, PGN, and H2O2. In this respect, injury is perceived as distinct from and even opposite to microbial challenge. These responses are especially antithetical for the compact group of genes 6 to 21 (Fig. 3). The genes in this group are particularly strongly down-regulated upon in vitro bacterial challenge or PGN treatment and also end their septic injury-induced activation prematurely (before 24 h), while remaining activated in sterile injury. The composition of cluster 1A is unusual (Table 1), as genes of the carbohydrate metabolism class predominate (6/26 HO genes). They encode enzymes involved in glycolysis-related processes (glucosidases, trehalase, lactate dehydrogenase), an enzyme essential for the anaplerotic reactions (phosphoenolpyruvate carboxykinase), and a key enzyme for the pentose phosphate pathway, uridine kinase; no enzymes directly involved in the citric acid cycle are represented in this cluster. Other notable features are the total absence of immunity genes, the scarcity of redox/mitochondrial genes, and the presence of three transport genes and two cytoskeletal genes (Table 1). Furthermore, aside from the 10 major classes, this cluster includes nine miscellaneous genes, six of which have Drosophila homologues of unknown function. The response to H2O2 highlights two apparent groupings: genes 1–5 show stronger down-regulation than after bacterial challenge, and genes 22 to 26 are weakly up-regulated.
Cluster 1B.
In strong contrast to 1A, this cluster represents a response that is activated not only by injury but also by in vitro microbial challenge. Interestingly, cluster 1B can be distinguished into two gene groups following H2O2 challenge: genes 27–34 are weakly or transiently up-regulated by oxidative stress, whereas genes 35 to 42 are clearly down-regulated. In contrast, the latter group shows particularly strong activation by LPS. The composition of cluster 1B is radically different from that of cluster 1A and encompasses genes of only three major classes. One of the four redox/mitochondrial genes encodes a ferredoxin reductase, a FAD flavoprotein that belongs to the family of pyrimidine nucleotide disulfide oxidoreductases and may affect the redox state of the internal milieu. The immunity class is represented by three diverse genes, two of which may serve “mopping-up” functions only indirectly related to immunity: a chitin-binding domain gene, and a putative apoptotic corpse-scavenger gene, homologous to Drosophila croquemort (14). Also included in the 1B cluster are eight miscellaneous genes, four of which are of unknown function in Drosophila (see Table 1).
Cluster 2.
This cluster, which is activated by septic but not sterile injury, represents the in vivo response to bacterial infection. Not surprisingly, it is dominated by immunity genes (10/25 HO; Table 1). They encode the homologue of the Drosophila pattern-recognition receptor PGRP-LB (15), the Gram−-binding protein, GNBP (4, 16), a member of the family of thioester-containing putative opsonins (α-TEPIV; ref. 17 and E. Levashina, personal communication), two serine proteases (Easter-like and 14D; ref. 18), and a homologue of the Caenorhabditis elegans adaptor protein, CED-6, which is implicated in phagocytosis of apoptotic corpses (19). Additional immunity genes encode two different fibrinogen domain (FBN) lectins reminiscent of the crustacean immunolectins that are involved in aggregation of bacteria (20) and of the mammalian ficolins that are capable of activating complement cascades and are implicated in phagocytosis (21). The two final members of this class correspond to well known immune related pathways: a putative receptor with extensive Leucine-rich repeats (LRR), that shows homology to the mammalian and Drosophila Toll-like receptors (22), and a putative inhibitor of the cactus/IκB family (23).
The septic cluster 2 also encodes four members of the protein metabolism class, three cytoskeletal and adhesion components, two members of the redox/mitochondrial class, and two putative transporters including a putative Na/Cl-dependent dopamine transporter (24). The latter may provide a link between the immune response and stimulation of the nervous system. Dopamine is the product of dopa decarboxylase (a member of cluster 1B) and is both a neurotransmitter and an intermediate in the oxidative pathway leading to the formation of melanin, a central component of insect defense.
Most of the cluster 2 genes are also activated in vitro by bacteria or PGN; an overlapping but not identical subset is activated by LPS. In contrast, most of these genes (53 to 64) are down-regulated by H2O2. An intriguing group of genes, 64–67, are strongly up-regulated by septic injury and in vitro challenge by bacteria and microbial elicitors but antithetically down-regulated by sterile injury. One of these, gene 65, is one of the most highly and consistently induced sequences in challenged cell cultures, showing up to 16-fold up-regulation, especially by PGN or Zymosan. The encoded protein has extended His stretches and resembles the product of the cats-up gene of Drosophila, encoding a negative regulator of the production of dopa and other catecholamines (25). This interesting group also encodes a putative cell-surface receptor with significant homology to EGF-domain proteins, 66, an FBN lectin, 64, and the LRR putative receptor, 67.
Cluster 3.
This cluster is defined by its down-regulation by injury, both sterile and septic. It is dominated by the redox/mitochondrial class (16/39 HO genes), including numerous components of the oxidative phosphorylation pathway (several subunits of the NADH dehydrogenase and cytochrome c oxidase complexes). The carbohydrate metabolism genes are also over-represented (5/39 HO genes), and here (unlike in cluster 1B) they encode components of the citric acid cycle. Additional notable components are ceramidase (down-regulated by PGN and slightly up-regulated by H2O2) and a Ser protease inhibitor (serpin) that is down-regulated by both injury and H2O2.
Cluster 3 is largely unresponsive in cultured cells challenged by bacteria. Intriguingly, however, partially overlapping gene groups are induced by PGN (genes 71–78) and H2O2 (genes 73–84). Two prominent groups are strongly up-regulated (genes 99–106) or down-regulated (genes 82–86) by LPS. A subcluster of genes, 82–86, is strongly down-regulated by LPS. Although it is premature to infer their functions, coregulated groups clearly suggest the existence of differentially regulated pathways, which were not obvious from the in vitro data alone, but were highlighted when filtered through the injury-induced down-regulation responses.
Malaria-Induced Genes.
For a preliminary comparison of the responses to infection by bacteria and malaria, matched sets of mosquitoes were fed normal mouse blood or blood infected with Plasmodium berghei, and 24 h later, when the parasite is known to cross the midgut epithelium and elicit an immune reaction (3, 4, 8), differential expression profiles were determined. Comparison of the septic response profiles clearly demonstrated that the malaria response of adult mosquito of this strain extensively overlaps with but is not identical to the response to bacteria (Fig. 3). Malaria up-regulates 13 HO genes and down-regulates 5 HO genes (24 and 10 TO genes, respectively; cluster M in Fig. 3, Table 4). The up-regulated genes are mostly also induced by bacteria, PGN and LPS in vitro. However, some are down-regulated by bacteria, PGN and H2O2 in vitro, and by septic and sterile injury (genes 69 and 70); by septic and sterile injury (gene 74); by PGN, sterile injury, and H2O2 (gene 64); or by sterile injury (gene 67). A prominent coregulated group, including genes 32, 83, and 84, is down-regulated by both LPS and malaria.
The M cluster includes a variety of functional classes but is especially enriched in immunity-class genes. Genes of this class that are activated by both bacterial and malaria challenge encode the PGRP-LB receptor, the GNBP opsonin, an FBN lectin, a thioester-containing putative opsonin, the serine protease 14D, the CED-6-like phagocytic adaptor, and the LRR putative receptor. Other HO genes that are specifically activated by malaria but not bacteria encode an isocitrate dehydrogenase, a dsRNA binding RNase 3, and a mitochondrial phosphate carrier.
Conclusion.
Unlike previous gene-by-gene studies of mosquito innate immunity (reviewed in ref. 26) this study is a genome-scale analysis made possible by a pilot EST discovery project. Cross-correlation of in vivo and in vitro experiments proved informative, identifying numerous previously uncharacterized immunity-related genes and the following broad expression features:
A distinctive immune response to bacteria and microbial elicitors can be separated readily from the responses to injury in the mosquito and H2O2-induced stress in the cell culture.
Gene expression and putative function are well correlated: the innate immunity class (I) is the most prominent component of the microbially activated response. Only a few atypical members of this class are induced by H2O2 or injury.
However, the immune response engages additional physiological systems. Taken together, five distinct classes of genes (R, TP, TR, PS, and S) are as well represented as the I class in the microbially activated gene clusters.
Within the expression clusters, smaller groups of tightly coregulated genes are evident, especially in response to PGN, LPS, and H2O2. They may reflect specific regulatory pathways.
The responses to injury and malaria are also complex, involving characteristic and diverse sets of genes. The malaria response overlaps extensively with the reaction to bacteria. In the injury response, the down-regulated cluster is enriched in genes that function in the mitochondria and are mainly related to the oxidative phosphorylation and citric acid cycle, whereas the activated cluster includes genes related to glycolysis.
Evidently, the full mosquito genome sequence will help extend and refine our view of the responses to bacteria, malaria, and injury. An early example is a gene up-regulated more than 75-fold upon challenge with Zymosan and PGN in cultured cells. The EST sequence alone was inadequate to detect homology, but when extended by using the genome sequence traces (http://trace.ensembl.org/perl/ssahaview), it was found to include leucine-rich repeats and to be homologous to Drosophila Toll-like genes. Beyond gene identification, experimental manipulation such as RNA interference will be required to detect the functions of specific genes and to define the regulatory pathways. Preliminary experiments have shown that RNAi-mediated silencing of gene 67 (Table 1) prevents the induction of certain immunity genes upon challenge with specific microbial elicitors (G.K.C., unpublished results).
Supplementary Material
Acknowledgments
We thank the entire Anopheles Genome Consortium, and especially Celera Genomics, Genoscope, and F. H. Collins for A. gambiae genome information, and W. Ansorge, P. Bork, D. Göttel, P. Lichter, A. Richter, E. Wabeck, and J. Zimmermann for advice. G.K.C. was supported by a Marie-Curie Fellowship. This research was supported by European Union Network Grants EU-TMR (ERBFMRXCT960017) and EU-RTN (HPRN-CT-2000-00080) and by National Institutes of Health Grant U01 AI48846.
Abbreviations
- EST
expressed sequence tag
- PGN
peptidoglycan
- LPS
lipopolysaccharide
- SOM
self-organizing maps
- TO
total of sequenced cluster and nonsequenced clones
- HO
gene clusters showing homology to genes of other organisms
References
- 1.Hoffmann J A, Reichhart J M. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 2.Dimopoulos G, Casavant T L, Chang S, Scheetz T, Roberts C, Donohue M, Schultz J, Benes V, Bork P, Ansorge W, et al. Proc Natl Acad Sci USA. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E S, Golub T R. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann J A, Reichhard J-M, Hetru C. Curr Opin Immunol. 1996;8:8–13. doi: 10.1016/s0952-7915(96)80098-7. [DOI] [PubMed] [Google Scholar]
- 6.Dimopoulos G, Richman A, Mueller H-M, Kafatos F C. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller H-M, Dimopoulos G, Blass C, Kafatos F C. J Biol Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 8.Samakovlis C, Asling B, Boman H G, Gateff E, Hultmark D. Biochem Biophys Res Commun. 1992;188:1169–1175. doi: 10.1016/0006-291x(92)91354-s. [DOI] [PubMed] [Google Scholar]
- 9.Kanzok S M, Fechner A, Bauer H, Ulschmid J K, Mueller H-M, Botella-Munoz J, Schneuwly S, Schirmer R, Becker K. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 11.Nappi A J, Ottavani E. BioEssays. 2000;22:469–480. doi: 10.1002/(SICI)1521-1878(200005)22:5<469::AID-BIES9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Richman A M, Dimopoulos G, Seeley D, Kafatos F C. EMBO J. 1997;16:6114–6119. doi: 10.1093/emboj/16.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimopoulos G, Seeley D, Wolf A, Kafatos F C. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franc N C, Heitzler P, Ezekowitz R A, White K. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- 15.Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. Proc Natl Acad Sci USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S Y, Wang R, Söderhäll K. J Biol Chem. 2000;275:1337–1343. doi: 10.1074/jbc.275.2.1337. [DOI] [PubMed] [Google Scholar]
- 17.Oduol F, Xu J, Niare O, Natarajan R, Vernick K D. Proc Natl Acad Sci USA. 2000;97:11397–11402. doi: 10.1073/pnas.180060997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman M J, Andreeva O V, Paskewitz S M. Insect Biochem Mol Biol. 2000;30:35–46. doi: 10.1016/s0965-1748(99)00095-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Z, Hartwieg E, Horvitz H R. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 20.Gokudan S, Muta T, Tsuda R, Koori K, Kawahara T, Seki N, Mizunoe Y, Wai S N, Iwanaga S, Kawabata S. Proc Natl Acad Sci USA. 1999;96:10086–10091. doi: 10.1073/pnas.96.18.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsushita M, Endo Y, Fujita T. J Immunol. 2000;164:2281. doi: 10.4049/jimmunol.164.5.2281. [DOI] [PubMed] [Google Scholar]
- 22.Imler J L, Hoffmann J A. Trends Cell. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- 23.Geisler R, Bergmann A, Hiromi Y, Nusslein-Volhard C. Cell. 1992;71:613–621. doi: 10.1016/0092-8674(92)90595-4. [DOI] [PubMed] [Google Scholar]
- 24.Porzgen P, Park S K, Hirsh J, Sonders M S, Amara S G. Mol Pharmacol. 2001;59:83–95. doi: 10.1124/mol.59.1.83. [DOI] [PubMed] [Google Scholar]
- 25.Stathakis D G, Burton D Y, McIvor W E, Krishnakumar S, Wright T R, O'Donnell J M. Genetics. 1999;153:361–382. doi: 10.1093/genetics/153.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimopoulos G, Mueller H-M, Levashina E A, Kafatos F C. Curr Opin Immunol. 2001;13:79–88. doi: 10.1016/s0952-7915(00)00186-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.