Abstract
The presence of homologues of Salmonella enterica sv. Typhimurium LT2 genes was assessed in 22 other Salmonella including members of all seven subspecies and Salmonella bongori. Genomes were hybridized to a microarray of over 97% of the 4,596 annotated ORFs in the LT2 genome. A phylogenetic tree based on homologue content, relative to LT2, was largely concordant with previous studies using sequence information from several loci. Based on the topology of this tree, homologues of genes in LT2 acquired by various clades were predicted including 513 homologues acquired by the ancestor of all Salmonella, 111 acquired by S. enterica, 105 by diphasic Salmonella, and 216 by subspecies 1, most of which are of unknown function. Because this subspecies is responsible for almost all Salmonella infections of mammals and birds, these genes will be of particular interest for further mechanistic studies. Overall, a high level of gene gain, loss, or rapid divergence was predicted along all lineages. For example, at least 425 close homologues of LT2 genes may have been laterally transferred into Salmonella and then between Salmonella lineages.
Salmonellae are divided taxonomically into two species, Salmonella enterica, and Salmonella bongori. S. enterica is comprised of more than 2,000 serovars assigned to seven subspecies (ssp): I, II, IIIa, IIIb, IV, VI, and VII. S. bongori, formerly referred to as ssp. V, was assigned a separate species based initially on its different biotype and phylogenetic data acquired by multilocus enzyme electrophoresis (MLEE) (1, 2).
The Salmonella Reference Collection C (SARC) contains 16 strains used in DNA sequence studies encompassing two strains each from all seven subgroups of S. enterica and S. bongori (3). Data from MLEE analysis of 24 loci and from the combined sequences of five housekeeping genes both are consistent in their prediction of S. bongori as an outgroup. The common ancestor of ssp. I, II, IIIb, and VI acquired the mechanism of flagellar antigen shifting, which is thought to play an important role in adaptation of the salmonellae to warm-blooded hosts (4). Subsequently, ssp. I became highly specialized for mammals and birds, with some serovars adapting to a single host species (e.g., S. enterica sv. Typhi in humans). Overall, ssp. I accounts for more than 99% of enteric and systemic infections in humans (5).
DNA microarray technology is an emerging technique to investigate genetic relationships between closely related bacterial strains. Species analyzed with this method include Helicobacter pylori (6–8), Campylobacter (9), mycobacteria (10), Staphylococcus aureus (11), obligate endosymbionts of the tsetse fly (12, 13), Shewanella oneidensis (14), Pseudomonas species (15), and Vibrio cholerae (16).
The completed genome sequence of S. enterica sv. Typhimurium (STM) LT2 (17) allowed us to construct an LT2 microarray of PCR-amplified whole ORFs representing over 97% of the 4,596 coding sequences assigned to the bacterium. With this LT2 chip we compared the genetic content of the Salmonella Ames strains (18). In addition, this chip allows the genetic content of the entire Salmonella clade to be surveyed with respect to the Typhimurium LT2 genome at single-gene resolution. The results of our analysis are in agreement with the phylogenetic relationships predicted for the salmonellae from sequence data of housekeeping or invasion genes. But we now are able to predict which genes, or close homologues, were acquired at various stages in Salmonella evolution and therefore may be associated with the acquisition of phenotypes shared with LT2. Such genes of known taxonomic distribution also may prove useful markers for classification of Salmonella. The comparisons presented here illustrate a high degree of genetic exchange between salmonellae.
Materials and Methods
Strains, Culture Conditions, and DNA Extraction.
All Salmonella strains used in this study are described in Table 1. Strains were maintained by standard methods (19) and grown to stationary phase at 37°C in Luria broth. Genomic DNA was prepared from 4 ml of overnight culture by using the DNEasy kit (Qiagen, Chatsworth, CA) according to manufacturer instructions.
Table 1.
Bacterial strains used in this study and number of LT2 genes absent
| Strain | Abbreviation | Salmonellassp. | Source/ref. | No. of CDS missing* | No. in Fig. 1 |
|---|---|---|---|---|---|
| Y. pestis CO92 | YPE | — | (23) | 2326† | 1 |
| K. pneumoniae MGH 78578 | KPN | — | (17) | 1642† | 2 |
| E. coli K12 | ECH | — | (21) | 1491† | 3 |
| E. coli O157:H7 | ECO | — | (22) | 1425† | 4 |
| S. bongori S-1399 | SBO S-1399 | (V) | SGSC‡ | 776 | 5 |
| S. bongori SA4410 | SBO SA4410 | (V) | SGSC | 773 | 6 |
| S. bongori SARC11 | SBO SARC11 | (V) | SGSC | 710 | 7 |
| S. enterica SARC16 | — | VII | SGSC | 734 | 8 |
| S. enterica SARC15 | — | VII | SGSC | 740 | 9 |
| S. enterica SARC14 | — | VI | SGSC | 677 | 10 |
| S. enterica SARC13 | — | VI | SGSC | 599 | 11 |
| S. enterica SARC10 | — | IV | SGSC | 766 | 12 |
| S. enterica SARC9 | — | IV | SGSC | 742 | 13 |
| S. enterica SARC8 | — | IIIb | SGSC | 657 | 14 |
| S. enterica SARC7 | — | IIIb | SGSC | 684 | 15 |
| S. enterica sv Arizonae | — | IIIa | SGSC | 706 | 16 |
| S. enterica SARC6 | — | IIIa | SGSC | 787 | 17 |
| S. enterica SARC5 | — | IIIa | SGSC | 754 | 18 |
| S. enterica SARC4 | — | II | SGSC | 564 | 19 |
| S. enterica SARC3 | — | II | SGSC | 619 | 20 |
| S. enterica sv. Paratyphi A SARB42 | SPA SARB44 | I | SGSC | 480 | 21 |
| S. enterica sv. Typhi IN15 | STY IN15 | I | SGSC | 430 | 22 |
| S. enterica sv. Typhi CT18 | STY CT18 | I | (20) | 426 | 23 |
| S. enterica sv. Paratyphi B SARB44 | SPB SARB44 | I | SGSC | 307 | 24 |
| STM SL1344 | STM SL1344 | I | D. G. Guiney, San Diego | 171 | 25 |
| STM TA97 | STM TA97 | I | D. M. DeMarini, Research Triangle Park, NC | 15 | 26 |
| STM LT2 | STM LT2 | I | (17) | 0 | — |
Determined by microarray (for cutoffs see Materials and Methods) unless stated otherwise.
Determined by sequence comparison (for cutoffs see Materials and Methods).
Salmonella Genetic Stock Center, University of Calgary, Calgary, AB, Canada.
Salmonella typhimurium ORF Microarray Construction.
The present annotation of the STM LT2 genome contains 4,596 annotated S. enterica sv. Typhimurium LT2 coding sequences, (http://genome.wustl.edu/gsc/Projects/S.typhimurium/), 4,483 (97.5%) of which were PCR-amplified successfully without any byproduct. The details of primer and PCR amplification conditions and microarray design are described elsewhere, as are labeling and hybridization conditions, data acquisition, and normalization (18).
Thresholds for Determining Presence and Absence of Genes.
The available genome sequence of Escherichia coli K12, E. coli O157:H7, Klebsiella pneumoniae MGH 78578, and Yersinia pestis CO92 were obtained (17, 21–23), and gene presence was evaluated with the following parameters, which were determined to maximize the accuracy with which reciprocal orthologs were predicted as present (17): E. coli K12, >70% sequence identity on the DNA level; E. coli O157:H7, >70%; K. pneumoniae MGH 78578, >65%; and Y. pestis CO92, >60%. Values lower by less than 5% from these cutoffs were scored as uncertain. Genes with less homology than this were scored as absent.
For unsequenced genomes, a predictor for presence and absence of genes based on median of microarray hybridization ratios and standard deviation of data points for each genome was created to determine each gene status in each genome (for details, see Predictor for Presence and Absence of Genes, which is published as supporting information on the PNAS web site, http://www.pnas.org). This predictor was superior to results obtained by choosing any fixed arbitrary ratio cutoff for all data sets (data not shown).
By using sequence information for LT2, S. enterica sv. Typhi CT18 and Paratyphi A (17, 20), this method correctly classified almost all genes that had over 90% identity with a gene in LT2 as present and genes with under 80% identity as absent. The few seemingly incorrect calls, 0.9% of genes in S. enterica sv. Typhi and 1.8% in S. enterica sv. Paratyphi A, were primarily because of the microarray data correctly reporting the presence of genes that were highly homologous to only part of an LT2 gene. Nevertheless, readers are cautioned that the data generated from sequence comparisons (i.e., for E. coli K12, E. coli O157:H7, K. pneumoniae MGH 78578, and Y. pestis CO92) are not necessarily congruent with the microarray data.
Phylogenetic Tree.
The final data set consisting of four different values (0 = absent, 1 = uncertain, “?” = missing data, and 2 = present) was incorporated into the PAUP* software program (http://paup.csit.fsu.edu). Phylogenetic trees of the Salmonella clade were generated under a variety of different assumptions including genetic distance using neighbor joining and parsimony analysis using equal weighting or 2:1 weighting against acquisition of the gene to minimize the number of times a gene appears to be recruited on different parts of the tree. Uncertain assignments were considered missing data when making such predictions.
Results
Microarray Analysis.
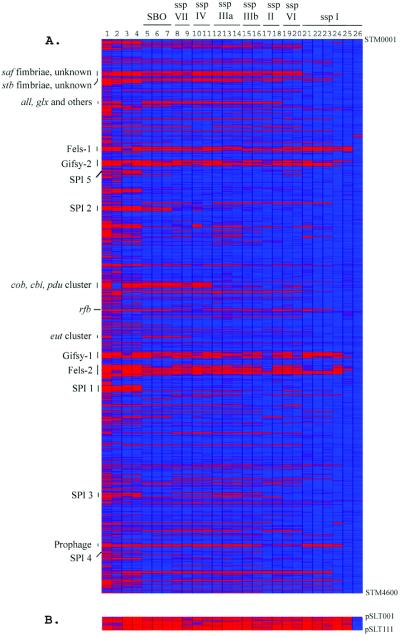
We compared the genomic ORF content of representative strains of S. enterica and S. bongori with STM LT2. The complete data set on presence and absence of each individual LT2 gene can be viewed in Tables 4 and 5, which are published as supporting information on the PNAS web site. The absence/presence calls for each investigated strain are visualized in Fig. 1.
Figure 1.
Presence and absence of STM LT2 protein coding sequence homologues in 26 other enterobacterial strains including representatives of all Salmonella ssp and S. bongori (SBO). The numbers on top correspond to strain numbers in Table 1, column 6. Strains are sorted from left to right with ascending relatedness to LT2. The gene status is color-coded: blue, present; purple, uncertain; red, absent. For cutoffs of absence and presence predictions, refer to Materials and Methods. Some prominent regions are indicated. (A) The genes on the chromosome are represented in order of position in LT2 from STM 0001 to 4600. (B) The genes of the LT2 virulence plasmid pSLT.
There were 1,424 LT2 genes (32% of those examined) called absent or diverged too extensively to be detected in at least one of the other Salmonella genomes. As expected, all the pSLT plasmid genes represented on the chip fell into this category, as did all of the genes from the five prophages identified in the LT2 genome. Table 1 illustrates the number of LT2 genes found to be absent or diverged in each of the Salmonellae used in this assay. One must presume that a similar number of genes are present in these strains and absent in LT2, but the current microarray cannot monitor such genes.
Salmonella-Specific Genes.
By using genomic data from two E. coli strains (K12 and O157:H7) (21, 22), a sample sequence from K. pneumoniae MGH 78578 (17), and the Y. pestis CO92 sequence (23) we identified 935 genes present in LT2 and consistently absent in the four genomes of the other enterobacterial genera. These are possibly genes recruited to the Salmonella clade from different organisms or genes that have been under selective pressure to diverge more quickly than average.
In some cases, homologues of the 935 LT2 genes may be present in other strains of Escherichia, Klebsiella, or Yersinia but not the strains that were sequenced. Of 935 “Salmonella” genes, 224 were called “present” or “uncertain” but never “absent” in any of the 22 Salmonella strains investigated. The most consistent were 56 genes always detected as present in all 22 Salmonellae investigated (Table 6, which is published as supporting information on the PNAS web site). This short list of Salmonella “signature” genes contains seven genes from Salmonella pathogenicity island (SPI)1, a well studied pathogenicity island of Salmonella, and the DNA helicase gene res. In addition, the tetrathionate reductase complex ttr, the asr operon encoding an anaerobic sulfide reductase complex, and phsB and phsC, involved in hydrogen sulfide production, belong to this group. With this subset of genes, Salmonella can use tetrathionate as an electron acceptor during anaerobic respiration, which is reduced to H2S (24, 25). Furthermore, three genes possibly involved in tricarboxylic transport are part of the Salmonella signature genes. There is no assigned function to 34 of the 56 genes with homologues in all Salmonellae investigated, indicating that this group of genes remained largely unstudied.
Metabolic Gene Clusters.
Whereas the majority of the missing genes in the different salmonellae are of plasmid or phage origin in LT2, some metabolic operons are absent also. One of the genetic elements not detected in S. bongori and ssp. VII, the most distant of the S. enterica ssp. from LT2, is the eut cluster, encoding proteins involved in utilization of ethanolamine as a carbon, energy, and nitrogen source. The cob, cbi, and pdu clusters are missing in S. bongori and S. enterica ssp. VII and IV. The pdu operon is responsible for propanediol utilization as a carbon and energy source under aerobic conditions. Proteins encoded by the immediately adjacent cob/cbi cluster are necessary for de novo biosynthesis of adenosyl-cobalamin (Ado-CBL) under anaerobic conditions, required for enzymatic activities of ethanolamine ammonia-lyase EutBC and propanediol dehydratase Pdu (26, 27).
Other metabolic gene clusters are not universally present in all salmonellae, exemplified by the fuc, dgo, and cai/fix operons. Overall, a considerable number of sugar transport and/or metabolism genes are distributed unevenly throughout the salmonellae, suggesting redundancy of several sugar compounds for survival of the bacterium in its hostile environment.
Fimbrial and Flagellar Genes.
Fimbriae are a heterogeneous group of surface hair-like structures intimately involved in binding to external features, particularly on host cells. The 12 fimbrial operons of the chaperone-usher class detected in LT2 are not distributed uniformly throughout the salmonellae but display very specific patterns of presence and absence, which are summarized in Table 2.
Table 2.
Homologues of LT2 chaperone-usher fimbrial clusters
| Fimbrial clusters | STM no. | YPE | KPN | E. coli | SBO | ssp. VII | ssp. IV | ssp. IIIa | ssp. IIIb | ssp. VI | ssp. II | SPA | STY | SPB | STM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fim | 0543–0552 | − | − | − | − | + | + | + | + | + | + | + | + | + | + |
| lpf | 3636–3640 | − | − | − | + | − | − | − | − | − | − | − | − | + | + |
| bcf | 0021–0028 | − | − | − | + | − | − | + | + | + | + | + | + | + | + |
| stb | 0336–0340 | − | ? | − | − | − | − | − | + | − | + | + | + | + | + |
| std | 3027–3029 | − | − | − | − | − | − | − | + | + | − | + | + | + | + |
| saf | 0299–0302 | − | − | − | − | − | − | − | − | − | − | + | + | + | + |
| stc | 2149–2152 | − | − | − | − | − | − | − | − | − | − | − | + | + | + |
| stf | 0195–0201 | − | − | − | − | − | − | − | − | − | − | + | − | + | + |
| sti | 0174–0177 | − | − | − | − | − | − | − | − | − | − | − | − | + | + |
| sth | 4591–4597 | − | − | − | + | + | + | − | − | + | + | + | + | + | + |
| stj | 4571–4575 | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| pef | PSLT013–019 | − | − | − | − | − | − | − | − | − | − | − | − | − | +/− |
+, Most of the genes of the cluster are present in all investigated strains. See Table 1 for number of strains considered in each column and whether data were based on sequence comparison or microarray analysis. −, Most of the genes of the cluster are absent; +/−, most genes of the cluster are present in one but not all investigated strains; ?, cluster is partly present and partly absent; YPE, Y. pestis; KPN, K. pneumoniae; SBO, S. bongori; SPA, S. enterica sv. Paratyphi A; STY, S. enterica sv. Typhi; SPB, S. enterica sv. Paratyphi B.
Most of the genes of the flagellar apparatus were found to have homologues in all salmonellae. An exception is the gene coding for the filament cap protein FliD, which was not found in strains SARC04 and SARC06, and doubtful in several others, including the ssp. VII, IV, and IIIa strains. The fljA and hin genes encode a transcriptional repressor of the phase 1 flagellin, FliC, and an invertase necessary for site-specific inversion of the fljAB operon, enabling expression of the phase 2 flagellin FljB. These genes required for the diphasic switch are present in most diphasic strains (ssp. I, II, IIIb, and VI) but not in monophasic Salmonella ssp. or S. bongori. A few strains in the diphasic lineage have reverted also to monophasic, for example S. enterica sv. Typhi and the ssp. II strain SARC04.
The SPIs.
The status of homologues of the five SPIs and the Salmonella outer membrane proteins (Sops) is variable. Homologues of most of the 50 genes of SPI1 are universally present in almost all Salmonellae investigated. Most of the 44 ORFs in SPI2 are absent in S. bongori, confirming previous observations (28). Homologues are generally present in S. enterica including the ssa, ssc, and sse gene clusters. However, STM 1379–1382, four ORFs encoding genes of uncertain functions, have close homologues in most S. bongori, but are primarily absent or too divergent to be detected in the S. enterica ssp. VII, IIIa, and IIIb. Of the genes that comprise SPI3, a phosphotransferase system seems to be absent or divergent in almost all salmonellae except ssp. I and II. A small cluster of five genes including the magnesium transporter genes mgtBC, are present in all salmonellae, whereas others have close homologues only in ssp. I and II and S. bongori. Finally, the “middle” part of SPI3 with 10 genes is partly absent or divergent only from ssp. IIIa and partly missing in both IIIa and IIIb. The six genes of SPI4, STM 4257–4262, are absent or uncertain in ssp. IIIa and IIIb. Among the eight SPI5 representatives, two (pipA and pipB) are absent or divergent in ssp. VI and II.
Overall, only ssp. I has a full complement of the genes of all five pathogenicity islands excluding three genes of SPI2 (STM 2901–2903) absent in S. enterica sv. Typhi and S. enterica sv. Paratyphi A, avrA STM absent in S. enterica sv. Paratyphi A, S. enterica sv. Paratyphi B, and possibly in S. enterica sv. Typhi, and sugR, absent in S. enterica sv. Typhi.
Prophages.
There are at least five prophage genomes present on the LT2 chromosome: Gifsy-1, Gifsy-2, Fels-1, Fels-2, and the region from STM 4196–4219 (17). Most of these genes are absent or highly diverged in the bacterial strains of our study (Fig. 2 and Table 7, which is published as supporting information on the PNAS web site). Surprisingly, despite being quite distant from LT2, S. enterica ssp. VI SARC13 contained homologues to a considerable portion (over 33%) of genes from all five of these phage regions. A Gifsy-1 region present in SARC04 overlaps almost completely the region present in SARC13, suggesting a common phage related to Gifsy in those strains.
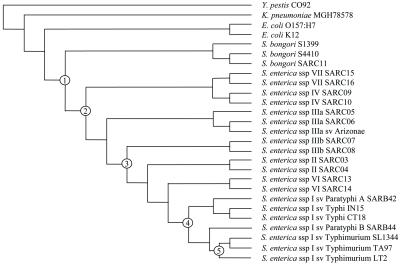
Figure 2.
Phylogenetic tree of the Salmonella clade. The cladogram was constructed with PAUP* software (Sinauer) by using maximum parsimony, equal weight, and 1,000 bootstraps with Y. pestis as the outgroup. Five crucial stages in Salmonella evolution are indicated: 1, divergence of Salmonella from E. coli; 2, separation of S. enterica from S. bongori; 3, evolution of the diphasic S. enterica strains; 4, partition of S. enterica ssp. I; and 5, development of STM. The number of genes predicted to be recruited at each node is shown in Table 3.
As more phage genomes are sequenced it will be possible to add genes from these phages to the array and use microarrays to “phage-type” an increasing diversity of phage families in Salmonella genomes.
Other Features.
IS200, present in six copies on the LT2 genome, displayed notably lower hybridization signals in S. enterica ssp. II–VII. A region of 41 genes from STM 0958 to 0999 was ≈2-fold overrepresented in strain SARC13. Apparent duplications of large parts of a genome have been observed previously in a number of Typhimurium strains (ref. 18; and unpublished data)
Phylogenetic Tree.
Based on the microarray data predicting the presence and absence of LT2 genes, we built bifurcating trees illustrating possible phylogenetic relationships between the different salmonellae (Fig. 2). Trees of very similar topology were found by using various other methods including genetic distance with neighbor joining and parsimony with 2:1 weighting against gain of genes. We confirmed clustering of S. enterica from S. bongori, monophasic S. enterica's from diphasic ones, and ssp. I from all the other ssp. We analyzed data of genome sequencing of two E. coli strains (K12 and O157:H7), Y. pestis CO92, and the partially sequenced K. pneumoniae MGH 78578 by using the tree-building program with cutoffs for presence and absence of genes as indicated in Materials and Methods.
Genes and operons predicted to be gained from the ancestor at certain stages in Salmonella evolution were determined. We predicted which genes were first to appear (i) when the salmonellae were formed, (ii) when S. enterica split from S. bongori, (iii) when the diphasic diverged from the monophasic salmonellae, (iv) when ssp. I separated from the other ssp, and (v) when STM evolved. However, we observed a mosaic distribution of presence/absence patterns of many genes in the different ssp, species, and the other enterobacteria. Thus, the phylogenetic data are consistent with previous pairwise comparisons among enterobacterial genomes (22, 29–32). This mosaic appearance indicates that although the phylogenetic tree may be indicative of the history of most of the genes, during the divergence of these taxa, the most parsimonious solution implies that many genes have been acquired multiple times on multiple lineages. However, gene conversion of different rates of divergence may be the explanation in some cases. The most parsimonious prediction of the number of genes that may have been acquired at each stage in Salmonella evolution is illustrated in Table 3 together with prominent examples.
Table 3.
Acquisition of genes during the evolution of LT2 as predicted by parsimony
| No. | Evolution stage | No. of genes gained
|
Examples | |
|---|---|---|---|---|
| Total | Unnamed | |||
| 1 | → Salmonella | 513 | 313 (61%) | mod/res, SPI1, bcf, sth, hof, rfa, mgt, ttr, scr, cit, phs, asr |
| 2 | → S. enterica | 111 | 65 (59%) | SPI2, fim, iro |
| 3 | → Diphasic Salmonella | 105 | 53 (50%) | fljAB, hin, stb, std, pgt |
| 4 | → S. entericassp. I | 216 | 128 (59%) | hsdMR, mrr, rfb, stc, saf, stf, sinR, phn |
| 5 | → STM | 144 | 128 (89%) | stj, hsdS, rtc |
The first column refers to the nodes in the cladogram in Fig. 2.
Gene Acquisitions at Different Evolutionary Time Points.
We identified 513 genes possibly gained at the formation point of Salmonella. This list includes most of the hil, spa, and inv genes of SPI1 and the bcf and sth fimbrial genes. Furthermore, some Salmonella-specific lipopolysaccharide core biosynthesis genes rfa were modified profoundly from E. coli rfa genes.
When S. enterica diverged from S. bongori, it recruited the second type III secretion mechanism: the SPI2 ssa, sse, and ssr genes. Moreover, it also gained the fim genes encoding another subset of fimbriae.
As diphasic Salmonella evolved they added, together with the fljAB/hin system, the stb and std fimbrial genes and a phosphoglycerate transport system pgt. The latter was found also in K. pneumoniae, suggesting lateral transfer from one bacterial species to another or loss in the E. coli and monophasic Salmonella lineages.
As ssp. I separated from the other ssp, it recruited the stf, saf, and stc fimbrial operons and a transcriptional regulator sinR. A specific form of the rfb cluster was recruited, which is important in the lipopolysaccharide side chain synthesis, and thus the envelope of the bacterium was changed, possibly to adapt to its new warm-blooded host environment. Some genes of this cluster were identified in Y. pestis, but none were within the presence similarity threshold in any of the other three enterobacteria included in this study. The ssp. 1 salmonellae also may have added the envF gene at this stage, encoding an envelope lipoprotein, and the shdA gene, a Peyer's patch colonization and shedding factor (33). Cluster STM 3251–3256 encodes components of sugar activation and transport (fructose-specific phosphotransferase system component IIA and putative sugar kinases), specific for ssp. I salmonellae. The phn operon encoding a 2-aminoethylphosphonate transporter, was recruited. This operon is shared with K. pneumoniae. Of the 216 genes probably gained during separation of S. enterica ssp. I, 16 ORFs including the three fimbrial operons are predicted by PSORT (34) to be located in the outer membrane and thus may be accessible for therapy. In addition to the 10 outer membrane proteins encoded in the fimbrial operons, this list includes STM 0280, 2816, 3026 (no gene names), 2423 (yfeN), ratB, and sinI. However, yfeN and stc are absent in the S. enterica sv. Paratyphi A strain investigated, stf and STM 0280 are absent from the S. enterica sv. Typhi strains, and STM 3026 was not found in the S. enterica sv. Paratyphi B strains, which excludes them from being potential universal ssp. I therapeutic targets. Of all 216 ssp. I signature genes, 74 (including some rfb genes, the saf fimbriae, sinI, and ratB) are present or possibly present in all the ssp. I strains investigated and absent or probably absent in all the other salmonellae or enterobacteria studied (Table 8, which is published as supporting information on the PNAS web site).
Lastly, as STM was formed, the stj fimbrial operon was gained together with a number of prophage and a very large number of unknown genes. Of 144 genes acquired at this step, 128 have no name, illustrating the gaps in our information as to which gene functions led to the evolution of Typhimurium.
Homoplasy in the Phylogenetic Tree.
Many LT2 genes or homologues are distributed among the taxa in a manner indicating they either were acquired once and later lost or diverged substantially in many subsequent lineages, or acquired by horizontal transfer on multiple independent occasions. According to the cladogram illustrated in Fig. 2, 1,976 genes were acquired at least once on the lineage to LT2. Most of the genes are in clusters of two or more adjacent genes. However, hundreds of genes apparently were recruited individually. Approximately 425 genes or close homologues were predicted to have been recruited by two or more Salmonella lineages or diverged at substantially different rates on multiple lineages, perhaps because of gene conversion events. Presumably multiple recruitment events often involved the transfer of the gene or gene cluster into one Salmonella and then the transfer of these genes to one or more different clades of Salmonella. The complete data set is presented in Table 5, which is published as supporting information on the PNAS website.
Discussion
The evolution of Salmonella has been studied extensively by using MLEE (3), sequence information from both housekeeping and invasion genes (4, 35), and rRNA sequences (36). Results from MLEE revealed somewhat different relationships from those identified by sequence analysis: whereas ssp. I, IIIa, IIIb, and VI clustered as a group separate from ssp. II, VII, and IV in the MLEE data, sequence information determined that relationships were following the order I–II/VI/IIIb–IV/VII–IIIa, with S. bongori being an outgroup (5).
Using a whole-genome microarray chip, we analyzed the genomic content of representatives of all species and ssp. within the Salmonella at single-gene resolution compared with S. enterica sv. Typhimurium LT2. Bifurcating trees were constructed to identify potential phylogenetic relationships between the different groups. The topologies were extremely similar whether generated by neighbor joining (a genetic-distance method) or parsimony, using either equal weighting of gene loss and gain or a 2:1 bias in favor of gene loss to simulate the fact that a gene is less likely to be gained twice. The topology of these trees supported the sequence-based phylogenetic trees, with only slight differences in clustering. ssp. IIIa was grouped closer to ssp. 1 than ssp. IV and VII, but the other relationships were as predicted from sequencing.
Approximately 10–15% of the genes in LT2 lack close homologues in other ssp. (Table 1), consistent with observations gained from recent comparisons of fully or partially sequenced S. enterica ssp. I genomes (17, 30). It is likely that a similar number of genes are found in other strains but not in LT2, although these genes are not on the microarray and thus were not monitored in the present study.
Most striking is the fact that many gene clusters are distributed in a manner indicating that they were either acquired and then lost in many subsequent lineages or acquired by lateral transfer on multiple independent occasions. This phenomenon results in a patchy distribution pattern and illustrates the significant degree of genetic fluidity between the different Salmonella strains. Most of these clusters are unnamed genes with mostly unknown functional properties. They demonstrate the ability of Salmonella to exchange genetic material within their genus or beyond.
In a previous study estimating the extent of the genetic flux between E. coli and LT2, codon usage was used as an indicator for recent additions to the E. coli genome (37). However, our ongoing analysis indicates that the genes identified by this method do not correlate well with genes predicted to be acquired recently as revealed by whole-genome sequencing, indicating that differences in codon usage among genes might often have a different cause (unpublished data).
Some LT2 genes that probably were acquired by lateral transfer at a certain stage in the evolution of the salmonellae have close homologues in some of the other related enterobacterial species of our study. Given that transfers, including gene conversion events, over shorter phylogenetic distances probably are more frequent than transfers over long distances, an enterobacterium is a likely source of many of these acquisitions in Salmonella. Such genes include those in the cluster STM 0514–0532, encoding proteins for transport, allantoin, and glyoxylate metabolism. Close homologues are present in E. coli and S. enterica ssp. II and I but not the other salmonellae, K. pneumoniae MGH 78578, or Y. pestis CO92. Although it is possible that the cluster was eliminated or diverged in all other Salmonella ssp. after acquisition, it seems more likely to have been acquired at a later stage in Salmonella evolution (immediately before splitting of ssp. II and I). A second example is region STM 0761–0765, with close homologues in K. pneumoniae MGH 78578, S. enterica sv. Paratyphi B, and LT2 but absent in all others. Other examples for possible lateral transfer events within the Salmonella include the ydi cluster from STM 1350–1362 and the cbi, pdu, pgt, eut, and dgo operons. In some of these cases, though, loss of the cluster from several Salmonella ssp. may be an explanation for the observed pattern. Taking all these possible routes into account, a large proportion of LT2 genes (935) still remains that are not a standard part of the Y. pestis, K. pneumoniae, or E. coli genomes or have diverged much more than typical genes.
Fifty-six genes with homologues in all Salmonella strains were not detected in the sequences of any of the four other enterobacteria in the study. It would be interesting to determine the effects of mutagenesis on any of these Salmonella signature genes. Their ubiquitous presence in Salmonella points toward necessity of their gene products in the natural Salmonella life cycle. Mutation analyses might yield insights into specific traits of this bacterium compared with its relatives.
It is intriguing to note that the majority of genes specific for S. enterica ssp. I have no known function. The subset of 74 uniquely and constantly present genes in all ssp. I strains contains only 20 genes with a name based on a putative function. Of this list, six candidates, safC, safD, stcA, sinI, ratB, and STM 2816 (a putative glycoporin), are predicted to reside in the outer membrane. Because these homologues may be important components of ssp. I and are predicted to be on the surface, they might be suitable targets for therapeutic treatments of all Salmonella strains capable of infecting warm-blooded animals including humans. The ubiquitous presence of homologues for some genes in the rfb cluster (and rfc) in ssp. I (also observed in ref. 38) might be indicative of the importance of genes in these clusters for the adaptation of the bacterium to its new hosts.
In summary, this work presents an examination of the changes in gene content associated with an evolutionary bacterial adaptation process to a new environment, the adaptation of Salmonella to warm-blooded hosts. The most striking observation is the scale of gene flux among and within the various ssp. Given this flux, it is likely that many of the genes responsible for the development of particular stages in the evolution of Salmonella are no longer present in all descendants and have been replaced by genes more adapted to their particular niches. Nevertheless, many genes found in all or most descendants of a particular stage in Salmonella evolution were determined in our analyses. These genes are likely to be fruitful targets for further experiments to determine their effect on the biology of Typhimurium and its relatives in ssp. I. In particular, this information will facilitate speculation on the importance of individual gene clusters in the process of adaptation of the bacterium to warm-blooded animals.
Supplementary Material
Acknowledgments
We thank Ken Sanderson for bacterial strains and helpful comments, J. Frye for constructive criticism and support during the microarray fabrication process, G. Gebauer and J. Welsh for critical reading of the manuscript, and E. Proctor for technical expertise. This work was supported by National Institutes of Health Grant AI34829 (to M.M.).
Abbreviations
- MLEE
multilocus enzyme electrophoresis
- ssp
subspecies
- SPI
Salmonella pathogenicity island
- STM
Salmonella enterica sv. Typhimurium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Beltran P, Musser J M, Helmuth R, Farmer J J, 3rd, Frerichs W M, Wachsmuth I K, Ferris K, McWhorter A C, Wells J G, Cravioto A, et al. Proc Natl Acad Sci USA. 1988;85:7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves M W, Evins G M, Heiba A A, Plikaytis B D, Farmer J J., 3rd J Clin Microbiol. 1989;27:313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd E F, Wang F S, Whittam T S, Selander R K. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selander R K, Li J, Nelson K. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2691–2707. [Google Scholar]
- 6.Israel D A, Salama N, Krishna U, Rieger U M, Atherton J C, Falkow S, Peek R M., Jr Proc Natl Acad Sci USA. 2001;98:14625–14630. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorkholm B, Lundin A, Sillen A, Guillemin K, Salama N, Rubio C, Gordon J I, Falk P, Engstrand L. Infect Immun. 2001;69:7832–7838. doi: 10.1128/IAI.69.12.7832-7838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell N, Mangan J A, Laing K G, Hinds J, Linton D, Al-Ghusein H, Barrell B G, Parkhill J, Stoker N G, Karlyshev A V, Butcher P D, Wren B W. Genome Res. 2001;11:1706–1715. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald J R, Sturdevant D E, Mackie S M, Gill S R, Musser J M. Proc Natl Acad Sci USA. 2001;98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akman L, Aksoy S. Proc Natl Acad Sci USA. 2001;98:7546–7551. doi: 10.1073/pnas.131057498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akman L, Rio R V, Beard C B, Aksoy S. J Bacteriol. 2001;183:4517–4525. doi: 10.1128/JB.183.15.4517-4525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray A E, Lies D, Li G, Nealson K, Zhou J, Tiedje J M. Proc Natl Acad Sci USA. 2001;98:9853–9858. doi: 10.1073/pnas.171178898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho J C, Tiedje J M. Appl Environ Microbiol. 2001;67:3677–3682. doi: 10.1128/AEM.67.8.3677-3682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dziejman M, Balon E, Boyd D, Fraser C M, Heidelberg J F, Mekalanos J J. Proc Natl Acad Sci USA. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClelland M, Sanderson K E, Spieth J, Clifton S W, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Nature (London) 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 18.Porwollik S, Wong R M, Sims S H, Schaaper R M, DeMarini D M, McClelland M. Mutat Res. 2001;483:1–11. doi: 10.1016/s0027-5107(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Parkhill J, Dougan G, James K D, Thomson N R, Pickard D, Wain J, Churcher C, Mungall K L, Bentley S D, Holden M T, et al. Nature (London) 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 21.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 22.Perna N T, Plunkett G, 3rd, Burland V, Mau B, Glasner J D, Rose D J, Mayhew G F, Evans P S, Gregor J, Kirkpatrick H A, et al. Nature (London) 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill J, Wren B W, Thomson N R, Titball R W, Holden M T, Prentice M B, Sebaihia M, James K D, Churcher C, Mungall K L, et al. Nature (London) 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 24.Hensel M, Hinsley A P, Nikolaus T, Sawers G, Berks B C. Mol Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 25.Price-Carter M, Tingey J, Bobik T A, Roth J R. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Toole G A, Rondon M R, Trzebiatowski J R, Suh S-J, Escalante-Semerena J C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 710–720. [Google Scholar]
- 27.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hensel M, Shea J E, Baumler A J, Gleeson C, Blattner F, Holden D W. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sebaihia M, Thomson N, Holden M, Parkhill J. Trends Microbiol. 2001;9:579. doi: 10.1016/s0966-842x(01)02275-2. [DOI] [PubMed] [Google Scholar]
- 30.Edwards R A, Olsen G J, Maloy S R. Trends Microbiol. 2001;10:94–99. doi: 10.1016/s0966-842x(01)02293-4. [DOI] [PubMed] [Google Scholar]
- 31.McClelland M, Florea L, Sanderson K, Clifton S W, Parkhill J, Churcher C, Dougan G, Wilson R K, Miller W. Nucleic Acids Res. 2000;28:4974–4986. doi: 10.1093/nar/28.24.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochman H, Jones I B. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingsley R A, van Amsterdam K, Kramer N, Baumler A J. Infect Immun. 2000;68:2720–2727. doi: 10.1128/iai.68.5.2720-2727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakai K, Horton P. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 35.Boyd E F, Li J, Ochman H, Selander R K. J Bacteriol. 1997;179:1985–1991. doi: 10.1128/jb.179.6.1985-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen H, Nordentoft S, Olsen J E. Int J Syst Bacteriol. 1998;48:605–610. doi: 10.1099/00207713-48-2-605. [DOI] [PubMed] [Google Scholar]
- 37.Lawrence H G, Ochman H. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S J, Romana L K, Reeves P R. J Gen Microbiol. 1992;138:1843–1855. doi: 10.1099/00221287-138-9-1843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.