Abstract
The creation of novel enzymatic function is of great interest, but remains a challenge because of the large sequence space of proteins. We have developed an activity-based selection method to evolve DNA polymerases with RNA polymerase activity. The Stoffel fragment (SF) of Thermus aquaticus DNA polymerase I is displayed on a filamentous phage by fusing it to a pIII coat protein, and the substrate DNA template/primer duplexes are attached to other adjacent pIII coat proteins. Phage particles displaying SF polymerases, which are able to extend the attached oligonucleotide primer by incorporating ribonucleoside triphosphates and biotinylated UTP, are immobilized to streptavidin-coated magnetic beads and subsequently recovered. After four rounds of screening an SF library, three SF mutants were isolated and shown to incorporate ribonucleoside triphosphates virtually as efficiently as the wild-type enzyme incorporates dNTP substrates.
The process of in vitro evolution has proven to be a powerful approach for generating proteins that have desired physical or biological functions. Many techniques have been reported that can produce large libraries of mutant proteins, such as cassette mutagenesis (1, 2), error-prone PCR (3, 4), staggered extension process PCR (5), and gene shuffling (6, 7). Methods to identify proteins with desired activities include phage display (8, 9), ribosomal display (10), and complementation (11). Phage display has emerged as a particularly powerful technique to select desired mutants from large libraries based on affinity for a given target molecule. In conjunction with techniques to attach both enzymes and substrates to a single phage particle, phage display is also expected to be able to select mutants based on enzymatic activities. Using this idea, we have developed an activity-based selection method to evolve mutants of the Stoffel fragment (SF) of Thermus aquaticus DNA polymerase I with novel catalytic activities.
The manipulation of DNA polymerase activity has attracted a great deal of attention because of the central roles of polymerases in biological processes as well as their utility in biotechnology applications. Earlier efforts to modify polymerase activity have focused largely on the rational design of site-directed mutants. For example, significant effort has been directed toward mutagenizing a DNA polymerase into an RNA polymerase (RNAP) (12, 13). Mutants that extend DNA primers by the incorporation of single ribonucleoside triphosphates (rNTPs) have been constructed; however, mutants that efficiently add successive rNTPs have proven more difficult to isolate. Moreover, in all reported cases, the mutant enzyme still prefers the dNTP substrates. The limited success of the rational approach likely results from the limited sequence space of the polymerases examined in these experiments. In vitro evolution strategies in which large populations of mutants are sampled for those with the desired activities are more likely to be successful, especially for rare activities. For example, with water-in-oil emulsion technology (14), Holliger and coworkers (15) evolved DNA polymerases that were either more thermally stable or more resistant to an inhibitor. Winters and coworkers (16) demonstrated that phage pIII-displayed DNA polymerases could be isolated based on their activity-dependent modification of an attached substrate. However, the substrate was attached to the major phage coat protein, pVIII, raising the concern of cross-reactivity between a polymerase on one phage and a substrate attached to another phage. Cross-reactivity will compromise the association of genotype with phenotype and inhibit the successful evolution of desired function (see below).
We have developed an activity-based selection method, in which a DNA polymerase and its substrate are both attached to the minor phage coat protein, pIII. The pIII proteins are localized to only one end of the phage particle (Fig. 1), which is expected to favor intramolecular reaction of the enzyme with its associated substrate, thus ensuring the linkage of genotype and phenotype. Selection of desired mutants depends on the enzyme-catalyzed labeling of the attached substrate (e.g., biotinylation), which allows for the selective isolation of phage particles displaying active polymerases. Here we report the use of this method to evolve SF DNA polymerase mutants with RNAP activity. Three mutants are isolated and shown to efficiently catalyze the incorporation multiple rNTP substrates with efficiencies approaching those of the wild-type enzyme with dNTP substrates.
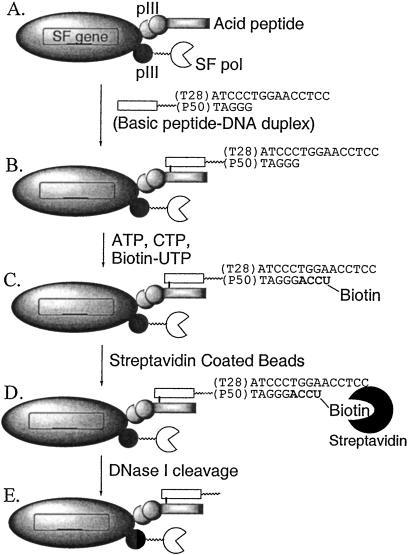
Figure 1.
The phage selection scheme. (A) Phage particle with displayed polymerase and acidic peptide. (B) Ribonucleotide substrate attachment. (C) Biotinylation of phage particles displaying active polymerases. (D) Immobilization of biotin-tagged phage particles with strepavidin beads. (E) Release of immobilized phage by DNase I cleavage.
Materials and Methods
Plasmid Construction.
The SF of Taq DNA polymerase was amplified from plasmid pWB254b (ATCC no. 69244) with primers 5′-TTT AAG CTT CAT ATG GCC CAG CCG GCC AGC CCC AAG GCC CTG GAG GAG GCC CC and 5′-GAA TCC GCG GCC GCC CTC CTT GGC GGA GAG CCA GTC CTC C, and cloned into pET23b(+) to generate pET-SF. The internal SfiI site in the SF gene was eliminated by using a QuikChange mutagenesis kit (Stratagene). The SfiI/NotI-treated SF was ligated into a modified pFAB5c.His vector to generate pFAB-SF. The ampicillin resistance gene in the original pFAB5c.His vector was replaced with the spectinomycin resistance gene to increase the stability of the phagemid.
Phage Preparation.
Construction and isolation of the helper phage X30 has been described (8). Briefly, X30 was derived from M13 helper phage by fusing of gIII to DNA coding for an “acidic” peptide that is used to attach substrate (see below). All phage discussed in this article were grown in Escherichia coli XL1-blue MRF′ (Stratagene) at 30°C to minimize the toxicity of expressed SF protein. Wild-type SF displaying phage was prepared by using standard protocols (17). To produce SF library phage, 300 μl of XL1-blue MRF′ competent cells was electroporated with 10 μg of SF library phagemid. The transformed cells were diluted in 150 ml of 2× yeast extract:tryptone (YT), containing 7.5 μg/ml tetracycline, and 50 μg/ml spectinomycin and grown at 30°C until the OD600 reached 0.6. The culture was inoculated with 75 μl of 1013 colony-forming units/ml X30 helper phage (M13 phage modified with a pIII-acidic peptide fusion, see below) and incubated at 37°C for 1 h without shaking. Next, the infected cells were collected and resuspended in 300 ml of fresh 2× YT with 7.5 μg/ml tetracycline, 50 μg/ml spectinomycin, 50 μg/ml kanamycin, and 0.4 mM isopropyl β-d-thiogalactoside. The resuspended culture was grown at 30°C for 16 h before the supernatant, containing the phage particles, was separated from the host cells by centrifugation. Phage particles were purified by two polyethylene glycol precipitations.
Synthesis of Basic Peptide-P50 DNA Conjugate.
The basic peptide, C(GGS)4AQLKKKLQALKKKNAQLKWKLQALKKKLAQGGC, whose affinity for the phage displayed acidic peptide allows for coupling of the RNAP substrate to the phage particle, contains a photoprotected cysteine (bold) and a free cysteine (underlined). The peptide was synthesized with Boc-protected amino acids (18) (Calbiochem-Novabiochem), and the crude product was purified by reverse-phase HPLC and characterized by matrix-assisted laser desorption ionization–time of flight MS. The peptide was coupled to N,N′-bis(3-maleidopropiryl)-2-hydroxy-1,3-propanediamine as described (8). The 5′ thiol-modified oligonucleotide (5′-S-TTATG TATGT ATTTT CGACG TTTGC TAACA AGATA CGACT CACTA TAGGG) was synthesized by using a 5′ thiol reagent (S = 5′-Thiol-Modifier C6, Glen Research, Sterling, VA), manually deprotected by using manufacturer's protocols, and purified on a 12% denaturing polyacrylamide gel. This oligonucleotide is referred to as P50, and after attachment to phage, it serves to prime polymerase synthesis during the selection experiment. To conjugate P50 to the basic peptide, 300 μg of basic peptide dissolved in 150 μl of water was mixed with 75 μl of 1 M sodium phosphate (pH 7.0), 30 μl of 5 M NaCl, and 22 μl of 3 mM P50 oligonucleotide. The mixture was incubated at 23°C for 14 h. The procedure was performed anaerobically to avoid oxidation. The peptide-P50 DNA conjugate was purified by anion exchange FPLC (Amersham Pharmacia).
DNA Substrate Attachment to Phage.
In an 80-μl reaction, 20 μl of 15 μM peptide-P50 DNA conjugate was mixed with 20 μl of PBS and 40 μl of 50 μM T28 template (5′-CCTCC AAGGT CCCTA TAGTG AGTCG TAT-NH2 (NH2 = 3′ amino modified, Glen Research). The amino group was included to prevent the 3′ end of the template from priming synthesis). The mixture was heated rapidly to 85°C, cooled slowly to anneal primer P50 and template T28, and deprotected under 365-nm UV light (3 cm from a 4-W light source) for 45 min. The deprotected peptide-DNA conjugate was diluted with 300 μl of attachment buffer (TBS containing 2.5 mM KCl, 1 mM EDTA, and 1 mM cystamine). To attach DNA substrate to phage, 100 μl of deprotected peptide-DNA conjugate was mixed with 700 μl of dilution buffer and 200 μl of 1.5 × 1013 colony-forming units/ml phage and incubated for 1 h at 37°C. The substrate-attached phage was polyethylene glycol-precipitated once to remove the free peptide-DNA conjugate.
Phage Selection.
Model selection experiments were performed with deoxyribonucleotide substrates (dNTP substrates were used, as opposed to rNTP substrates, which were used in the actual selection experiments described below, because the model selections involved wild-type polymerase). In the first round of screening, 400 μl of substrate attached phage (3 × 1012 colony-forming units) was added to a final reaction volume of 1.5 ml, containing 20 μM Tris⋅HCl (pH 8.0), 10 mM MgCl2, 40 μM CTP, 40 μM ATP, and 2 μM biotin-UTP. The reaction was initiated by transferring the mixture from ice to a 50°C water bath. After 5 min of incubation, 100 μl of 0.5 M EDTA was added to stop the reaction, and the phage particles were polyethylene glycol-precipitated twice to remove unreacted biotin-UTP and subsequently resuspended in 400 μl of TBS. After removing residual precipitate by centrifugation (20,800 × g for 10 min), the supernatant was added to 200 μl of streptavidin-coated magnetic beads (Dynal, Oslo). The beads were then washed 10 times with TBS containing 0.5% Tween-20. To release the bound phage, the beads were resuspended in 50 μl of TBS containing 10 mM MgCl2 and 1 mg/ml DNase I and incubated at 37°C for 30 min. The phage present in the supernatant was titered and subjected to further amplification.
Construction of Mutant SF Libraries.
Six regions of SF were randomly mutated based on the crystal structure of Taq with bound primer/template and triphosphate substrates (19). A focused library was created at each region of the polymerase by using two-step overlapping extension PCR to generate mutant genes. The first PCR products with primer-introduced random mutations at one end were used as primers in the second PCR amplification, so that the library fragment could be ligated into the SF gene at available restriction sites. The doping ratio for the mutagenic primers ranged from 8% to 45% depending on the size of the regions to be mutated (Table 1). Specifically, two regions of the polymerase metal binding site were mutated, including residues 597–615 (8% doping ratio) and 783–786 (45%); two regions of the template binding site, residues 728–734 (45%) and 746–754 (45%); the duplex binding site, residues 568–587 (8%); and the O-helix, residues 665–676 (8%). The mutant gene PCR products were then cloned into pFAB-SF. To minimize the wild-type polymerase gene background, vectors used in ligations contained an amber stop codon mutation within the O-helix region of the wild-type gene.
Table 1.
SF mutant libraries
| Directed library | Residues | Library size |
|---|---|---|
| Duplex binding site | A568-R587 | 2 × 107 |
| Metal binding site 1 | A597-E615 | 4 × 107 |
| O-helix | P656-H676 | 5 × 106 |
| Template interaction site 1 | R728-E734 | 6 × 107 |
| Template interaction site 2 | R746-Q754 | 7 × 107 |
| Metal binding site 2 | V783-E786 | 2 × 105 |
The library sizes were estimated by counting colonies after transformation of E. coli or by calculating the theoretical library size, whichever is smaller.
Mutant Polymerase Characterization.
After four rounds of selection, SF inserts from recovered phagemids were cloned into a modified pET23b(+) vector for overexpression of SF in E. coli BL21(DE3) (Novagen). Single colonies were used to inoculate 10 ml of 2× YT containing 100 μg/ml ampicilin and 34 μg/ml chloramphenicol. The inoculated culture was grown at 37°C until the OD600 reached 0.4. The cells were pelleted and resuspended in 10 ml of fresh 2× YT supplemented with ampicillin, chloramphenicol, and 0.4 mM isopropyl β-d-thiogalactoside. After 4 additional h of growth, the induced cells were pelleted and resuspended in 0.5 ml of 50 mM Tris⋅HCl buffer, pH 7.6. Cells were lysed by sonication, and cellular debris was removed by centrifugation at 20,800 × g for 15 min. For the partial purification of SF mutant proteins by heat treatment, the clarified cell lysates were incubated in a 75°C water bath for 30 min and subsequently centrifuged to remove cellular proteins. The partially purified SF mutants were present in the clarified lysates at concentrations of 0.05–0.1 μg/μl and were used in primer extension assays to examine RNAP activity.
Mutant Polymerase Purification.
The SF mutants with RNAP activity were prepared on a larger scale. Freshly grown cells were diluted in 300 ml of 2× YT containing 100 μg/ml ampicilin and 34 μg/ml chlorophenicol and grown at 37°C until the OD600 reached 0.4. Then, isopropyl β-d-thiogalactoside was added to a final concentration of 0.4 mM to induce expression. The overexpressed SF proteins were partially purified by heat treatment and further purified with nickel chromatography. Proteins eluted from nickel columns were buffer-exchanged into 50 mM Tris⋅HCl (pH 7.6), 0.5 mM EDTA, and 1 mM DTT and concentrated to 0.5 mg/ml by using Centricon concentrators (Millipore). With this method, about 100–300 μg of the SF mutant proteins (purity greater than 90%) was routinely obtained from 300 ml of culture.
Mutant Polymerase Activity Assay.
The SF activity was assayed by using a standard primer-template extension assay (20). A typical reaction mixture contained 40 nM radiolabeled primer/template duplex, varying concentrations of triphosphates, and 0.1–10 nM SF enzyme. The reaction mixtures were incubated in a 50°C water bath for 3–30 min. The products were separated on a 15% denaturing polyacrylamide gel and analyzed by phosphorimaging (Storm 860, Molecular Dynamics). The steady-state kcat and KM values were obtained by fitting the data to the Michaelis–Menten equation with kaleidagraph (Synergy Software, Reading, PA).
Results
Selection System.
We have developed a selection method for DNA polymerase evolution that is based on the codisplay on a single phage particle of both enzyme and an acidic peptide that may be used to attach substrate. The phage-based activity selection system is based on one previously reported for the evolution of nuclease activity (8). The displayed acidic peptide is designed to form a leucine zipper and a disulfide bond with a second basic peptide that is added after isolation of the phage particles. The exogenous basic peptide is modified by attachment of an oligonucleotide substrate, and its specific association with the displayed peptide results in the substrate, polymerase mutant, and DNA encoding for the mutant, all displayed on a single phage particle.
Both the SF polymerase and the acidic peptide required for substrate attachment were displayed on phage as fusions with the phage minor coat protein pIII. To prevent any deleterious effects on polymerase activity caused by a C-terminal fusion to the pIII protein, a synthetic DNA fragment, encoding a flexible peptide linker (GGSGGS) was introduced between the SF gene and the truncated gIII in the pFAB-SF phagemid. Superinfection of bacteria harboring the pFAB-SF phagemid with X30 helper phage results in phage particles containing the phagemid DNA, but whose coat consists almost entirely of proteins encoded by the acid-helper phage genome. During packaging of the phage, both polymerase-pIII fusion and peptide-pIII fusion are produced. The phage particles obtained from a typical preparation carry either one or zero copies of the polymerase-pIII fusion plus 3–5 copies of the acidic peptide-pIII fusion. Western blots and phage ELISA indicated that polymerases were successfully displayed on phage, with a display level of 0.1–1% (ratio of phage with displayed polymerase to the total number of phage particles). The displayed polymerases retained their DNA polymerase activity in a primer/template extension assay using radiolabeled primer (data not shown). Basic peptide-DNA duplex substrates were attached to the phage particles as described in Materials and Methods.
The selection system is illustrated in Fig. 1. The isolation of desired polymerase mutants is based on their ability to catalyze nucleic acid synthesis by using the primer/template substrate attached to the same phage particle. Primer extension results in the incorporation of a biotinylated nucleoside tag that can be used to isolate the corresponding phage with immobilized streptavidin. Therefore, libraries of DNA polymerase mutants may be enriched for mutants with the desired activity by incubating the phage particles first with suitable biotin-labeled substrates and then with streptavidin-coated magnetic beads. After sufficient enrichment, through multiple rounds of reinfection, selection, and isolation, the phage are isolated and their displayed polymerases are characterized.
Model Study.
To optimize the activity-based selection, model selection experiments were conducted by using phage with no displayed polymerase (X30 helper-phage particles) and phage displaying wild-type SF polymerase [polymerase phage (pol/phage) particles], both with attached DNA substrate. After incubation with 50 μM dATP, 50 μM dCTP, and 2 μM biotin-dUTP, the phage titers were measured, both before binding to the streptavidin-coated magnetic beads and after their release from the beads. The recovery ratio was calculated as the number of recovered phage particles divided by the number of input phage particles. As a positive control, X30 phage particles were incubated with exogenously added wild-type polymerase (0.5 μM SF) and all required substrates, and then isolated on streptavidin beads. With the positive control, a high yield of the phage (≈40%) was released from the solid support upon treatment with DNase I. Recovery of X30 phage without added polymerase was very low (≈0.002%), as was recovery of pol/phage particles in the absence of biotin-dUTP (≈0.001%). Pol/phage with SF displayed and with DNA substrates attached were efficiently recovered (≈4%) in the presence of biotin-dUTP. The presence of displayed SF and biotin-dUTP results in at least a 1,000-fold increase in phage recovery.
In principle, cross-reactivity between a polymerase displayed on one phage particle and a substrate attached to another phage particle could circumvent the coupling of enzyme activity to phage isolation. To assess the impact of cross-reactivity, substrate-bearing wild-type SF phage and X30 phage particles (both with attached substrates) were mixed at different ratios to mimic varying stages of active polymerase enrichment and subjected to the biotin-dUTP incorporation selection. When SF pol/phage and X30 phage were mixed in a 1:1 ratio, no significant enrichment of the SF phage particles was observed. However, when SF and X30 phage were mixed 1:100 and 1:10,000, the recovered phage were enriched 12- and 450-fold in pol/phage particles, respectively. These results demonstrate that even in the presence of cross-reactivity and nonspecific phage binding, the selection system is capable of enriching libraries based on polymerase activity.
Library Construction.
To create SF libraries, we randomly mutated polymerase residues that contact the substrates or metal cofactors. Six directed libraries based on the Taq polymerase ternary complex crystal structure were generated (19). Each directed library contained random mutations focused to a specific region of the polymerase including two regions of the polymerase metal binding site, two regions of the template binding site, the duplex binding site, and the O-helix. The location and size of each library is presented in Table 1. Forty-eight clones from each focused library were chosen randomly and sequenced to assess the size and quality of the library. Approximately 50–80% of the clones sequenced contained inserts with correct reading frames and no stop codons. The six focused SF libraries were combined according to their sizes so that each was equally represented in the final library.
Polymerase Selection and Evaluation.
As an initial target we chose to evolve a DNA polymerase into an RNAP. Based on the library size (≈2 × 108) and polymerase display efficiency (≈0.1%), we used 3 × 1012 phage particles in the first round of screening to ensure that the average mutant will be displayed on 10 phage particles.
After four rounds of selection, mutant SF genes were isolated and cloned into a pET vector for overexpression. From 20 randomly chosen colonies, six clones were found to express SF. After partial purification (see Materials and Methods), four of these proteins were identified that could efficiently extend a DNA primer by insertion of ATP opposite dT in the template. The corresponding plasmids were isolated and sequenced (Table 2). One sequence (SFR1) was isolated twice and contained nine mutations. The two additional clones, SFR2 and SFR3, contained two and five mutations, respectively. Two mutations were found to be conserved in each isolated clone A597T and E615G. Two additional sites were found to be mutated in two of the three isolated clones Trp-604 to Gly or Arg, and I614T.
Table 2.
Alignment of wild-type and selected mutants
| 531 | 596 | 601 | 606 | 611 | ||
|---|---|---|---|---|---|---|
| SF-WT | K | RAFIA | EEGWL | LVALD | YSQIE | |
| SFR1 | E | —T——T | ———G— | ——SV— | ———TG | |
| SFR2 | — | —T——— | ————— | ————— | ————G | |
| SFR3 | — | —T——— | ———RQ | ————— | ———TG |
Dashes indicate identical residues to the wild-type protein.
His-tagged mutant proteins were further purified by nickel affinity chromatography. To quantitate the RNAP activity of each clone, we used a steady-state single nucleotide incorporation assay with a 23-mer DNA primer and 45-mer DNA template (Table 3 and Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). The kinetics revealed a remarkable increase in incorporation efficiency, with SFR3 showing increased rates of 103-104 for each rNTP relative to wild-type SF. The kcat/KM for UTP insertion opposite dA, which has proven especially difficult for DNA polymerase mutants to catalyze because of the strong selectivity against UTP (23), is increased by 103-fold in each mutant. With the same template, the rate of incorporation of each rNTP by SFR3 was only 2- to 9-fold slower than the incorporation of the corresponding dNTP by the wild-type enzyme. At least for single incorporation, SFR3 is as efficient with rNTP substrates as SF is with dNTP substrates. SFR1 and SFR2 were nearly as active (Table 3).
Table 3.
SFR1–SFR3 kinetic data
| 5′-TAATACGACTCACTATAGGGAGA |
| 3′-ATTATGCTGAGTGATATCCCTCTXGCTAGGTTACGGCAGGATCGC |
| ↓ dNTP or rNTP |
| 5′-TAATACGACTCACTATAGGGAGAN |
| 3′-ATTATGCTGAGTGATATCCCTCTXGCTAGGTTACGGCAGGATCGC |
| X | Single incorporation of correct dNTP kcat/KM, min−1⋅M−1 | Single incorporation of correct rNTPkcat/KM, min−1⋅M−1 | |
|---|---|---|---|
| SF | dT | 2.77 × 105 | 8.3 |
| dG | 4.42 × 106 | 1.2 × 103 | |
| dC | 3.03 × 106 | 1.5 × 103 | |
| dA | 8.5 × 105 | 8.0 | |
| SFR1 | dT | 6.1 × 104 | 1.4 × 105 |
| dG | 2.8 × 105 | 7.0 × 105 | |
| dC | 2.4 × 105 | 6.7 × 105 | |
| dA | 1.5 × 105 | 1.0 × 105 | |
| SFR2 | dT | 3.1 × 104 | 7.9 × 104 |
| dG | 2.5 × 105 | 5.7 × 105 | |
| dC | 1.7 × 105 | 3.8 × 105 | |
| dA | 1.0 × 105 | 5.9 × 104 | |
| SFR3 | dT | 4.6 × 104 | 1.4 × 105 |
| dG | 5.0 × 105 | 1.3 × 106 | |
| dC | 6.7 × 105 | 1.3 × 106 | |
| dA | 1.6 × 105 | 9.7 × 104 |
Assay conditions: 40 nM template-primer, 0.11–1.34 nM SF, 20 mM Tris (pH 8.0), 10 mM MgCl2, 1 mM DTT, 50 μg/mL BSA. Reactions were initiated by adding the DNA-SF mixture to an equal volume (5 μl) of a 2 × NTP stock solution, incubated at 50 °C, and quenched by the addition of 20 μl 95% formamide, 20 mM EDTA. The reaction mixture (5 μl) was analyzed by 15% PAGE containing 8 M urea. Data shown are the average of three independent determinations.
We also examined processive RNA synthesis. Each mutant was able to efficiently extend the DNA primer up to 6–8 ribonucleotides, before synthesis stopped (Fig. 2). The reaction conditions were typical of wild-type enzyme-mediated synthesis and did not contain Mn2+. Moreover, dG, dC, dA, and dT were each efficiently transcribed, as predicted by the single rNTP incorporation kinetic analysis. The transcription efficiency is demonstrated by the complete conversion of 23-mer primer to extended products within the shortest reaction times examined (minutes). Truncated synthesis after the incorporation of eight rNTPs may result from a requirement for a long template, or alternatively, from a conformational change in the hybrid duplex after the RNA chain grows to eight ribonucleotides in length.
Figure 2.
RNA synthesis by SFR3 mutant. The reactions contained 7 nM polymerase (wild type or SFR3 mutant) and 70 nM of primer/template duplex (see Table 3, X = dA), 200 μM each of rNTP, 10 mM Tris⋅HCl (pH 8.5), 5 mM MgCl2, 50 mM KCl, and 0.1% Triton X-100. The mixtures were incubated at 50°C, and sample aliquots were taken at 5, 15, and 60 min. The sequence corresponding to the incorporated ribonucleotides is shown on the right. The leftmost lane shows unextended primer.
Discussion
SF has significant structural and mechanistic similarities with T7 RNAP (21, 22). However, there is little sequence homology between the DNA and RNA polymerases, and both classes of enzyme show high selectivity for the correct nucleoside triphosphate, including high selectivity for the correct sugar structure. Typically, the DNA polymerases, including SF, favor the incorporation of dNTPs by 103- to 105-fold, relative to the corresponding rNTPs (13, 23). Analysis of sequence and structural data suggests that specific side chains may contribute to the specificity for the sugar ring by occupying the position required for the 2′-OH group of the incoming rNTP, thus favoring dNTP incorporation (13). For example, mutation of Klenow fragment residue Glu-710 to Ala increased the efficiency (kcat/Kd) for rCTP and UTP incorporation by 23- and 1,200-fold, respectively (12). The corresponding residues in the reverse transcriptases from HIV-1 and Moloney murine leukemia virus are thought to play a similar role in substrate discrimination (24). Despite the more efficient incorporation of single rNTPs, these mutations do not allow for processive RNA synthesis, indicating that additional, less apparent factors control the extension of RNA/DNA hybrid primers. Mutating a DNA polymerase into a true, processive RNAP must involve additional, and potentially more subtle, mutations than those required for efficient single rNTP incorporation.
Loeb and coworkers (11, 23) have used a combination of selection and screening to identify interesting members of polymerase libraries. Taq polymerase libraries consisting of 104-105 mutants were generated and preselected for wild-type activity by genetic complementation of E. coli with a temperature-sensitive host polymerase I. Mutants that supported bacterial growth at the nonpermissive temperature were isolated and then screened for interesting activity, including altered fidelity and RNAP activity. Libraries focused to motif A (residues 605–617) or the O-helix (residues 659–671) of Taq polymerase were generated. From the O-helix library, Suzuki et al. (11) isolated and characterized a mutant, A661E, that increased the efficiency of rATP incorporation by 30-fold, but at the expense of DNA synthesis fidelity. One mutant from the motif A library, I614K, had an increased efficiency of rNTP incorporation (200- to 500-fold) compared with wild-type Taq (23). However, the enzyme still preferentially inserted dNTPs by 20- to 4,000-fold. Incorporation of UTP was especially problematic and multiple rNTPs were only incorporated in the absence of dA in the template, and only then under error-prone conditions (i.e., Mn2+ and high concentrations of mutant enzymes were used). Patel and Loeb (23) have also reported that the mutation of the corresponding mutant in KF to Phe results in a polymerase that can synthesize full-length RNA. However, the reported steady-state data demonstrated that the mutant still preferred dNTP substrates by 1–3 orders of magnitude, and under the conditions used the wild-type DNA polymerase could also synthesize RNA, although less processively (25). The low efficiency of RNA synthesis, relative to DNA synthesis, in these previously isolated mutants, was likely a result of the absence of selection pressure for the desired activity: despite the preselection for natural DNA polymerase activity, the mutants were identified by individually screening each mutant for RNAP activity. In contrast, the phage-based activity selection used herein applies selection pressure directly for the desired activity. The selection conditions are fulfilled only by those polymerase mutants that efficiently incorporate multiple rNTPs, which is a prerequisite for the incorporation of the biotinylated UTP.
The three mutants isolated from the phage display-based activity selection all can efficiently insert each rNTP. In fact, each mutant can synthesize RNA virtually as efficiently as the wild-type DNA polymerase can synthesize DNA (Table 3). This remarkably increased efficiency (kcat/KM for rNTP insertion increased up to 104-fold) likely resulted from both the application of selection pressure directly for RNAP activity, as well as the large library used in the selection. Moreover, each rNTP is inserted with similarly efficient rates (within a factor of 10). Although full-length transcripts were not detected (primers were extended by the addition of only up to eight rNTPs), good processivity is demonstrated by the full conversion of primer to elongated products in a reasonable amount of time and under normal reaction conditions (Fig. 2). It is interesting to note that the selection pressure used in the evolution experiments was completely satisfied by the incorporation of four rNTPs: each evolved mutant was sufficiently processive to allow for its isolation. It is also interesting to note that the synthesis of short RNAs is very common during DNA-directed RNA synthesis. For example, primase synthesizes RNA primers of up to 12 ribonucleotides in length for lagging strand DNA synthesis (26). The primary product of wild-type RNAP-mediated transcription is a short transcript of no more than nine ribonucleotides in length (27). Even during processive full-length RNA transcription, the RNA remains hybridized to the DNA over only 3 bp (28). These examples imply that the synthesis of RNA using DNA as a template may contain an intrinsic barrier to extension past approximately 8 nt of RNA/DNA hybrid duplex. The barrier may include a hybrid duplex conformational change as the length of the RNA is increased past approximately 8 nt. Whatever the origin of the compromised processivity in the mutants, the evolution of a truly processive RNAP may require directly selecting for this particular activity.
Interestingly, all of the mutations, except E531K in SFR1, were from one focused library, corresponding to the 19-aa region of the polymerase metal binding site. (Glu-531 was not included in the mutagenic primers and mutation at this site must have occurred spontaneously.) Mutations in this region of the protein were not over-represented before selection as confirmed by sequencing 48 clones from the initial library. The mutations were found at nine different sites. Five sites were mutated only once, and four of these were present in SFR1: K531E, A600T, A608S, and L609V. One unique mutation, L605Q, was present in SFR3. Leu-605, Ala-608, and Leu-609 all are known to be highly mutable to both hydrophobic or hydrophilic residues without loss of natural activity in Taq (29).
Two sites were mutated in two of the three isolated proteins. Trp-604, which is not conserved in other type I polymerases (the corresponding residue is tyrosine in KF), was mutated to either Gly (SFR1) or Arg (SFR3). A second mutation, I614T, was present in both SFR1 and SFR3. Ile-614 is known to be highly mutable without loss of wild-type activity (25, 29). Previously, mutation of Ile-614 to Lys was found to increase the efficiency of rNTP incorporation (200- to 600-fold) compared with wild-type Taq (23) but also to decrease fidelity of DNA synthesis (30). These studies led to the hypothesis that substitutions at Ile-614, especially hydrophobic substitutions, lead to a more “open” active site that is more accommodating to nucleotides with altered structures (30).
Two mutations, A597T and E615G, were conserved among all three of the selected polymerases. Both Ala-597 and Glu-615 are conserved among type I polymerases and Glu-615 was not highly mutable in vivo (25, 29). It has been suggested by Gao and Goff (24) and Joyce (13) that Glu at this position acts to prevent the incorporation of rNTP substrates by occupying the space required for the 2′-OH group. Mutation of Glu-615 to smaller residues was found to facilitate rNTP incorporation. Presteady-state kinetics (12) showed that in KF, the corresponding mutation to Ala resulted in a polymerase that incorporates CTP only 4-fold slower than dCTP; however, steady-state kinetics (25) showed a stronger preference for the dNTP substrates (700 to 20,000). Nonetheless, the mutation of Glu-615 to Gly in each selected mutant is consistent with the steric exclusion model.
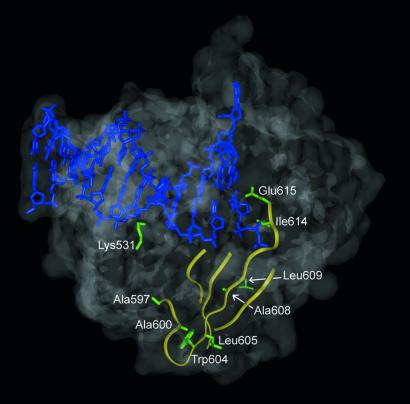
Structurally, the mutations in residues 596–615 occur in or near the third strand of the four-stranded anti-parallel β sheet of the “palm” region of the polymerase (Fig. 3) (19). This strand is of obvious functional relevance as it contains Asp-610, which chelates one of the two catalytic Mg2+ ions. The strand forms part of the floor and wall of the binding site proximal to the primer terminus and incoming triphosphate. Surprisingly, Ala-597 is removed from the substrate by at least 14 Å. Nonetheless, the conservation of the A597T mutation in each selected mutant implies that Thr-597 is required to fulfill the selection criteria. Residues Ile-614 and Glu-615, at the C terminus of the strand, pack on the sugar ring of the incoming triphosphate and are located near the N terminus of the O-helix, which is also known to make functionally important contacts with the triphosphate substrate (23). The isolation of mutations along the full length of this strand implies that higher-order interactions between the strand and the duplex or other parts of the protein may be important.
Figure 3.
Structure of ternary complex of Taq DNA polymerase with bound primer/template DNA and deoxycytidine triphosphate. The DNA is colored blue. The trace of the four stands of the palm-domain β-sheet is shown in yellow. The residues where mutations were identified are labeled and the corresponding side chains (wild-type sequence) are illustrated in green.
In summary, the mutants isolated from the activity-based selection system fulfill the selection criteria by efficiently inserting each rNTP. In fact, SFR1, SFR2, and SFR3 are the most efficient unnatural RNAPs isolated to date, significantly exceeding the activity of other known mutants. The kinetics of each step of SFR1–SFR3-mediated RNA synthesis need to be analyzed further to fully evaluate the efficiency and fidelity of each enzyme. Proteins containing the individual mutations must be prepared to determine the role that each selected mutation plays in the evolved RNAP activity. The beneficial mutations can then be used as a framework or “new wild-type sequence” upon which to construct new libraries to subject to further selection such as increased processivity.
Supplementary Material
Acknowledgments
We thank Philip Dawson for aid with peptide synthesis. Funding was provided by the National Institutes of Health (Grant GM 60005 to F.E.R.) and the Skaggs Institute for Chemical Biology (to F.E.R. and P.G.S.).
Abbreviations
- SF
Stoffel fragment
- RNAP
RNA polymerase
- rNTP
ribonucleoside triphosphate
- pol/phage
polymerase phage
- YT
yeast extract:tryptone
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Oliphant A R, Nussbaum A L, Struhl K. Gene. 1986;44:177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- 2.Hutchison C A, 3rd, Swanstrom R, Loeb D D. Methods Enzymol. 1991;202:356–390. doi: 10.1016/0076-6879(91)02019-6. [DOI] [PubMed] [Google Scholar]
- 3.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell R C, Joyce G F. PCR Methods Appl. 1994;3:S136–S140. doi: 10.1101/gr.3.6.s136. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Giver L, Shao Z, Affholter J A, Arnold F H. Nat Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 6.Stemmer W P. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 7.Stemmer W P. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen H, Holder S, Sutherlin D P, Schwitter U, King D S, Schultz P G. Proc Natl Acad Sci USA. 1998;95:10523–10528. doi: 10.1073/pnas.95.18.10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clackson T, Hoogenboom H R, Griffiths A D, Winter G. Nature (London) 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 10.Roberts R W, Szostak J W. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki M, Avicola A K, Hood L, Loeb L A. J Biol Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- 12.Astatke M, Ng K, Grindley N D, Joyce C M. Proc Natl Acad Sci USA. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce C M. Proc Natl Acad Sci USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths A D, Tawfik D S. Curr Opin Biotechnol. 2000;11:338–353. doi: 10.1016/s0958-1669(00)00109-9. [DOI] [PubMed] [Google Scholar]
- 15.Ghadessy F J, Ong J L, Holliger P. Proc Natl Acad Sci USA. 2001;98:4552–4557. doi: 10.1073/pnas.071052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jestin J-L, Kristensen P, Winter G. Angew Chem Int Ed. 1999;38:1124–1127. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1124::AID-ANIE1124>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 17.Kay B K, Winter J, McCafferty J. Phage Display of Peptides and Proteins: A Laboratory Manual. San Diego: Academic; 1996. [Google Scholar]
- 18.Schnolzer M, Alewood P, Jones A, Alewood D, Kent S B. Int J Pept Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Korolev S, Waksman G. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Tae E, Wu Y, Xia G, Schultz P G, Romesberg F E. J Am Chem Soc. 2001;123:7439–7440. doi: 10.1021/ja010731e. [DOI] [PubMed] [Google Scholar]
- 21.Sousa R. Trends Biochem Sci. 1996;21:186–190. [PubMed] [Google Scholar]
- 22.Arnold E, Ding J, Hughes S H, Hostomsky Z. Curr Opin Struct Biol. 1995;5:27–38. doi: 10.1016/0959-440x(95)80006-m. [DOI] [PubMed] [Google Scholar]
- 23.Patel P H, Loeb L A. J Biol Chem. 2000;275:40266–40272. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- 24.Gao G, Goff S P. J Virol. 1998;72:5905–5911. doi: 10.1128/jvi.72.7.5905-5911.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinkai A, Patel P H, Loeb L A. J Biol Chem. 2001;276:18836–18842. doi: 10.1074/jbc.M011472200. [DOI] [PubMed] [Google Scholar]
- 26.Rowen L, Kornberg A. J Biol Chem. 1978;253:770–774. [PubMed] [Google Scholar]
- 27.Reznikoff W S, Siegele D A, Cowing D W, Gross C A. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- 28.Rice G A, Kane C M, Chamberlin M J. Proc Natl Acad Sci USA. 1991;88:4245–4249. doi: 10.1073/pnas.88.10.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel P H, Loeb L A. Proc Natl Acad Sci USA. 2000;97:5095–5100. doi: 10.1073/pnas.97.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel P H, Kawate H, Adman E, Ashbach M, Loeb L A. J Biol Chem. 2001;276:5044–5051. doi: 10.1074/jbc.M008701200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.