Abstract
For the design of potent subunit vaccines, it is of paramount importance to identify all antigens immunologically recognized by a patient population infected with a pathogen. We have developed a rapid and efficient procedure to identify such commonly recognized antigens, and here we provide a comprehensive in vivo antigenic profile of Staphylococcus aureus, an important human pathogen. S. aureus peptides were displayed on the surface of Escherichia coli via fusion to one of two outer membrane proteins (LamB and FhuA) and probed with sera selected for high Ab titer and opsonic activity. A total of 60 antigenic proteins were identified, most of which are located or predicted to be located on the surface of the bacterium or secreted. The identification of these antigens and their reactivity with individual sera from patients and healthy individuals greatly facilitate the selection of promising vaccine candidates for further evaluation. This approach, which makes use of whole genome sequence information, has the potential to greatly accelerate and facilitate the formulation of novel vaccines and is applicable to any pathogen that induces Abs in humans and/or experimental animals.
Infectious diseases are the second leading cause of death worldwide, and the acquisition of antibiotic resistance by many pathogenic bacteria has recently spurred interest in generating vaccines to cure or prevent disease. Although vaccines composed of polysaccharides have proven valuable for the prevention of bacterial infections, in general they induce serotype-specific immune responses and contribute to serotype redistribution (1). Therefore, antigenic proteins that show little sequence variation in diverse clinical isolates will likely be superior antigens for the development of a broadly effective vaccine against a particular pathogen. To design potent and universally applicable subunit vaccines, it is necessary to identify those antigens that are recognized as nonself by the immune systems of many individuals of a wide patient population during infection. The availability of complete genomic sequences of bacterial pathogens has greatly facilitated the search for these antigens among all genome-encoded proteins. By applying bioinformatics to predict surface-exposed or exported proteins from Neisseria meningitides and Streptococcus pneumoniae, it was possible to express and purify a limited number of antigens to identify potential vaccine candidates (2, 3). However, the identification of relevant proteins by predictive algorithms and the subsequent need for expression and purification of full-length proteins in a heterologous host restrict the final selection of candidates for vaccine development. A number of novel vaccine candidates were also identified from Helicobacter pylori and Mycobacterium tuberculosis by combining genomics and proteomics (4, 5). Yet the proteomic approach is severely limited by the number of proteins expressed by the pathogen under in vitro growth conditions. The described approaches determined the immunogenicity of recombinant proteins in animal models (2, 3) or have used animal models to produce mAbs for selection (4).
To overcome these constraints and to identify vaccine candidates that are truly recognized by the human immune system, we developed an approach based on genomic peptide libraries in combination with well characterized human sera. Expression libraries for large polypeptides (6, 7) or small peptides encoded by random synthetic oligonucleotides of identical length (8) have frequently been used before. We have generated small diversely sized peptide libraries, encoded by randomly fragmented genomic DNA, to ensure that all potential antigens encoded by the genome of the pathogen can be identified. In addition, a preselection procedure was established to eliminate clones, which are not expressed in-frame with the fusion protein used to display the peptide on the surface to enrich for naturally occurring sequences. The comprehensive screening of these libraries by magnetic cell sorting (MACS) determines the profile of antigens, which are expressed in vivo and elicit an immune response in humans. This strategy was applied to the methicillin-resistant Staphylococcus aureus strain (MRSA) COL, because MRSA is one of the most common causes of hospital-acquired infections, and clinical strains have been isolated that are resistant to both methicillin and vancomycin (9). More than 60 antigenic proteins were identified and their Ab-binding sites mapped. Importantly, most of these proteins are predicted to be secreted or located on the cell surface of S. aureus.
Materials and Methods
Frame Selection Vector pMAL4.1.
The vector pMAL4.1 was constructed to allow frame selection of randomly generated genomic DNA fragments inserted into the gene encoding β-lactamase fused with the sequence encoding OmpA signal peptide. The β-lactamase gene was amplified by PCR with oligonucleotides ICC.162 (GGCCGGCCTCCCGGGGGCGGCCGCCACCCAGAAACGCTGGTGAAAG) and ICC.140 (TGCAGAATTCTTACCAATGCTTAATCAGTGAGG) from pBluescript KS- and the fragment encoding the OmpA signal peptide with oligonucleotides ICC.001 (ACGTCCATGGAAAAGACAGCTATCGCGATTG) and ICC.163 (GGCCGCCCCCGGGAGGCCGGCCGCGGCCTGCGCTACGGTAGCGAAAC) by using pEV218 (10) as template. The fusion was generated with oligonucleotides ICC.001 and ICC.140 by using both PCR products as templates. The resulting PCR product was cleaved with NcoI and EcoRI and ligated into plasmid pEH1 (11) digested with the same restriction enzymes. The resulting construct was verified by DNA sequence analysis.
Construction of Genomic Libraries.
S. aureus genomic DNA fragments were mechanically sheared into fragments ranging in size from 150 to 300 bp by using a cup-horn sonicator (Bandelin Sonoplus UV 2200 sonicator equipped with a BB5 cup horn, 10-sec pulses at 100% power output) or into fragments of approximately 50 bp in size by mild DNase I treatment by using a shotgun cleavage kit (Novagen, Madison, WI). Genomic DNA digestion was performed in the presence of 10 mM MnCl2 in a volume of 60 μl at 20°C for 5 min to ensure double-stranded cleavage by the enzyme. Reactions were stopped with 2 μl of 0.5 M EDTA, and fragmentation efficiency was evaluated on a 2% Tris-Acetate-EDTA–agarose gel. Fragments were blunt-ended twice by using T4 DNA polymerase in the presence of 100 μM dNTPs. Fragments were used immediately for ligation reactions into SmaI-digested pMAL4.1. Transformed DH10B cells (Life Technologies, Grand Island, NY) were plated on LB plates containing 50 μg/ml each of ampicillin and kanamycin. LSA50 and LSA250 were generated from ≈50- and 250-bp fragments inserted into pMAL4.1, respectively. Between 107 and 108 transformants were obtained per microgram of vector DNA used for the ligation.
DNA fragments were excised from pMAL4.1 libraries LSA50 and LSA250 by using restriction enzymes FseI and NotI and transferred into vectors pMAL9.1 and pHIE11 (12) encoding the display platforms LamB and FhuA, respectively. The resulting libraries were termed LamB-LSA50 and FhuA-LSA250.
Human Sera.
Human serum samples were obtained from patients suffering mostly from S. aureus wound- and catheter-related infections, causing local soft tissue infection and bacteraemia or septicaemia, respectively. The sera were taken in the acute phase of the S. aureus infection. Therefore, S. aureus-specific Abs might be partly preexisting or induced by the current infection and synthesized by previously primed B cells. Normal sera were donated by healthy adults. The healthy donors were free of any obvious infection by S. aureus. Control sera were obtained from infants between 6 and 24 mo of age hospitalized for noninfectious diseases.
ELISA.
ELISA plates were coated with 2–10 μg/ml of antigen in coating buffer (100 mM sodium carbonate, pH 9.2). Serial dilutions of sera (100–100,000) were made in TBS/1% BSA. Highly specific anti-human IgG secondary Abs coupled to horseradish peroxidase (Southern Biotechnology, Birmingham, AL) were used according to the manufacturer's recommendations (dilution 1:2,000). Antigen–Ab complexes were quantified by measuring the conversion of the substrate (ABTS) to colored product based on OD405 readings in an automated ELISA reader (Wallace Victor 1420, Perkin–Elmer). The titers were compared at those dilution steps where the response was linear. The individual normal and patient sera were ranked on the basis of IgG titer, then the scores (1–8) for ELISA values against the individual antigens were summed and the highest ones selected for further analysis.
MACS Screening.
Between 107 and 108 cells of the respective library were used for an individual experiment. Protein expression was induced with 1 mM isopropyl β-D-thiogalactoside for 30 (FhuA) or 90 min (LamB). Cells were harvested and washed twice with LB medium and incubated with 10 μg of IgGs from patient (P1) or infant sera in 50 μl of LB medium overnight at 4°C. The sera were preadsorbed against E. coli cells expressing the relevant wild-type platform protein. The cells were then washed and incubated with biotinylated goat–anti-human IgG Abs (Southern Biotechnology Associates) at 0.2 μg/sample in LB medium for 30 min at 4°C. After another wash with LB medium, 10 μl of MACS microbeads coupled to streptavidin (Miltenyi Biotech, Bergisch Gladbach, Germany) was added and the incubation continued for 20 min at 4°C. Thereafter, 900 μl of LB medium was added, and the MACS microbead cell suspension was loaded onto the equilibrated MS column (Miltenyi Biotech), which was attached to the magnet. The column was washed once with 3 ml of LB medium and the elution performed by removing the magnet and washing with 2 ml of LB medium. After washing the column with 3 ml of LB medium, the loading, washing, and elution process was repeated twice, resulting in a final eluate of 1 ml. Cells were plated onto LB plates containing kanamycin to identify selected clones. Two cycles of MACS selection with patient and normal high-titer sera resulted in approximately 30- to 50-fold more recovered clones in comparison to infant sera.
Opsonophagocytosis Assay.
Pooled high-titer sera were heat inactivated and anti-E. coli Abs removed by incubation with whole E. coli cells. Protein G affinity-purified IgG preparations were tested for opsonic activity in an in vitro FACS-based opsonophagocytosis assay. FITC-labeled 8325–4 spa- (protein A deficient) S. aureus cells (107) were preopsonized with 100 μg of IgG in the absence or presence of 2% hamster serum as complement source. The bacteria were then added to 106 freshly isolated human polymorphonuclear cells (PMNs) and incubated for 30 min at 37°C. Bacterial uptake was registered as an increase in mean fluorescence intensity of the phagocytic PMNs measured by FACS analysis.
Animal Experiments.
Bacterial cultures were grown in LB medium supplemented with 50 μg/ml of kanamycin at 37°C to midlogarithmic phase. For protein expression, cells were induced with 1 mM isopropyl β-D-thiogalactoside for 3 h at 37°C. Cells were harvested by centrifugation, washed twice with PBS, and resuspended in PBS at 1010 cells/ml. The bacteria were disrupted by sonication on ice to yield less than 10 living cells per 109 cells before injection. The protein concentration of the sonicated extracts was determined by Bradford assay. Degradation of the proteins because of the harsh sonication steps was not observed, as determined by Coomassie gel electrophoresis and Western blotting with anti-FhuA Abs (12). An equivalent of 109 cells in 100 μl of PBS (1 mg of total protein) was injected intravenously into BALB/c mice. Serum samples were taken before injection and 2 and 4 wk after injection. All sera from mice were adsorbed with E. coli cell extracts expressing wild-type FhuA protein to remove Abs recognizing any E. coli protein.
Results and Discussion
Generation of Frame-Selected Genomic S. aureus Libraries.
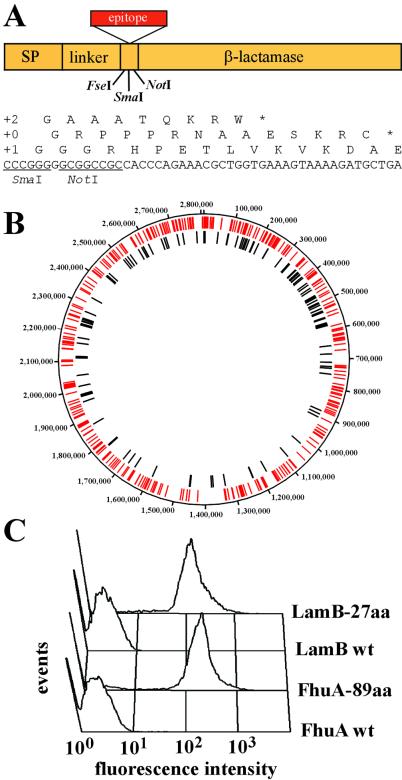
To generate libraries potentially expressing all genome-encoded amino acid sequences, S. aureus genomic DNA was randomly fragmented by DNase I treatment or sonication to an average size of 50 or 250 bp, because small peptide sequences have a much higher success rate of expression compared with full-length proteins. The genomic fragments were cloned into plasmid pMAL4.1 to select only those sequences that are in-frame with the β-lactamase gene (Fig. 1A). Elimination of those clones, which would disrupt the frame of the fusion protein used for display, will reduce the number of clones in the expression library by a factor of up to 18, resulting in a much lower background during MACS selection from negative cells. Sequencing of approximately 600 randomly picked clones from the 50-bp library in pMAL4.1 (LSA50) and alignment of these sequences with the genome of S. aureus confirmed the random distribution of these fragments throughout the genome (Fig. 1B). The frame-selected S. aureus genomic fragments were then transferred into vectors encoding the bacterial surface display platform proteins LamB and FhuA, maintaining the correct reading frame and generating two small fragment S. aureus genomic libraries, termed LamB-LSA50 (50 bp) and FhuA-LSA250 (250 bp). It was previously shown that the model epitopes myc and T7⋅tag were efficiently displayed on the surface of E. coli by both platform proteins (Fig. 1C), enabling recovery of cells by MACS with the respective mAb (12). Two different outer membrane proteins were used to display the S. aureus peptide libraries on the cell surface to reduce the possible bias imposed by inefficient expression, export, or incorporation of the fusion protein into the outer membrane.
Figure 1.
Surface display of peptides derived from random genomic fragments by E. coli. (A) Genomic fragments were frame-selected by using the β-lactamase selection vector pMAL4.1. Insertion of fragments into the SmaI site fulfilling the 3n + 1 length requirement restore the reading frame of β-lactamase, whereas the other two reading frames (3n + 0, 3n + 2) result in premature termination. SP, signal peptide. (B) The sequence of 600 randomly picked clones from the LSA50 library was determined and aligned with the chromosome of S. aureus. Red and black bars correspond to matches to ORFs and to noncoding sequences, respectively. (C) Efficient surface display of the T7⋅tag in loop 5 of LamB (27 aa) and of six myc epitopes in loop 5 of FhuA (89 aa) is shown by FACS analysis. Cells were stained with mAb T7⋅Tag (Novagen) and mAb α-myc 9E10, respectively.
Characterization of S. aureus-Specific Immune Sera.
Bacterial infections of hypo- and agammaglobulinemic patients are most frequently caused by S. aureus, but S. aureus infections are also part of the disease spectrum characteristic for patients with defects in neutrophil granulocyte functions. This notion suggests that humoral immunity is a cornerstone of host defense and that opsonizing Abs are crucial to combat S. aureus. Human sera were therefore selected for high levels of antistaphylococcal Abs to enable identification of antigens that are recognized by the human immune system. The identification of S. aureus proteins as antigens by Abs from human sera is a strong indication that these antigens are expressed in vivo and can elicit an immune response, which is a requirement for a potential vaccine candidate. A collection of 150 serum samples obtained from three major groups of individuals was analyzed: patients with S. aureus infections, healthy adults, and infants. The first two groups served as sources of antistaphylococcal Abs, whereas the 6- to 24-mo-old infants provided naïve human serum samples (negative controls). Although Abs in patients may not be considered protective because they could not prevent disease, the lack of protection may be due to an insufficient level of the respective Abs. Induction of these Abs by vaccination may well lead to protection. In that respect, it is interesting to note that vaccination of animals with surface proteins (13) and secreted toxins (14) resulted in protection of mice in various models of infection, although Abs against, e.g., FnbpA/B can also be detected in human sera from healthy individuals and patients without vaccination (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). On the other hand, antigens identified by human sera from healthy individuals may be especially important, because it could be speculated that these may protect those individuals from infection, whereas patients were not capable of mounting an efficient Ab response against these antigens.
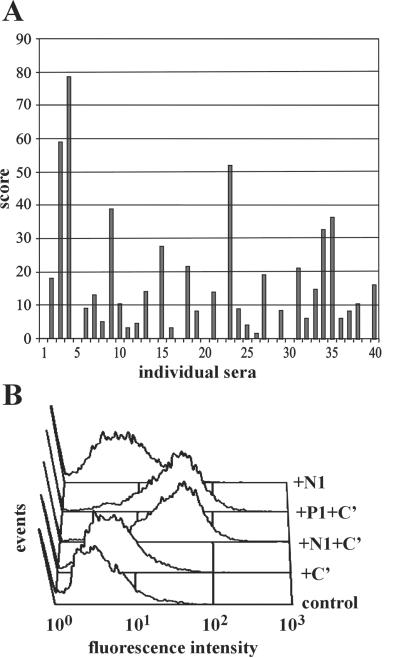
Sera with high levels of antistaphylococcal Abs from both patients and healthy adult donors were selected by measuring IgG Ab levels against multiple antigenic preparations of S. aureus (Fig. 2A). On the basis of the cumulative titer, we classified ≈10–15% of the normal and ≈30% of the patient sera as having high titers of antistaphylococcal Abs. Five individual selected sera were pooled and their purified IgG preparations tested for functional activity in an in vitro opsonophagocytosis assay. Phagocytosis of bacteria by freshly isolated human neutrophils was increased in the presence of complement (Fig. 2B) and was IgG concentration dependent (unpublished data). IgG preparations from patients and healthy individuals induced a similar level of opsonization.
Figure 2.
Characterization of human serum collections. (A) Cumulative Ab levels were determined by ELISA by using multiple S. aureus antigens. Antistaphylococcal Ab levels were measured individually with total S. aureus lysate, lipoteichoic acid, peptidoglycan, and 13 recombinant proteins: clumping factor A and B (ClfA, ClfB), Fibronectin-binding protein (FnBPA), SD-repeat proteins (SdrC, SdrE), MHC class II analogous protein (Map-w), elastin-binding protein, enolase, iron transport lipoproteins (LP309, LP342), Sortase, Coagulase, and extracellular fibrinogen-binding protein. Individual sera from healthy individuals were ranked on the basis of their IgG titer against each antigen. The total score was calculated as a sum of the ranking scores against individual antigens. The selection of patient and normal sera was based on the same criteria. (B) IgGs from five pooled high-titer healthy normal (N1) or patient (P1) sera were tested for opsonic activity in an in vitro opsonophagocytosis assay. FITC-labeled 8325–4 protein A deficient S. aureus cells (107) were preopsonized with 100 μg of human IgGs in the absence or presence of complement (C′). Bacteria were incubated with 106 freshly isolated human polymorphonuclear cells (PMNs), and opsonophagocytosis was measured by FACS analysis. (C) Control: PMNs incubated with labeled bacteria in the absence of IgG or C′.
Identification of Antigenic Epitopes by Bacterial Surface Display.
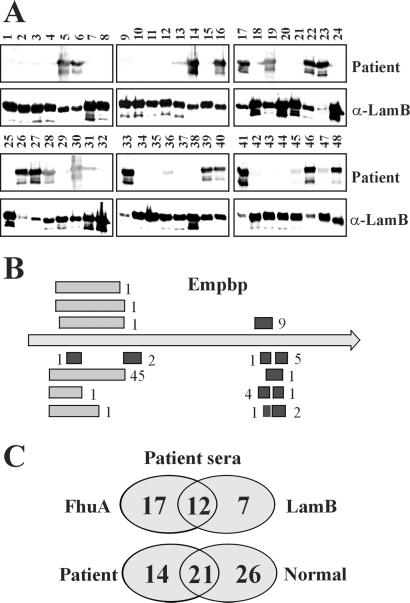
The two different peptide expression libraries (LamB-LSA50 and FhuA-LSA250) were screened with two serum pools applying MACS for fast and efficient recovery of positive cells (Table 1, Fig. 3). A representative number of recovered clones (500–1,000) were randomly selected for sequencing. More than 93% of positive clones were derived from predicted ORF sequences. Because of the genomic nature of the libraries, a small number of epitopes were also derived from nonpredicted ORFs. This observation shows that the technology may also identify novel protein-encoding sequences. The bioinformatic analysis also revealed that individual clones were recovered with different frequencies (Table 1). Assuming equal representation of the epitopes in the library and knowing that cells are amplified only to a very limited extent by our procedure, the antigenic epitopes selected most frequently will likely be those against which serum Abs are most abundant and/or have the highest affinity. Recovered clones were verified by Western blot analysis for reactivity with the same serum pool used for screening (Fig. 3A), showing that up to 50% of cells were recovered specifically. As exemplified by the Empbp protein, antigenic regions are often identified by an overlapping set of sequences because of the random nature of the libraries (Fig. 3B). Those antigenic epitopes that were recovered most frequently were identified by both platform protein screens, for example, Empbp, SA2581 (15), IsaA (16), SA2291, Lipase, Autolysin, and Protein A (Table 1, Fig. 3; for more information on ORFs, see: http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database = gsa). LPXTGp5 was frequently identified in the LamB-LSA50 screen, whereas SA0723, SA1472, and Coagulase were discovered only in the FhuA-LSA250 screen. Approximately 40 additional proteins were identified with intermediate or low recovery rates (unpublished data). This analysis confirmed that the use of multiple platforms would lead to a more comprehensive identification of antigens (Fig. 3C). It was of interest to compare the results from screens performed with serum pools from patients and healthy individuals. Although SA2291, SA2006, SA1472, Autolysin, SA0723, Empbp, SA2581, and Protein A were identified frequently with both serum pools, SA1781, IsaA, and Lipase were recovered mainly with the patient serum pool. This analysis suggests that Ab levels against the latter proteins are preferentially raised during a S. aureus infection. SA0470, Coagulase, and SAA0001 were among those identified only with the high-titer serum pool from healthy individuals, suggesting that these proteins may not be able to induce a strong Ab response in patients. In contrast, exposure of healthy individuals with the pathogen seems to result in a high level of Abs against these antigens, which thus may contribute to the protection of healthy individuals from disease.
Figure 3.
Identification of antigenic proteins from S. aureus by MACS. (A) The LSA50-LamB library was screened with an IgG preparation from patients (P1) by MACS selection. Forty-eight randomly picked clones were tested with patient serum P1 (1 μg/ml IgG) and anti-LamB mAbs at a dilution of 1:2,000. (B) Epitopes recovered from the LSA50-LamB screen (black bars) and the LSA250-FhuA screen (gray bars) with P1 serum and aligned with the sequence of the gene encoding Empbp are localized mainly in two regions. Numbers next to bars represent the frequency of recovery of these epitopes. (C) Comparison of the number of proteins identified in screens with human patient serum in the two platforms LamB and FhuA and of those from screens performed with patient sera and high-titer sera from healthy individuals.
In Vitro and in Vivo Characterization of Immunogenic Proteins.
The presence of Abs against a particular antigen in a large number of patients is indicative of the expression of the respective protein in vivo and demonstrates that most individuals are able to mount an immune response against this protein; both are requirements for a successful vaccine candidate. Therefore, the reactivity of 40 patient sera as well as 30 sera from healthy individuals was assessed with the identified epitopes (Fig. 4, Table 1). Although many antigens reacted with a limited number of individual sera, 15 antigens showed reactivity with more than 70% of all tested sera. Thus, the idiosyncratic reaction of individual sera with the spectrum of identified antigens represents a valuable tool to select the most promising vaccine candidates among all antigens. In addition to the data obtained from the analysis with individual sera, the presence of the respective gene in 30 different S. aureus strains was analyzed by PCR by using oligonucleotides amplifying sequences encoding at least one of the identified immunogenic regions of the proteins (Table 1). The analysis of 13 immunogenic proteins showed that the genes encoding nine of these are present in all tested strains, whereas only four genes were absent in 5–30% of all strains analyzed. Together with the results obtained with the individual sera, these data suggest that a potential vaccine against S. aureus based on these antigens could be effective against a wide spectrum of clinical isolates.
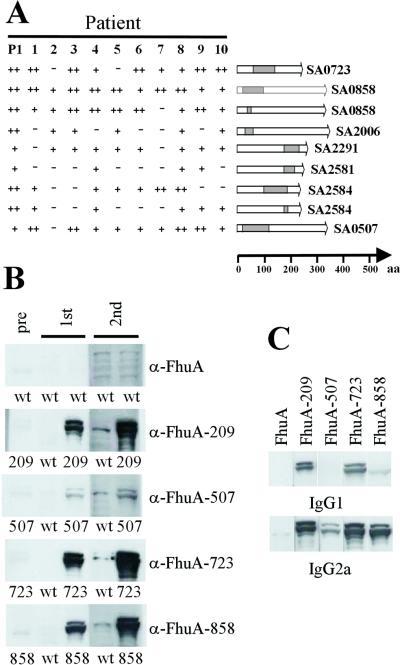
Figure 4.
Evaluation of antigenic epitopes. (A) Reactivities of a selection of 10 individual patient sera and patient pool P1 with 9 identified epitopes. The location of the epitope within the protein is shown schematically. (B) Approximately 109 lysozyme-treated and sonicated bacteria expressing FhuA or a fusion of the antigenic epitope and FhuA (e.g., FhuA-209) were injected intravenously into BALB/c mice. The four epitopes embedded in FhuA were expressed at comparable levels in E. coli on the basis of Western blot analysis with polyclonal anti-FhuA Abs. Serum was taken 2 wk after the first and second injections and depleted for Abs directed against E. coli or FhuA protein with a cellular lysate from E. coli expressing wild-type FhuA. FhuA fusion proteins were detected by Western blot analysis in lysates from an equivalent of 5 × 106 bacteria. Sera from mice injected with FhuA-858, 209, and 507 were diluted 1:5,000, sera from animals injected with FhuA-723 were diluted 1:50,000, and preimmune serum 1:1,000. Horseradish peroxidase (HRP)-coupled anti-mouse Igs were used at a dilution of 1:5,000. (C) The same lysate as above was assessed for reactivity with IgG type-specific Abs by using biotinylated anti-mouse IgG1 and IgG2a as secondary Abs at a concentration of 0.2 and 0.05 μg/ml, respectively. HRP-coupled streptavidin was diluted 1:5,000.
To facilitate the analysis of the immunogenicity and protective value of the identified antigens in an animal model, we used E. coli cells expressing single immunogenic epitopes from S. aureus as a vehicle for the delivery of antigens in mice (17). This possibility makes our approach very powerful in that epitopes identified by bacterial surface display can be directly tested for immunogenicity in vivo. As an example, four individual epitopes were selected representing clones recovered with different frequencies in our screens. Among these four clones, the one encoding the epitope derived from SA0723 was identified most frequently, those from Empbp and Coagulase with intermediate frequencies, and the one from SA0507 with low frequency. On i.v. injection of 109 dead bacterial cells into mice, serum samples were taken after 2 and 4 wk. The levels of Abs induced clearly correlated with the frequency of clones recovered from the screen. The strongest response was detected for SA0723, an intermediate response for Empbp and Coagulase, and a weak response for SA0507 (Fig. 4B). Using this vaccination strategy, it could be shown that the Th1-type IgG2a subclass of murine Abs was induced to a higher level than the IgG1 subclass (Fig. 4C). Because the presence of opsonizing and complement fixing Th1-type IgG1a and IgG3 human Abs is thought to be of central importance to fight S. aureus-caused disease, these results indicate that Abs induced by vaccination with these epitopes may be beneficial for the clearance of an infection by S. aureus.
Anti-LPXTGp5 Abs Mediate Opsonophagocytosis of S. aureus by Macrophages.
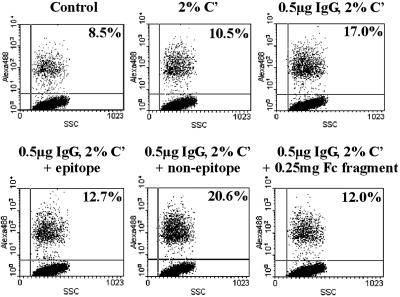
A large fraction of antigens that was identified by our method represent cell surface-localized and secreted proteins, among which known antigenic proteins such as FnbpA and B were found (18). One novel antigen, LPXTGp5, which is equipped with a typical Gram+ signal peptide and a C-terminal cell wall-sorting signal, the LPXTG motif was studied in more detail (19). On the basis of ELISA results with recombinant LPXTGp5 protein, it was apparent that patients with S. aureus infection produce significantly more anti-LPXTGp5 antibodies than healthy individuals, suggesting that the protein is expressed and immunogenic in vivo during infection (unpublished data). Bacterial surface display identified three different B cell epitopes with different recovery rates within LPXTGp5. Peptide ELISA confirmed the highest levels of Abs against the peptide that was selected most frequently (unpublished data). Using this highly immunogenic 15-aa-long peptide in affinity chromatography, Abs from human serum were purified. In the presence of these peptide-specific IgGs, an opsonizing agent, phagocytic cells were induced to take up labeled S. aureus (Fig. 5). These data suggest that the epitope is available for binding on the bacterial surface, and that the corresponding Abs are functionally relevant.
Figure 5.
Opsonization of S. aureus with B cell epitope-specific human Abs. IgGs were purified from human sera by affinity chromatography by using a 15-aa-long synthetic peptide (PPKDTNQTQPATQPA) representing one LPXTGp5 epitope. Alexa-Fluor-labeled S. aureus Wood 46 (STAW) cells (107) were preopsonized with 0.5 μg of epitope-specific anti-LPXTGp5 Ab in the presence of 5 μg of LPXTGp5-derived specific (epitope) or nonspecific (nonepitope) peptides. Uptake was registered as a percentage of phagocytic P388 mouse monocytic cells (106) with increased fluorescence after 15-min incubation with labeled STAW measured by FACS. The control sample represents P388 cells incubated with labeled bacteria in the absence of complement (C′).
The availability of the genomic sequence of S. aureus has allowed the development of a comprehensive and rapid approach for the identification of immunogenic epitopes/proteins in this pathogen. The identification of all those antigens that are recognized by the human immune system is a prerequisite for the selection of candidates for the development of a potent subunit vaccine. The described approach allows the comprehensive identification of antigens from genomic peptide expression libraries as well as the use of recovered clones for further analysis in vitro and in vivo without the need for expression and purification of recombinant proteins. It therefore not only allows the rapid identification of antigens but also enables them to be moved quickly into vaccine studies. The display of small genome-derived peptides is further advantageous because B cell epitopes are directly delineated, and the development of synthetic vaccines based on these epitopes may reduce or even abolish the risk of autoimmune reactions caused by vaccination with full-length bacterial proteins (20, 21). We have identified more than 60 immunogenic proteins from the pathogenic bacterium S. aureus and have shown that bacterial surface display is uniquely suited to assess the value of the antigenic epitopes without further manipulation for the development of a vaccine against this important pathogen. Furthermore, we have extended our approach to the related bacterium Staphylococcus epidermidis, which causes similar nosocomial infections, to select proteins that may lead to a protective vaccine.
Supplementary Material
Acknowledgments
We are grateful to S. Grill and M. Dandonneau for technical assistance. We thank T. Foster for S. aureus strains Newman, 8325–4 and its protein A-deficient derivative. This work was supported by the Wiener Wirtschaftsförderungsfond and the Forschungförderungsfond.
Abbreviation
- MACS
magnetic cell sorting
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pelton S I. Vaccine. 2000;19:S96–S99. doi: 10.1016/s0264-410x(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 2.Pizza M, Scarlato V, Masignani V, Giuliani M M, Arico B, Comanducci M, Jennings G T, Baldi L, Bartolini E, Capecchi B, et al. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 3.Wizemann T M, Heinrichs J H, Adamou J E, Erwin A L, Kunsch C, Choi G H, Barash S C, Rosen C A, Masure H R, Tuomanen E, et al. Infect Immun. 2001;69:1593–1598. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravarti D N, Fiske M J, Fletcher L D, Zagursky R J. Vaccine. 2000;19:601–612. doi: 10.1016/s0264-410x(00)00256-5. [DOI] [PubMed] [Google Scholar]
- 5.Jungblut P R, Schaible U E, Mollenkopf H J, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufmann S H, et al. Mol Microbiol. 1999;33:1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- 6.Kemp D J, Coppel R L, Cowman A F, Saint R B, Brown G V, Anders R F. Proc Natl Acad Sci USA. 1983;80:3787–3791. doi: 10.1073/pnas.80.12.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereboeva L A, Pereboev A V, Wang L F, Morris G E. J Med Virol. 2000;60:144–151. [PubMed] [Google Scholar]
- 8.Folgori A, Tafi R, Meola A, Felici F, Galfre G, Cortese R, Monaci P, Nicosia A. EMBO J. 1994;13:2236–2243. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchese A, Schito G C, Debbia E A. J Chemother. 2000;12:459–462. doi: 10.1179/joc.2000.12.6.459. [DOI] [PubMed] [Google Scholar]
- 10.Freudl R. Gene. 1989;82:229–236. doi: 10.1016/0378-1119(89)90048-6. [DOI] [PubMed] [Google Scholar]
- 11.Hashemzadeh-Bonehi L, Mehraein-Ghomi F, Mitsopoulos C, Jacob J P, Hennessey E S, Broome-Smith J K. Mol Microbiol. 1998;30:676–678. doi: 10.1046/j.1365-2958.1998.01116.x. [DOI] [PubMed] [Google Scholar]
- 12.Etz H, Bui D M, Schellack C, Nagy E, Meinke A. J Bacteriol. 2001;183:6924–6935. doi: 10.1128/JB.183.23.6924-6935.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rennermalm A, Li Y H, Bohaufs L, Jarstrand C, Brauner A, Brennan F R, Flock J I. Vaccine. 2001;19:3376–3383. doi: 10.1016/s0264-410x(01)00080-9. [DOI] [PubMed] [Google Scholar]
- 14.Stiles B G, Garza A R, Ulrich R G, Boles J W. Infect Immun. 2001;69:2031–2036. doi: 10.1128/IAI.69.4.2031-2036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang S, Livesley M A, Lambert P A, Littler W A, Elliot T S J. FEMS Immunol Med Microbiol. 2000;29:213–220. doi: 10.1111/j.1574-695X.2000.tb01525.x. [DOI] [PubMed] [Google Scholar]
- 16.Lorenz U, Ohlsen K, Karch H, Thiede A, Hacker J. FEMS Immunol Med Microbiol. 2000;29:145–153. doi: 10.1111/j.1574-695X.2000.tb01517.x. [DOI] [PubMed] [Google Scholar]
- 17.Leclerc C, Charbit A, Molla A, Hofnung M. Vaccine. 1989;7:242–248. doi: 10.1016/0264-410x(89)90237-5. [DOI] [PubMed] [Google Scholar]
- 18.Casolini F, Visai L, Joh D, Conaldi P G, Toniolo A, Hook M, Speziale P. Infect Immun. 1998;66:5433–5442. doi: 10.1128/iai.66.11.5433-5442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarre W W, Schneewind O. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamula M J. Immunol Rev. 1998;164:231–239. doi: 10.1111/j.1600-065x.1998.tb01223.x. [DOI] [PubMed] [Google Scholar]
- 21.Regner M, Lambert P-H. Nat Immunol. 2001;2:185–188. doi: 10.1038/85228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.