Abstract
N-ethylmaleimide (NEM) modification of a lactose permease mutant containing a single-Cys in place of Ala-122 (helix IV) abolishes active lactose transport. Moreover, lactose, melibiose, and β,d-galactopyranosyl 1-thio-β,d-galactopyranoside protect against NEM inactivation of lactose transport and/or alkylation of Cys-122 by [14C]NEM. Remarkably, however, d-galactose transport is relatively unaffected by NEM, and the monosaccharide affords no protection against NEM inactivation of lactose transport. Consistently, competitive inhibition of [14C]galactose transport by lactose, melibiose, or β,d-galactopyranosyl 1-thio-β,d-galactopyranoside is drastically reduced after NEM modification, whereas inhibition by unlabeled galactose is unaffected. The results indicate that alkylation of Cys-122 selectively inhibits binding and transport of disaccharides, whereas transport of the monosaccharide galactose remains largely unaffected. In addition, although the conservative mutation Ala-122 → Ser causes only mild inhibition of lactose transport, the mutations Ala-122 → Phe and Ala-122 → Tyr lead to marked inhibition. In contradistinction, none of these replacements has a marked effect on galactose transport. The results demonstrate that Ala-122 is a component of the ligand-binding site and provide a strong indication that the side chain at position 122 abuts on the nongalactosyl moiety of d-galactopyranosides. This is in contrast to Cys-148, a neighboring residue in helix V, that interacts with the hydrophobic face of the galactosyl moiety of d-galactopyranosides.
The lactose permease (LacY) (1) of Escherichia coli is a paradigm for a major class of membrane transport proteins that transduce free energy stored in electrochemical ion gradients into solute concentration gradients (reviewed in refs. 1–4). LacY is a 12-transmembrane-helix bundle with the N and C termini on the cytoplasmic face of the membrane (5–7), and the 417-residue polypeptide has been solubilized, purified, reconstituted into proteoliposomes, and shown to be solely responsible for galactoside/H+ symport (reviewed in ref. 8). Several lines of evidence indicate that LacY is both functionally (9) and structurally a monomer (10–12). Analysis of an extensive library of mutants, particularly Cys replacement mutants (13), with a battery of site-directed biophysical and biochemical techniques has led to the formulation of a helix-packing model including tilts, as well as the working model for the mechanism of LacY (reviewed in ref. 14).
LacY is selective for disaccharides containing a D-galactopyranosyl ring, as well as D-galactose (15–17), but has no affinity for D-glucopyranosides or D-glucose (17, 18). The specificity of LacY is directed toward the galactosyl moiety of the substrate, and although the C-4 OH is clearly the major determinant for ligand binding, the C-2, C-3, and C-6 OH groups also participate in H-bonding interactions (17, 19).
Glu-126 and Arg-144, which are charge-paired, are irreplaceable determinants for substrate recognition and located at the interface between helices IV and V, respectively (20–23). Although Cys-148 (helix V) is not irreplaceable, it was the first residue shown to interact directly with substrate (18, 24–28). Alkylation with N-ethylmaleimide (NEM) abolishes lactose transport and p-nitrophenyl α,D-galactopyranoside binding, and substrates that include D-galactose and D-galactopyranosides afford protection against reaction with NEM and other thiol reagents (see ref. 14). Furthermore, replacement of Cys-148 with small hydrophobic residues (e.g., Ala, Val) increases apparent affinity for lactose (i.e., Km decreases), whereas hydrophilic replacements (e.g., Ser, Thr) increase Km. In addition, hydrophilic replacements decrease transport of D-galactose relative to galactopyranosides (26). The results indicate that Cys-148 interacts with the hydrophobic face of the galactopyranosyl moiety of the disaccharides or galactose.
Site-directed NEM labeling of single-Cys mutants in helices IV and V (29) reveals that mutant A122C (helix IV) exhibits properties similar to those of native Cys-148. Thus, NEM inactivates lactose transport and ligand affords protection against inactivation, as well as alkylation by NEM. Moreover, Ala-122 is located at about the same level in the membrane as Cys-148 (23). Therefore, it was suggested that Ala-122 may be a component of the substrate-binding site.
In this study, we determined the effect of site-directed chemical modification of single-Cys A122C LacY and site-directed mutagenesis of Ala-122 on transport activity and/or substrate-binding affinity of the monosaccharide D-galactose and various disaccharides. The results demonstrate clearly that alkylation of mutant A122C or replacement of A122 with bulky side chains selectively inactivates binding and transport of disaccharides with relatively little effect on D-galactose transport. It is concluded that Ala-122 is a component of the ligand-binding site and in close proximity to the nongalactosyl portion of D-galactopyranosides. Implications regarding the nature of the proposed binding site model (21) are discussed.
Materials and Methods
Materials.
[D-glucose 1-14C]lactose and [1-14C]galactose were purchased from American Radiolabeled Chemicals (St. Louis). N-([14C]ethyl)maleimide was purchased from Dupont NEN. Immobilized monomeric avidin was from Pierce. Sugars were obtained from Sigma.
Construction of LacY Mutants.
All mutants were constructed in a cassette devoid of Cys residues lacY gene encoding functional LacY (30) and containing a C-terminal biotin domain (CBD) (31). Plasmid pKR35/A122C/CBD with Ser in place of Cys-148 and plasmid pKR35/single-Cys-148/CBD were described (27, 29). Plasmid pKR35/A122C/C148A/CBD was generated by oligonucleotide-mediated, site-directed mutagenesis by using two-step PCR with pKR35/A122C/CBD as template. Plasmids pKR35/A122S/C148A/CBD, pKR35/A122F/C148A/CBD, and pKR35/A122Y/C148A/CBD were also constructed by two-step PCR with pKR35/A122C/C148A/CBD as template. All mutants were confirmed by DNA sequencing.
Growth of Cells.
E. coli T184 [lacI+O+Z−Y−(A) rpsL,met−,thr−,recA,hsdM,hsdR/F′,lacIqO+ZD118(Y+A+)] containing given mutants were grown in Luria–Bertani broth with 100 μg/ml of ampicillin. Overnight cultures were diluted 10-fold and allowed to grow for 1.5–2 h at 37°C before induction with 1 mM isopropyl-1-thio-β,D-galactopyranoside. After additional growth for 2–3 h at 37°C, cells were harvested by centrifugation.
Preparation of Right-Side-Out (RSO) Vesicles.
RSO membrane vesicles were prepared by osmotic lysis as described (32, 33), suspended in 100 mM potassium phosphate (KPi; pH 7.5)/10 mM MgSO4 at a protein concentration of about 10 mg/ml, frozen in liquid N2, and stored at −80°C until use.
Active Transport.
E. coli T184 containing given LacY mutants were washed with 100 mM KPi (pH 7.5)/10 mM MgSO4 and adjusted to an OD420 of 10.0 (0.7 mg protein/ml). Transport was assayed in the presence of [D-glucose 1-14C]lactose (10 mCi/mmol) or [1-14C]galactose (10 mCi/mmol) at a final concentration of 0.4 mM. When using vesicles or samples treated with NEM, transport was conducted in the presence of 20 mM ascorbate and 0.2 mM phenazine methosulfate under oxygen (34, 35). At given times, reactions were stopped with quenching buffer [100 mM KPi (pH 5.5)/100 mM LiCl/10 mM MgSO4] and assayed by rapid filtration and liquid scintillation spectrometry (35).
NEM Inhibition and Substrate Protection.
E. coli T184 cells expressing a given single-Cys mutant at a protein concentration of 0.7 mg/ml were preincubated without or with substrates, and then treated with NEM at a final concentration of 2 mM at 24°C. Reactions (0.25 ml) were stopped with 10 mM DTT as indicated, and cells were diluted immediately with 1.75 ml of ice-cold 100 mM KPi (pH 7.5)/10 mM MgSO4 and centrifuged. The pellet was resuspended in 2.0 ml of the same buffer, centrifuged, and washed once more. Finally, the pellet was resuspended in 0.25 ml of the same buffer and kept on ice until use. Rates of transport were measured at 0, 10, 20, and 30 sec as described. Transport rates are expressed as a percentage of a control samples to which 10 mM DTT was added before NEM and then treated as described.
Effect of NEM on Substrate Inhibition of d-Galactose Transport.
RSO membrane vesicles at a protein concentration of 4 mg/ml were incubated without or with 2 mM NEM at 24°C for 20 min, and 10 mM DTT was added to terminate the reaction. A given sugar was added at an indicated concentration, keeping the total volume of the samples constant. [1-14C]Galactose was then added and the reactions were quenched at 0, 5, 10, and 15 sec. Transport rates in the presence of sugar are expressed as a percentage of the rate obtained in the absence of unlabeled sugar. Ki values were determined with the ORIGIN computer program (Microcal Software, Northanmpton, MA) by using nonlinear least-squares curve fitting to the following user-defined equation: Y = (1 − P1)/(1 + X/P2) + P1, where P1 is the residual transport after full inhibition and P2 is the Ki.
[14C]NEM Labeling.
Reactivity of single Cys-122 with [14C]NEM was determined in situ in the absence and presence of β,D-galactopyranosyl 1-thio-β,D-galactopyranoside (TDG) (27, 36).
Results
LacY with a Single Cys at Position 122.
LacY with single-Cys A122C, Ala in place of Cys-148, and a biotin acceptor domain at the C terminus (A122C/C148A/CBD) transports lactose or galactose at about 36% the rate of wild-type LacY/CBD (data not shown). Cells transformed with plasmid pT7–5 devoid of a lacY insert grown under identical conditions do not transport galactose. Thus, all galactose transport observed under the conditions described is solely due to LacY and not to another symporter.
NEM Inactivation and Protection by Substrate.
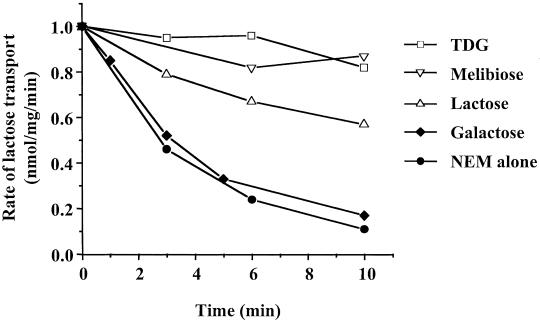
E. coli T184 expressing the single-Cys-122 mutant is almost fully inactivated by NEM with a half-time of about 3 min at 24°C (Fig. 1). When the cells are treated with NEM in the presence of a saturating concentration of various LacY substrates, essentially complete protection is observed with 10 mM TDG (Fig. 1) in confirmation of previous observations (29). Furthermore, melibiose at a concentration of 100 mM is almost as effective as 10 mM TDG, and lactose at a concentration of 500 mM also yields highly significant protection. In dramatic contrast, galactose at 500 mM elicits no protection whatsoever.
Figure 1.
NEM inactivation and substrate protection. E. coli T184 expressing mutant A122C/C148A/CBD at a protein concentration of 0.7 mg/ml were treated with 2 mM NEM in the absence or presence of 10 mM TDG; 100 mM melibiose, 500 mM lactose, or 500 mM galactose. Reactions were quenched with 10 mM DTT at indicated times, and the samples were washed and assayed for lactose transport as described in Materials and Methods.
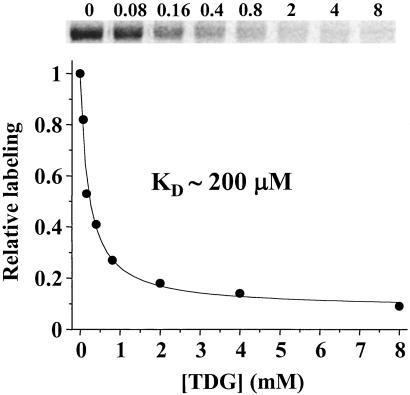
Protection by TDG against alkylation of Cys-122 by [14C]NEM was also determined quantitatively. Ligand exhibits essentially complete protection at sufficiently high concentrations with an Kd of about 200 μM (Fig. 2).
Figure 2.
Substrate protection against [14C]NEM labeling of single-Cys-122 LacY by TDG. RSO vesicles containing A122C/C148A/CBD were incubated with 100 mM KPi (pH 7.5) with 0.5 mM [14C]NEM for 10 min in the absence or presence of the given concentrations of TDG. Reactions were quenched with DTT, and biotinylated permease was solubilized in dodecyl β,D-maltopyranoside and purified by affinity chromatography on monomeric avidin. Samples were subjected to a NaDodSO4/12% polyacrylamide gel, and 14C-labeled protein was quantitated with a Storm 860 PhosphoImager. Labeling in the presence of a given concentration of sugar is expressed as percent labeling observed in the absence of the sugar.
Differential Effect of NEM on Lactose or Galactose Transport with a Single Cys at Position 122 or 148.
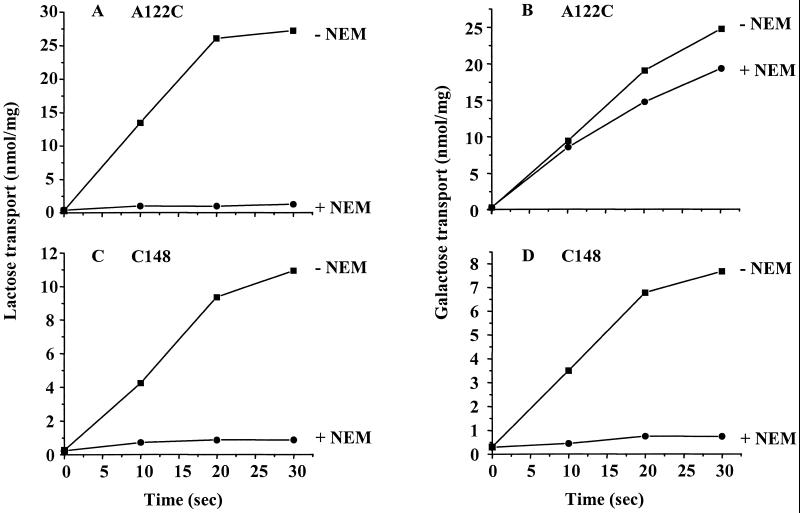
In vesicles containing LacY with single Cys-122, NEM completely inactivates lactose transport (Fig. 3A); however, alkylation of the mutant has minimal effects on galactose transport (Fig. 3B). In sharp contrast, when LacY contains a single Cys at position 148, both lactose (Fig. 3C) and galactose transport (Fig. 3D) are completely inactivated by NEM, which is consistent with the contention that Cys-148 interacts hydrophobically with the galactosyl moiety (see ref. 14). On the other hand, the nongalactosyl moiety of disaccharide substrates seems to be close to Ala-122.
Figure 3.
NEM inactivation of galactose or lactose transport in mutants with a single-Cys residue at position 122 or 148. RSO vesicles expressing A122C/C148A/CBD or Cys-148/CBD at a protein concentration of 4 mg/ml were incubated without or with 2 mM NEM and treated as described in Materials and Methods. Samples were then assayed for lactose or galactose transport as indicated and described in Materials and Method. (A) A122C, lactose; (B) A122C, galactose; (C) single-Cys-148, lactose; (D) single-Cys-148, galactose.
Effect of NEM on Disaccharide Inhibition of Galactose Transport.
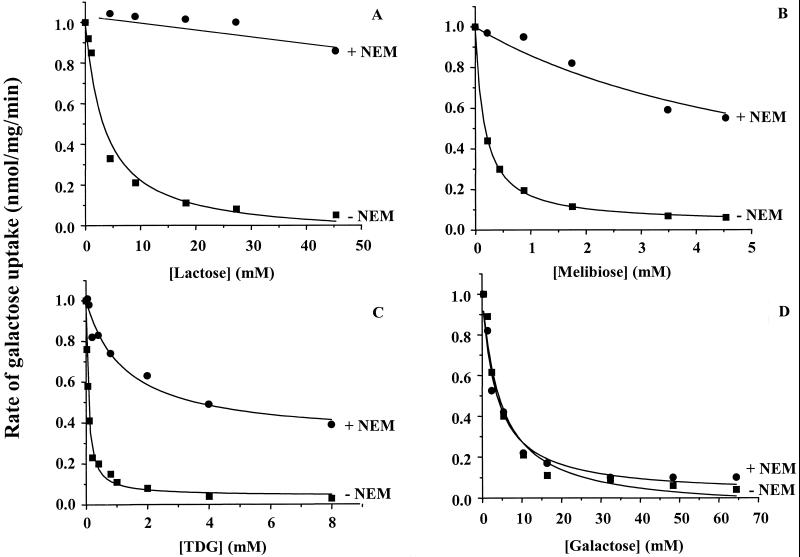
As shown in Fig. 4, with untreated RSO vesicles containing single-Cys-122 LacY, increasing concentrations of lactose (Fig. 4A), melibiose (Fig. 4B) or TDG (Fig. 4C) progressively inhibit the rate of galactose transport, exhibiting Ki values (the concentration at which half-maximal inhibition is observed) of ≈3.6 mM, 0.17 mM, or 0.06 mM, respectively. When the vesicles are treated with NEM, competitive inhibition of galactose transport by all three disaccharides is markedly diminished, and the Ki values increase to ≫50 mM, ≈10 mM, or ≈5 mM for lactose, melibiose, or TDG, respectively. Inhibition of [1-14C]galactose transport by unlabeled galactose is completely unaffected by alkylation with NEM (Fig. 4D), exhibiting a “Ki” of 4–5 mM.
Figure 4.
Inhibition of [1-14C]galactose transport by various sugars with mutant A122C. RSO vesicles with mutant A122C/C148A/CBD at a protein concentration of 4 mg/ml were incubated without or with 2 mM NEM for 20 min, the reaction was stopped with 10 mM DTT. A given sugar was then added, keeping the total volume of the samples constant, and initial rates of [1-14C]galactose transport were measured as described in Materials and Methods. Rates are expressed as a percentage of the rate obtained in the absence of substrate. (A) lactose; (B) melibiose; (C) TDG; (D) galactose.
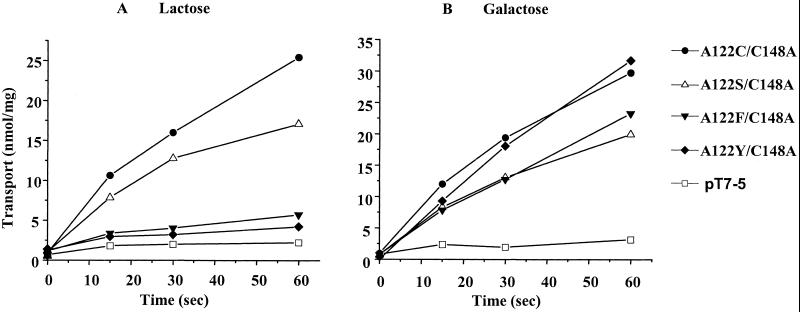
Ala-122 Replacements.
With mutant A122S, the rate of lactose transport is minimally affected (Fig. 5A). However, with mutant A122F or A122Y, lactose transport is almost abolished. In contradistinction, with galactose as substrate, Ser has essentially no effect, and replacement of Ala-122 with Phe or Tyr has only minor effects (Fig. 5B). Also, the Kd determined by TDG protection against alkylation of Cys-148 (37) increases from about 14 μM to ≈0.5 mM or 1.0 mM when Ala-122 is replaced with Val or Ile, respectively (data not shown). Thus, it seems that the bulk of the side chain at position 122 is important for disaccharides to bind rather than the polarity of the side chain. This conclusion is also supported by observations (not shown) demonstrating that treatment of single-Cys-122 LacY with methylmethanethiosulfonate, which forms a much less bulky adduct than NEM, causes only about 50% inhibition of lactose transport with no effect on galactose transport.
Figure 5.
Effect of given replacements for Ala-122 on rates of lactose (A) or galactose (B) transport. Indicated mutants were assayed for transport as described in Materials and Methods.
Discussion
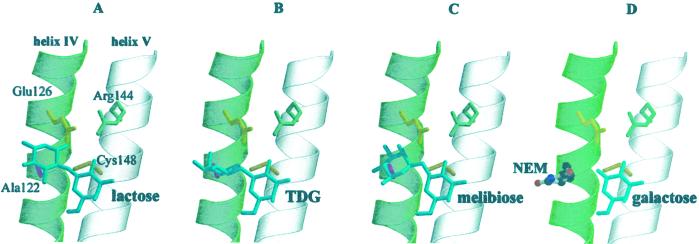
The findings presented here are readily explained by the structural models shown in Fig. 6 where the relevant portions of helices IV and V are shown with the Glu-126–Arg-144 charge pair, Ala-122 and Cys-148. With each disaccharide shown (Fig. 6 A, lactose; B, TDG; C, melibiose), the hydrophobic face of the galactosyl moiety interacts with Cys-148, whereas the nongalactosyl moiety is in close proximity to Ala-122. On the other hand, because galactose (Fig. 6D) lacks an anomeric substituent, only the hydrophobic interaction with the galactopyranosyl ring is operant, and increased bulk at position 122 (such as a Cys residue modified with NEM, as shown, or replacement of Ala-122 with Phe or Tyr) has little effect on binding or transport. Clearly, however, steric clash occurs with the nongalactosyl moiety of the disaccharides if a bulky adduct or side chain is present at position 122.
Figure 6.
Model for the binding site in LacY showing interactions of lactose (A), TDG (B), melibiose (C), or galactose (D). Also shown in D is the NEM adduct at position 122.
Although the results provide strong support for the contention that major determinants for substrate binding in LacY are located at the interface between helices IV and V, the findings also provide cause to question certain specific aspects of the model for the binding site suggested (21). A variety of experimental approaches indicates that a carboxyl group at position 126 (helix IV) and a guanidinium at position 144 (helix V) are irreplaceable with respect to substrate binding. Furthermore, strong evidence has been presented (reviewed in ref. 14) indicating that Glu-126 and Arg-144 form a salt bridge, which led to a model of the substrate-binding site with the following properties that are relevant to this discussion: (i) One of the guanidino-NH2 groups of Arg-144 was postulated to form an H bond with the OH group at the C-4 and/or C-3 position(s) of the galactosyl moiety of the substrate, an interaction thought to play a key role in substrate specificity. (ii) The other guanidino NH2 of Arg-144 was suggested to form a salt bridge with Glu-126, an interaction that would hold Arg-144 and Cys-148 in an orientation that allows specific interaction with the galactosyl moiety. (iii) One of the oxygen atoms of the carboxylate at position 126 could also act as an H bond acceptor from the C-6 OH of the galactosyl moiety.
In view of the orientation of the sugars in the models shown in Fig. 6, if the hydrophobic face of galactosyl moiety interacts with Cys-148 with the nongalactosyl moiety abutting Ala-122, the C-3, C-4, and C-6 OH groups are on the opposite side of the galactopyranoside ring from Arg-144 and Glu-126 and cannot therefore form the H bonds postulated. Although it is conceivable that the function of the salt bridge is to hold Cys-148 and Ala-122 in the correct position, thereby affecting ligand binding in an indirect fashion, R144K LacY seems to maintain the salt bridge, but substrate binding is strongly decreased (21, 37), whereas E126D LacY exhibits only a minor decrease in affinity (36). Possibly, Arg-144 H-bonds to the C-2 OH, but this position on the galactopyranosyl ring is relatively unimportant for affinity, and the orientation of the OH group at this position has essentially no effect on affinity (17).
Because the C-4 OH group is clearly the most important determinant for recognition (17, 19) and Arg-144 seems to be too distant to H bond with this position, a likely candidate for the role is Glu-269, which is located at the interface between helix VIII and helix V (14, 38, 39). Furthermore, evidence has been presented (21, 40–42) indicating that this residue is critical for substrate recognition. The only replacement for Glu-269 that exhibits binding or transport activity is Asp, and all other replacements completely inactivate LacY. Finally, studies (A. B. Weinglass, J. P. Whitelegge, K. F. Faull, and H.R.K., unpublished observations) with electrospray ionization mass spectrometry indicate that ligand binding blocks covalent modification of Glu-269 with carbodiimides.
Acknowledgments
We are indebted to Yonglin Hu for the computer graphics presented in Fig. 6. This work was supported in part by National Institutes of Health Grant DK51131:06.
Abbreviations
- LacY
lactose permease
- TDG
β,d-galactopyranosyl 1-thio-β,d-galactopyranoside
- NEM
N-ethylmaleimide
- KPi
potassium phosphate
- CBD
biotin acceptor domain at C terminus
- RSO
right side out
References
- 1.Kaback H R. J Cell Physiol. 1976;89:575–593. doi: 10.1002/jcp.1040890414. [DOI] [PubMed] [Google Scholar]
- 2.Kaback H R. J Membr Biol. 1983;76:95–112. doi: 10.1007/BF02000610. [DOI] [PubMed] [Google Scholar]
- 3.Poolman B, Konings W N. Biochim Biophys Acta. 1993;1183:5–39. doi: 10.1016/0005-2728(93)90003-x. [DOI] [PubMed] [Google Scholar]
- 4.Varela M F, Wilson T H. Biochim Biophys Acta. 1996;1276:21–34. doi: 10.1016/0005-2728(96)00030-8. [DOI] [PubMed] [Google Scholar]
- 5.Foster D L, Boublik M, Kaback H R. J Biol Chem. 1983;258:31–34. [PubMed] [Google Scholar]
- 6.Calamia J, Manoil C. Proc Natl Acad Sci USA. 1990;87:4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaback H R, Wu J. Q Rev Biophys. 1997;30:333–364. doi: 10.1017/s0033583597003387. [DOI] [PubMed] [Google Scholar]
- 8.Viitanen P, Garcia M L, Kaback H R. Proc Natl Acad Sci USA. 1984;81:1629–1633. doi: 10.1073/pnas.81.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahin-Tóth M, Lawrence M C, Kaback H R. Proc Natl Acad Sci USA. 1994;91:5421–5425. doi: 10.1073/pnas.91.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costello M J, Escaig J, Matsushita K, Viitanen P V, Menick D R, Kaback H R. J Biol Chem. 1987;262:17072–17082. [PubMed] [Google Scholar]
- 11.Sun J, Kaback H R. Biochemistry. 1997;36:11959–11951. doi: 10.1021/bi971172k. [DOI] [PubMed] [Google Scholar]
- 12.Guan L, Murphy F D, Kaback H R. Proc Natl Acad Sci USA. 2002;99:3475–3480. doi: 10.1073/pnas.052703699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frillingos S, Sahin-Tóth M, Wu J, Kaback H R. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 14.Kaback H R, Sahin-Tóth M, Weinglass A B. Nat Rev Mol Cell Biol. 2001;2:610–622. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 15.Sandermann H., Jr Eur J Biochem. 1977;80:507–515. doi: 10.1111/j.1432-1033.1977.tb11906.x. [DOI] [PubMed] [Google Scholar]
- 16.Olsen S G, Brooker R J. J Biol Chem. 1989;264:15982–15987. [PubMed] [Google Scholar]
- 17.Sahin-Tóth M, Akhoon K M, Runner J, Kaback H R. Biochemistry. 2000;39:5097–5103. doi: 10.1021/bi0000263. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Kaback H R. Biochemistry. 1994;33:12166–12171. doi: 10.1021/bi00206a020. [DOI] [PubMed] [Google Scholar]
- 19.Sahin-Tóth M, Lawrence M C, Nishio T, Kaback H R. Biochemistry. 2001;43:13015–13019. doi: 10.1021/bi011233l. [DOI] [PubMed] [Google Scholar]
- 20.Frillingos S, Gonzalez A, Kaback H R. Biochemistry. 1997;36:14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 21.Venkatesan P, Kaback H R. Proc Natl Acad Sci USA. 1998;95:9802–9807. doi: 10.1073/pnas.95.17.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, Zen K-C, Hubbell W, Kaback H R. Biochemistry. 1999;38:7407–7412. doi: 10.1021/bi9906524. [DOI] [PubMed] [Google Scholar]
- 23.Wolin C D, Kaback H R. Biochemistry. 2000;39:6130–6135. doi: 10.1021/bi0001269. [DOI] [PubMed] [Google Scholar]
- 24.Fox C F, Kennedy E P. Proc Natl Acad Sci USA. 1965;54:891–899. doi: 10.1073/pnas.54.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bieseler B, Prinz H, Beyreuther K. Ann NY Acad Sci. 1985;456:309–325. doi: 10.1111/j.1749-6632.1985.tb14882.x. [DOI] [PubMed] [Google Scholar]
- 26.Jung H, Jung K, Kaback H R. Biochemistry. 1994;33:12160–12165. doi: 10.1021/bi00206a019. [DOI] [PubMed] [Google Scholar]
- 27.Frillingos S, Kaback H R. Biochemistry. 1996;35:3950–3956. doi: 10.1021/bi952601m. [DOI] [PubMed] [Google Scholar]
- 28.le Coutre J, Whitelegge J P, Gross A, Turk E, Wright E M, Kaback H R, Faull K F. Biochemistry. 2000;39:4237–4242. doi: 10.1021/bi000150m. [DOI] [PubMed] [Google Scholar]
- 29.Kwaw I, Zen K-C, Hu Y, Kaback H R. Biochemistry. 2001;40:10491–10499. doi: 10.1021/bi010866x. [DOI] [PubMed] [Google Scholar]
- 30.van Iwaarden P R, Pastore J C, Konings W N, Kaback H R. Biochemistry. 1991;30:9595–9600. doi: 10.1021/bi00104a005. [DOI] [PubMed] [Google Scholar]
- 31.Consler T G, Persson B L, Jung H, Zen K H, Jung K, Prive G G, Verner G E, Kaback H R. Proc Natl Acad Sci USA. 1993;90:6934–6938. doi: 10.1073/pnas.90.15.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaback H R. Science. 1974;186:882–892. doi: 10.1126/science.186.4167.882. [DOI] [PubMed] [Google Scholar]
- 33.Short S A, White D C, Kaback H R. J Biol Chem. 1972;247:7452–7458. [PubMed] [Google Scholar]
- 34.Konings W N, Barnes E M, Jr, Kaback H R. J Biol Chem. 1971;246:5857–5861. [PubMed] [Google Scholar]
- 35.Kaback H R. Methods Enzymol. 1974;31:698–709. doi: 10.1016/0076-6879(74)31075-0. [DOI] [PubMed] [Google Scholar]
- 36.Sahin-Tóth M, le Coutre J, Kharabi D, le Maire G, Lee J C, Kaback H R. Biochemistry. 1999;38:813–819. doi: 10.1021/bi982200h. [DOI] [PubMed] [Google Scholar]
- 37.Sahin-Tóth M, Karlin A, Kaback H R. Proc Natl Acad Sci USA. 2000;97:10729–10732. doi: 10.1073/pnas.200351797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Perrin D, Sigman D, Kaback H. Proc Natl Acad Sci USA. 1995;92:9186–9190. doi: 10.1073/pnas.92.20.9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, Voss J, Hubbell W L, Kaback H R. Proc Natl Acad Sci USA. 1996;93:10123–10127. doi: 10.1073/pnas.93.19.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ujwal M L, Sahin-Tóth M, Persson B, Kaback H R. Mol Membr Biol. 1994;1:9–16. doi: 10.3109/09687689409161024. [DOI] [PubMed] [Google Scholar]
- 41.Franco P J, Brooker R J. J Biol Chem. 1994;269:7379–7386. [PubMed] [Google Scholar]
- 42.He M, Kaback H R. Biochemistry. 1997;36:13688–13692. doi: 10.1021/bi9715324. [DOI] [PubMed] [Google Scholar]