Abstract
CTCF, a conserved, ubiquitous, and highly versatile 11-zinc-finger factor involved in various aspects of gene regulation, forms methylation-sensitive insulators that regulate X chromosome inactivation and expression of imprinted genes. We document here the existence of a paralogous gene with the same exons encoding the 11-zinc-finger domain as mammalian CTCF genes and thus the same DNA-binding potential, but with distinct amino and carboxy termini. We named this gene BORIS for Brother of the Regulator of Imprinted Sites. BORIS is present only in the testis, and expressed in a mutually exclusive manner with CTCF during male germ cell development. We show here that erasure of methylation marks during male germ-line development is associated with dramatic up-regulation of BORIS and down-regulation of CTCF expression. Because BORIS bears the same DNA-binding domain that CTCF employs for recognition of methylation marks in soma, BORIS is a candidate protein for the elusive epigenetic reprogramming factor acting in the male germ line.
The ubiquitous, highly conserved, multivalent 11-zinc-finger (ZF) factor CTCF plays multiple roles in gene regulation, depending on the combinatorial utilization of different ZFs to bind varying CTCF target sites (1). CTCF–DNA complex formation can be regulated by CpG-specific DNA methylation (2, 3) if the involved CpG nucleotides are present within 50–60-bp-long CTCF target sequences precisely at DNA base positions required for recognition by the ZFs (2). Among the multitude of normal CTCF functions (1), its involvement in partitioning the genome into active or inactive domains by means of a chromatin insulator function (4) has received particular attention. This feature creates functionally autonomous gene cluster/expression domains by preventing unscheduled activation or silencing by neighboring regulatory elements. For example, the methylation-sensitive chromatin insulator within the imprinting control region (ICR) between IGF2 and H19 genes (5) regulates its interaction with CTCF in vivo (2), which likely results in maternal-specific repression of the IGF2 gene.
Strikingly, all vertebrate chromatin insulators identified so far interact with CTCF (1, 3, 6). Moreover, sites with proven or proposed potential to interact with CTCF include other epigenetic centers, such as the ICR within the Prader-Willi/Angelman syndrome locusm conserved regions within reciprocally imprinted Dlk1/Gtl2 genes (7, 8) and the Kcnq1 ICR (ref. 9; and G. Fitzpatrick, E.P., C.K., R.O., M. Higgins, and V.V.L., unpublished data). Finally, the choice/imprinting center of the XIST antisense gene, Tsix, contains several tandem CTCF-binding sites, which collectively function as CTCF-driven chromatin insulators (10).
Although the mechanism for reading of at least some imprinting marks in somatic cells may be explained by allele-specific interactions between CTCF and ICRs, there is as yet no explanation for the targeting mechanism by which the differentially methylated states of the CTCF-dependent ICRs are established de novo. By screening testes nuclear extracts for proteins binding to CTCF target sites, we identified and cloned a previously uncharacterized factor that shares all 11 ZFs with CTCF while being different in the regions flanking this DNA-binding domain. This CTCF-paralogue gene, termed Brother Of the Regulator of Imprinted Sites, or BORIS, may provide a conceptual framework for understanding epigenetic reprogramming events that go beyond the imprinting phenomenon.
Material and Methods
Nuclear Extracts (NE) and Electrophoretic Mobility Shift Assays (EMSA).
NE from tissues were prepared as described (11), and used in EMSA with DNA probes and antibodies as described (2, 12, 13).
PCR-Mediated Screening for CTCF-Like cDNA Sequences and Isolation of Human and Mouse BORIS cDNAs.
PCR-screening primers are listed in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. Following cloning and sequencing of PCR products obtained using the Marathon-Ready human testis cDNA (CLONTECH), one such fragment displayed a human cDNA sequence containing an ORF encoding CTCF-like ZFs. This was used to design the specific pairs of primers for PCR analyses of the Rapid Screen Arrayed human testis cDNA Library Panel (OriGene Technologies, Rockville, MD), as well as for the 5′ and 3′ rapid amplification of cDNA ends (RACE) with the Marathon-Ready testis cDNA and adaptor primers from the Marathon cDNA Amplification kit (CLONTECH). 5′ RACE was performed using the GeneRacer kit (Invitrogen). The mouse BORIS homologue was similarly amplified using the Marathon-Ready mouse testes cDNA library (CLONTECH).
RNA Isolation, Northern Blots, and Reverse Transcription (RT)-PCR.
Northern blots of isolated RNA (14) were probed with the NdeI–AccI fragment of the 5′ end of human CTCF cDNA clone p7.1 (14) and the XhoI–XhoI fragment of the BORIS cDNA (Fig. 2A). Expression patterns were determined by RT-PCR using human BORIS-specific primers (forward, 5′-caggccctacaagtgtaacgactgcaa-3′; reverse, 5′-gcattcgtaaggcttctcacctgagtg-3′) in parallel with amplification of human β-actin (primers from OriGene) as an internal control. Mouse BORIS-specific primers (forward, 5′-gagagacagacaagagagaagagaggttgctc-3′; reverse, 5′-cctgtgtgggtgttcacatggttcctaagaag-3′) were used together with the primers for mouse β-actin. For detection of CTCF mRNA, primers E5E6up (5′-tcgcaagtggacacccaaatc-3′) and E4E5down (5′-gaacccattcaggggaaaagc-3′) were used. For both CTCF and BORIS, the primers used for RT-PCR were chosen so as not to amplify genomic DNA.
Figure 2.
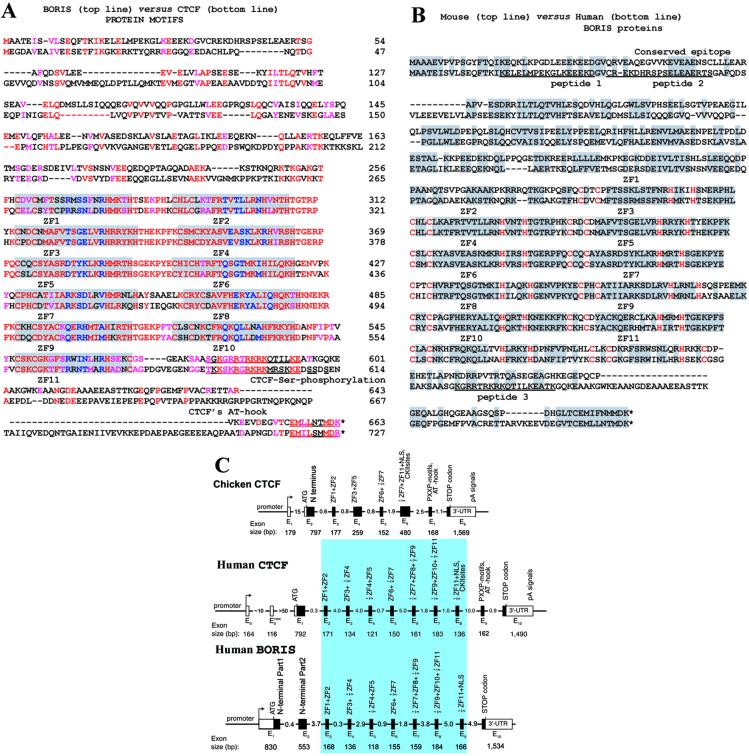
Human BORIS vs. CTCF and mouse BORIS amino acid sequences, and exon structure comparisons. (A) The best-fit alignment of the human CTCF and BORIS polypeptides produced by the GCG package of programs with zero-penalty for the gap extension. Identical and similar amino acids are shown by red letters; the near-identical residues in each ZF are highlighted by a gray background, and the Zn-coordinating C2 and H2 pairs (or the C2 and HC pair for the 11th ZF) of each ZF are shown by the outlined letters of a larger font. The critical base-recognizing residues at positions −1, 2, 3, and 6 within each ZF (reviewed in ref. 1) are shown by blue letters. Dashed lines show the gaps introduced to reveal minor similarities outside of the ZF domains. The major Ser-phosphorylation sites and the putative additional DNA-binding “AT-hook” motif (32), conserved in vertebrate CTCF proteins but absent in BORIS proteins, are indicated by outlined letters. The most likely shared epitopes in the C-terminal regions of both proteins, recognized by the anti-CTCF-C antibodies (for details, see discussion of Fig. 1 in Results) are underscored with red lines. (B) The optimal alignment of the mouse vs. human BORIS protein motifs. The identical and similar amino acids are shown by gray background, the other features are indicated as in the A. The sequences of the three peptides used to immunize chickens are underlined. (C) BORIS contains duplicated ZF-coding exons of mammalian, but not chicken, CTCF. Comparison of the overall exon–intron structures of CTCF and BORIS genes. The region of homology over mammalian exons encoding the 11 ZFs is shown by a blue box.
Expression of CTCF and BORIS in Pichia pastoris.
CTCF was purified as originally described (15) with modifications outlined recently (12). Expression of BORIS in yeast was accomplished using the Pichia Expression Kit (Invitrogen) according to the manufacturer's instructions, with chromatography steps similar to those described for CTCF (12).
Antibodies (Ab).
Rabbit antiserum against the bacterially produced N-terminal domain of CTCF (16) was produced as previously described for Ab against synthetic CTCF peptides (17). Chicken Ab against synthetic BORIS peptides (Fig. 2B) were produced by Aves Labs (Tigard, OR), purified, and characterized as described in Fig. 6, which is published as supporting information on the PNAS web site.
In Situ Analyses of mRNA, Protein, and Cytosine-Methylation.
Digoxygenin-11-dUTP or biotin-16-dUTP-labeled (Boehringer) 5′ end of the BORIS cDNA were hybridized to formaldehyde-fixed or frozen sections of mouse and human testis as described (18). For immunostaining, the slides were boiled for 10 min in 1× Citra (InnoGenex, San Ramon, CA), followed by incubation with affinity-purified chicken anti-BORIS, rabbit anti-CTCF, or sheep anti-5-methylcytosine (anti-5mC) Ab (Maine Biotechnology Services). The BORIS and CTCF epitopes were visualized by FITC-conjugated rabbit anti-chicken (Dako) and alkaline phosphatase (AP)-conjugated (using the 5-bromo-4-chloro-3-indolyl-phosphate substrate; Roche) goat anti-chicken or goat anti-rabbit (Promega) secondary Ab. For double-staining of chicken anti-BORIS and sheep anti-5mC Ab, a goat anti-chicken secondary Ab conjugated to biotin (Vector), with subsequent detection by avidin-Rhodamine (Vector), was used for BORIS detection, and rabbit anti-sheep Ab conjugated to FITC (Vector) for detection of 5mC.
Results
Detection of a CTCF-Like DNA-Binding Protein in Testes NE.
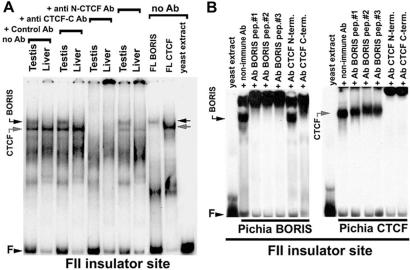
Using EMSA analyses of several well characterized CTCF-target sequences (1), we could document that testis NE produced a DNA–protein complex with a mobility slightly slower than that for the CTCF complex (Fig. 1A). This activity was not detected in NEs from a variety of somatic tissues from rats and mice (Fig. 1; data not shown). The binding activity in testis NEs could be competed with an excess of unlabeled DNA fragments bearing other CTCF targets, but not with the same fragments mutagenized at specific CTCF-contacting bases described previously (2, 3, 14, 17, 19). Fig. 1A also shows that the testis-specific factor could be “supershifted” in EMSAs with an excess of anti-CTCF-C Ab against the C-terminal region of CTCF downstream from the middle of the 11-th ZF. However, in contrast to DNA-bound CTCF, this testis-specific activity could not be supershifted by Ab against the N-terminal region upstream of the first ZF. These results suggested that in addition to CTCF, testis NE contained a different form of CTCF or a protein highly related to CTCF.
Figure 1.
EMSA analyses. (A) EMSA analyses of nuclear extracts (NEs) from rat liver versus testis tissues. no Ab, marked by downwards brackets, no antibodies were added to the EMSA reactions; anti CTCF-C Ab and anti CTCF-N, reactions contained affinity-purified antibodies against either the C-terminal CTCF region or the N-terminal CTCF region, respectively; Control Ab, equal amounts of the rabbit irrelevant polyclonal IgGs (in this case against GATA-1 protein). FL BORIS and FL CTCF, EMSA results with the full-length BORIS and CTCF proteins. Positions of protein-free FII DNA probe (F), and of the protein—CTCF- or BORIS-bound probe—are indicated by the arrowheads. (B) “Supershifting” EMSA with three anti-BORIS-peptide chicken antibodies and two anti-CTCF antibodies (marked at the top of the gel) and recombinant full-length (FL) CTCF and FL BORIS proteins produced in Pichia pastoris.
Cloning of Human cDNA Encoding BORIS.
Conserved regions within the vertebrate CTCF cDNAs were used to design a variety of primers for PCR amplification of CTCF-like sequence fragments from a human testis cDNA library that were then cloned and sequenced. Of more than 100 fragments, one PCR product contained a CTCF-like sequence encoding an amino acid sequence practically identical to that of ZF 4, 5, and 6 of CTCF. After additional PCR-mediated screening and 5′ rapid amplification of cDNA ends (RACE), we identified an ≈4-kb-long cDNA encoding a previously uncharacterized protein with all 11 CTCF-like ZFs (see Fig. 6). An alignment with CTCF sequence (Fig. 2A) revealed a remarkable similarity between the two 11-ZF domains, including most of the major DNA-base-recognition residues at positions −1, 2, 3, and 6 (1, 3) within each ZF. Referring to the CTCF function in reading imprinting marks in soma, we termed this previously unknown protein BORIS.
To verify that the cloned BORIS cDNA encodes the same CTCF-site-binding activity, and the same pattern of recognition by anti-CTCF Ab, as initially detected in testis NE by EMSA, the clone was used as a template for coupled in vitro transcription/translation and to produce a full-length recombinant BORIS in Pichia (12, 15). EMSA of the resulting full-length BORIS proteins demonstrated that recombinant BORIS forms a complex with the FII DNA, generating the same-mobility EMSA band as that produced by the endogenous BORIS from testis (Fig. 1A). Conversely, recombinant full-length CTCF produced the faster-migrating band present in NE from a variety of tissues (Fig. 1A; data not shown). DNA-bound recombinant BORIS interacted with an excess of anti-CTCF-C Ab, but not with anti-N-CTCF Ab (Fig. 1B Left), whereas DNA-bound recombinant CTCF was recognized by both of these Ab (Fig. 1B Right). The C-terminal regions of both proteins display two motifs of sufficient homology (shown underlined as shared epitope-CTCF-C in Fig. 2A), explaining the cross-reactivity of anti-CTCF-C Ab with BORIS. Fig. 1B also demonstrates that each of the three anti-BORIS chicken Ab, but not a control Ab, interacted specifically with DNA-bound recombinant BORIS, but not CTCF. These results prove that BORIS encoded by the human cDNA that we cloned corresponds to the endogenous testis-specific CTCF-target-binding activity, and that BORIS proteins are conserved in humans and rodents.
BORIS Is a Paralogue of CTCF.
We cloned the mouse BORIS counterpart with the entire ORF followed by the 3′-UTR with the poly(A)-tail (see Fig. 7, which is published as supporting information on the PNAS web site). The Bestfit alignment of mouse and human BORIS amino acid sequences (Fig. 2B) shows that, although all 11 ZFs are practically identical, the regions outside the ZFs are overall only similar but do contain several highly conserved motifs. Although CTCF is overall 93% conserved in avian and mammalian species (1), the regions of mouse and human BORIS and CTCF proteins outside of the “shared” 11 ZFs display no similarity to each other or to other known protein signature motifs.
A comparison of the overall genomic organization (Fig. 2C) and of the intron–exon junction sequences (see Table 1, which is published as supporting information on the PNAS web site) of avian (20), mouse, and human CTCF (13) with that of BORIS revealed striking similarities between BORIS and mammalian but not avian CTCF. This finding suggests that the CTCF and BORIS ZF exons have descended with divergence from a common ancestral gene. The idea that BORIS evolved through duplication and transposition events during mammalian radiation is further supported by our analyses of chromosome localizations of the two genes. A DNA fluorescence in situ hybridization (FISH) probe containing human BORIS exons revealed a single spot on chromosome segment 20q13.2 without any crosshybridization to other chromosomes (data not shown). This agrees perfectly with the Human Genome mapping data (http://genome.ucsc.edu/goldenPath/hgTracks.html), which placed contigs with BORIS exons (AL160176-AL035541) to the distal half of the chromosomal band 20q13.2. The conclusion that BORIS is a paralogue of CTCF is also supported by a “human genome paralogy” map (21) that relates a region flanking the BORIS-containing locus at 20q13 to the CTCF-containing region of chromosome 16q22 (ref. 30; http://u119.marseille.inserm.fr/Db/paralogy.html). A duplication event involving a larger chromosome region harboring several genes in addition to CTCF was likely to have occurred first, followed by further divergence of the initially duplicated region to yield BORIS with a testis-specific promoter (see below).
BORIS Expression Is Testis-Specific and Confined to CTCF-Negative Male Germ Cells Undergoing Genome Remethylation.
The pattern of BORIS expression was analyzed by Northern blots with poly(A)+RNA (Fig. 3A) and by RT-PCR with cDNAs from 24 human and 24 mouse tissues (Fig. 3 C and D). Internal controls were β-actin (Fig. 3 C Upper and D Upper) and CTCF (Fig. 3B; data not shown). These studies showed that expression of BORIS transcripts was testis-specific in both species and that even with the high sensitivity of RT-PCR, BORIS expression was below the limits of detection in mouse or human ovaries, and in tissues of 8.5-day to 19-day mouse embryos.
Figure 3.
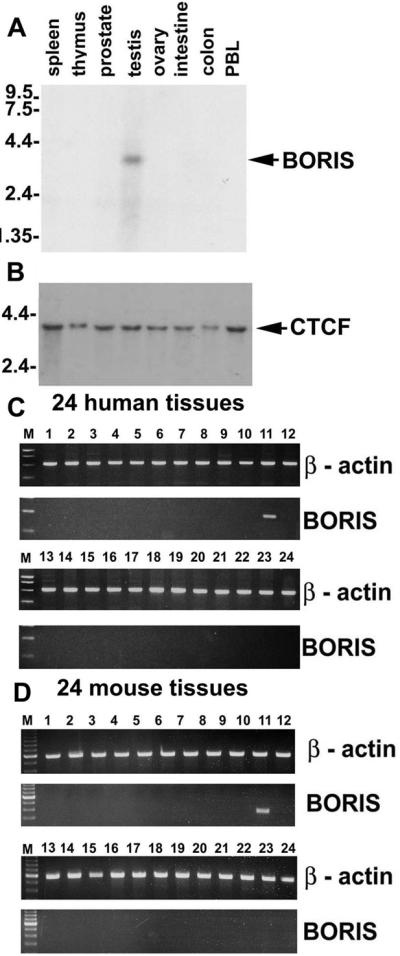
Normal expression of human and mouse BORIS mRNA is restricted to testis. (A) Northern blot containing 2 mg of poly(A)+RNA per lane from each tissue, indicated at the top of the panel, hybridized to a BORIS 5′-terminal probe. (B) The same blot probed with the human CTCF “no-ZF” probe. (C) Results of RT-PCR analyses performed using Rapid Scan Panel of cDNAs prepared from 24 human tissues including: 1, brain; 2, heart; 3, kidney; 4, spleen; 5, liver; 6, colon; 7, lung; 8, small intestine; 9, muscle; 10, stomach; 11, testis; 12, placenta; 13, salivary gland; 14, thyroid; 15, adrenal; 16, pancreas; 17, ovary; 18, uterus; 19, prostate; 20, skin; 21, PBL; 22, total bone marrow; 23, fetal brain; and 24, fetal liver. (D) The RT-PCR-mediated detection of mouse BORIS mRNA carried out with the Rapid Scan Panel of cDNAs from 24 mouse tissues including: 1, brain; 2, heart; 3, kidney; 4, spleen; 5, thymus; 6, liver; 7, stomach; 8, small intestine; 9, skeletal muscle; 10, lung; 11, testis; 12, skin; 13, adrenal gland; 14, ovary; 15, uterus; 16, prostate gland; 17, 8.5-day embryo; 18, 9.5-day embryo; 19, 12.5-day embryo/12.5; 20, 19-day embryo; 21, breast (virgin mice); 22, breast (pregnant mice); 23, lactating breast; and 24, involuting breast.
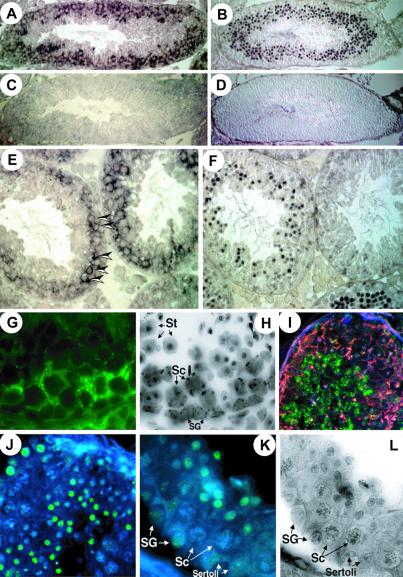
To identify BORIS-positive cells in testis, affinity-purified chicken Ab “ap-2-Ab” against the peptide 2, which contains a motif conserved in the mouse and human proteins (Fig. 2C), was generated. Western blot analyses showed that the ap-2-Ab bound specifically to the endogenous BORIS polypeptide in testis NE (see Fig. 8, which is published as supporting information on the PNAS web site). Visualization with alkaline phosphatase-conjugated secondary Abs showed that the BORIS- and CTCF-specific signals distribute to different populations of germ-line cells (Fig. 4 A, B, E, and F). The control slides showed only low background signals (Fig. 4 C and D). Whereas CTCF expression was detected in almost exclusively postmeiotic cells (round spermatids), expression of BORIS was restricted to primarily spermatocytes in adjacent sections (Fig. 4 E and F; see also Fig. 4 G and H). On these sections, most of the signal specific for the BORIS peptide-2 epitope appeared in cytoplasm rather than in nuclei (marked by arrowheads in Fig. 4E). Because DNA-binding activity of BORIS was initially detected in testis NE (Fig. 1A), we analyzed BORIS-positive cells for subcellular distribution of BORIS and CTCF by Western blotting of cytoplasmic and nuclear fractions obtained by the PEG-Dextran-Triton-extraction method that preserves natural nuclear–cytoplasmic partitioning patterns of fractionated proteins (described and illustrated in Fig. 8). These analyses indicated that approximately 10–30% of BORIS was present in nucleus with the rest in the cytoplasm. In contrast, CTCF was, as expected (9, 15) and shown for testis cells in Fig. 4F, found only in the nucleus.
Figure 4.
Mutually exclusive expression patterns of BORIS and CTCF correlate with epigenetic reprogramming in testis. (A and E) Immunostaining of thin (5-mm) sections of adult mouse testis with the affinity-purified anti-BORIS antibodies visualized by secondary alkaline phosphatase-conjugated rabbit anti-IgY antibodies. (B and F) Expression in adult mouse testis sections visualized using affinity-purified anti-N-CTCF antibodies and secondary alkaline phosphatase-conjugated antibodies. (C and D) Control staining obtained with normal chicken and rabbit serum, respectively. (G) BORIS-expressing cells in mouse tubule visualized by secondary FITC-labeled antibodies. (H) 4′,6-diamidino-2-phenylindole (DAPI)-inverted image of G. (I) Dual immunohistochemical staining by chicken ap-2-Ab anti-BORIS antibodies (Rhodamine fluorescence, red) and anti-5mC antibodies (FITC fluorescence, green). (J) Anti-5mC staining (FITC with DAPI counterstaining) at lower magnification. (K) Identification of the cell types with methylated DNA within the tubule, SG, Sertoli cells, and St. Note methylation free Sc. (L) An inverted DAPI image of K. (Magnification: A–D, 12-fold; E and F, 50-fold; G, H, K, and L, 106-fold; I and J, 44-fold.)
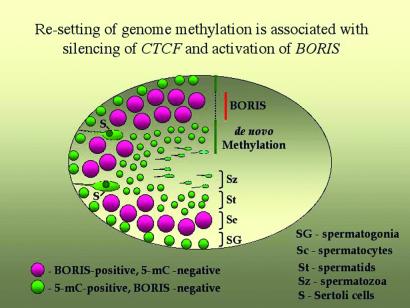
The same cell-type-specific pattern of BORIS expression as in mouse testes (Fig. 4) was also observed in human testis by in situ RT-PCR and by in situ hybridization with a “no-ZFs” cDNA probe (data not shown). Moreover, compared with mouse testes sections, the presence of nuclear-localized BORIS in spermatocytes was seen somewhat better in human samples (data not shown), perhaps because of a higher affinity of the ap-2-Ab for the region of human BORIS peptide-2 with amino acid sequence that is similar, but not identical, to the mouse sequence (Fig. 2B). Although dual immunostaining for CTCF and BORIS was not possible (because the same pretreatments that were necessary to reveal the BORIS epitope have also resulted in a severe loss of CTCF detection), it is clear that BORIS and CTCF expression patterns overlap very little, perhaps not at all. Moreover, because population of BORIS-positive cells includes primary spermatocytes (shown by the arrows in Fig. 4E), activation of BORIS transcription seems to be tightly linked with the final round of mitosis of the male germ-line cells. Fig. 5 summarizes the results as follows: BORIS is up-regulated in primary spermatocytes to become silenced on activation of CTCF in postmeiotic germ-line cells. Strikingly, this sequential up-regulation of BORIS (in CTCF-negative cells) and of CTCF (in BORIS-negative cells) takes place in association with erasure and re-establishment of methylation marks, respectively, as visualized by the Ab against 5mC (Fig. 4 I–L).
Figure 5.
Schematic representation of the overlapping patterns for BORIS-CTCF expression switch and for resetting of genome-wide DNA methylation during spermatogenesis.
Discussion
The major result of this work is the identification of a previously uncharacterized testis-specific gene that has the potential to reshape our thinking about site-specific epigenetic events in the male germ line. The gene has been named BORIS for Brother of the Regulator of Imprinted Sites. The sibling referred to is CTCF, the 11-ZF protein well known for its many roles in gene regulation (reviewed in ref. 1) including functional “reading” of allele-specific methylation marks (2, 10, 22, 23). BORIS shares an identical 11-ZF DNA-binding domain with CTCF as the basis for familial similarity while being completely divergent at the amino and carboxy termini. This finding alone indicates that nucleoprotein complexes generated by BORIS and CTCF bound to the same DNA sites are likely to have distinct functions. For the ZF-coding regions, we noted remarkable similarity in genomic sequence between BORIS and mammalian but not avian CTCF at each splice site, as well as similar overall arrangement of the ZF-coding exons. We therefore suggest that at some point of mammalian evolution, all of the ZF exons of CTCF were duplicated together with other surrounding regions and transposed to a different chromosome.
As a result of their stunning sameness in the ZF domain, BORIS and CTCF demonstrate currently indistinguishable DNA-binding specificity in vitro. In development, differentiation, and function of normal cells, this sharing of specificity cannot result in sibling rivalry because expression of BORIS is restricted to a select cell population in testis that is the only normal cell type known that does not express CTCF. Because inhibition of CTCF expression in cultured cells leads to apoptosis (S.V., D.I.L., E.P., H.M., H.C.M., and V.V.L., unpublished results), it is reasonable to assume that BORIS is activated to maintain some of the vital CTCF functions in these particular cells. Also, switching to BORIS may allow these cells to perform such specific functions that otherwise are inhibited in the presence of CTCF. With respect to the latter proposition, it is striking that the genome-wide erasure of methylation overlaps extensively with BORIS expression, and that remethylation of DNA in round spermatids is associated with the subsequent silencing of BORIS and reactivation of CTCF expression as depicted in Fig. 5. Thus, BORIS could be associated with demethylases that participate in the erasure of methylation marks.
It is also possible that the BORIS–CTCF switching is intimately linked with initiating (or regional targeting) de novo DNA methylation. In this case, reactivation of CTCF would serve to target de novo methylation to the paternal imprinting marks. Marking of chromatin for de novo DNA-methylation appears to involve histone H3 lysine methylases, such as the Suv39h1 and Suv39h2 (24). Because the expression pattern of testis-specific Suv39h (25) is remarkably similar to the pattern of BORIS–CTCF switching (Fig. 5), it would therefore be of great interest to test whether BORIS and CTCF may interact with the Suv39h histone H3 methyltransferases.
The finding of large amounts of BORIS protein in cytoplasm (Fig. 4), confirmed by Western blot analyses of sub fractionated cells (see Fig. 6), is in stark contrast to the strictly nuclear localization of CTCF. However, significant portion of DNA-binding BORIS is localized in nuclei as shown by EMSA with testes NE (Fig. 1A). Moreover, chromatin immunopurification (ChIP) experiments with cells from adult mouse testes showed that BORIS interacts in vivo with certain CTCF-target DNA sites (L. Liu, C.K., and R.O., unpublished observations). We propose therefore that the nuclear import of BORIS may have a regulatory role not shared with its sibling, CTCF; and that BORIS may be capable of selective in vivo occupation of CTCF sites in the maternally imprinted ICR rather than sites in the paternally imprinted ICRs. This important prediction is now being tested based on our knowledge of CTCF “footprints” within these two classes of ICRs exemplified by the Igf2/H19 (2) and Kcnq1 (ref. 9; G. Fitzpatrick, E.P., C.K., R.O., M. Higgins, and V.V.L., unpublished data) loci.
Finally, we showed here that human BORIS maps to 20q13.2. It has been shown that this chromosomal region is amplified in many cancers and may contain a dominant immortalizing or transforming gene(s) (26, 27). Our ongoing studies demonstrate that BORIS transcripts are abnormally activated in varying proportions of a wide variety of cancers (unpublished data). This defines BORIS as a previously unknown member of the cancer-testis (CT) gene family that comprises genes normally expressed only in testis but abnormally activated in different malignancies (28, 29). A conjunction of DNA-demethylation and CTCF-silencing in BORIS-positive cells that express other CT genes may indicate a common mechanism for transcriptional regulation of BORIS and other CT genes. However, BORIS is a unique member of the CT family because, unlike the others, it has a somatic counterpart with the same DNA-binding domain but with the properties of a tumor suppressor (13, 30, 31). Expression of BORIS in addition to CTCF, observed only in cancer cells, can set the stage for competition at IGF2/H19 ICR, and at some other CTCF target sites (1), resulting in cell growth deregulation (unpublished data). We suggest that BORIS and CTCF may act successively to govern epigenetic states in normal male germ cell development, while their rivalry caused by aberrant activation of BORIS in soma is associated with cancer. Therefore, characterization of mouse and human BORIS genes described here is likely to provide opportunities for understanding molecular mechanics of epigenetic reprogramming, both in normal development and in tumor genesis.
Supplementary Material
Acknowledgments
We thank Mrs. Anita Mattsson, Drs. Vladimir Dobrovitsky, David Lee, Zied Abdullaev, and Helena Malmikumpu for excellent technical assistance with preparing and analyzing human and mouse testis specimens; Mr. Hare Varnon, Mrs. Deborrah Curtis, and Dr. Nancy Wolford for the assistance with preparing this manuscript for publication; and Valentina Lobanenkova for great patience and encouragement during the course of the work presented here. This work was supported in part by National Institutes of Health Grants RO1 CA65145 and R37 CA54358 (to A.P.F.), NS30994 (to W.W.Q.), and HD-10793 (to M.K.), as well as grants from the Swedish Cancer Research Foundation and Swedish Pediatric Cancer Foundation (to R.O. and E.L.), the Association for International Cancer Research (to I.C. and E.M.K.), the UK Breast Cancer Campaign (to F.M.D. and E.M.K.), and the Deutsche Forschungsgemeinschaft (to R.R.).
Abbreviations
- ZF
zinc finger
- ICR
imprinting control region
- EMSA
electrophoretic mobility shift assay
- Ab
antibodies
- RT
reverse transcription
- NE
nuclear extracts
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. AF336042).
Ohta, T., Khadake, J. R., Rodriguez-Jato, S., Mione, C., Knepper, J. L., Svoboda, P., McCarrey, J. R., Schultz, R. M., Yang, T. P. & Nicholls, R. D. (2001) Am. J. Hum. Genet. 69, 52 (abstr.).
References
- 1.Ohlsson R, Renkawitz R, Lobanenkov V. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 2.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi C F, Wolffe A, Ohlsson R, Lobanenkov V V. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 3.Filippova G N, Thienes C P, Penn B H, Cho D H, Hu Y J, Moore J M, Klesert T R, Lobanenkov V V, Tapscott S J. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 4.Bell A C, West A G, Felsenfeld G. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 5.Holmgren C, Kanduri C, Dell G, Ward A, Mukhopadhya R, Kanduri M, Lobanenkov V, Ohlsson R. Curr Biol. 2001;11:1128–1130. doi: 10.1016/s0960-9822(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 6.West A G, Gaszner M, Felsenfeld G. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 7.Paulsen M, Takada S, Youngson N A, Benchaib M, Charlier C, Segers K, Georges M, Ferguson-Smith A C. Genome Res. 2001;11:2085–2094. doi: 10.1101/gr.206901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takada S, Paulsen M, Tevendale M, Tsai C E, Kelsey G, Cattanach B M, Ferguson-Smith A C. Hum Mol Genet. 2002;11:77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- 9. Kanduri, C., Fitzpatrick, G., Mukhopadhyay, R., Kanduri, M., Lobanenkov, V., Higgins, M. & Ohlsson, R. (2002) J. Biol. Chem., 10.1074/jbc.M200031200. [DOI] [PubMed]
- 10.Chao W, Huynh K D, Spencer R J, Davidow L S, Lee J T. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- 11.Lobanenkov V V, Nicolas R H, Adler V V, Paterson H, Klenova E M, Polotskaja A V, Goodwin G H. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 12.Vostrov A A, Taheny M J, Quitschke W W. J Biol Chem. 2002;277:1619–1627. doi: 10.1074/jbc.M109748200. [DOI] [PubMed] [Google Scholar]
- 13.Filippova G N, Qi C F, Ulmer J E, Moore J M, Ward M D, Hu Y J, Loukinov D I, Pugacheva E M, Klenova E M, Grundy P E, et al. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 14.Filippova G N, Fagerlie S, Klenova E M, Myers C, Dehner Y, Goodwin G, Neiman P E, Collins S J, Lobanenkov V V. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quitschke W W, Taheny M J, Fochtmann L J, Vostrov A A. Nucleic Acids Res. 2000;28:3370–3378. doi: 10.1093/nar/28.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klenova E M, Nicolas R H, U, S, Carne A F, Lee R E, Lobanenkov V V, Goodwin G H. Nucleic Acids Res. 1997;25:466–474. doi: 10.1093/nar/25.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klenova E M, Nicolas R H, Paterson H F, Carne A F, Heath C M, Goodwin G H, Neiman P E, Lobanenkov V V. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pack S D, Zbar B, Pak E, Ault D O, Humphrey J S, Pham T, Hurley K, Weil R J, Park W S, Kuzmin I, et al. Cancer Res. 1999;59:5560–5564. [PubMed] [Google Scholar]
- 19.Awad T A, Bigler J, Ulmer J E, Hu Y J, Moore J M, Lutz M, Neiman P E, Collins S J, Renkawitz R, Lobanenkov V V, Filippova G N. J Biol Chem. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- 20.Klenova E M, Fagerlie S, Filippova G N, Kretzner L, Goodwin G H, Loring G, Neiman P E, Lobanenkov V V. J Biol Chem. 1998;273:26571–26579. doi: 10.1074/jbc.273.41.26571. [DOI] [PubMed] [Google Scholar]
- 21.Popovici C, Leveugle M, Birnbaum D, Coulier F. Biochem Biophys Res Commun. 2001;288:362–370. doi: 10.1006/bbrc.2001.5794. [DOI] [PubMed] [Google Scholar]
- 22.Bell A C, Felsenfeld G. Nature (London) 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 23.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. Nature (London) 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 24.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 25.O'Carroll D, Scherthan H, Peters A H, Opravil S, Haynes A R, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, et al. Mol Cell Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanner M M, Tirkkonen M, Kallioniemi A, Collins C, Stokke T, Karhu R, Kowbel D, Shadravan F, Hintz M, Kuo W L, et al. Cancer Res. 1994;54:4257–4260. [PubMed] [Google Scholar]
- 27.Cuthill S, Agarwal P, Sarkar S, Savelieva E, Reznikoff C A. Genes Chromosomes Cancer. 1999;26:304–311. [PubMed] [Google Scholar]
- 28.Chen Y T, Gure A O, Tsang S, Stockert E, Jager E, Knuth A, Old L J. Proc Natl Acad Sci USA. 1998;95:6919–6923. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renkvist N, Castelli C, Robbins P F, Parmiani G. Cancer Immunol Immunother. 2001;50:3–15. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filippova G N, Lindblom A, Meincke L J, Klenova E M, Neiman P E, Collins S J, Doggett N A, Lobanenkov V V. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 31.Rasko J E, Klenova E M, Leon J, Filippova G N, Loukinov D I, Vatolin S, Robinson A F, Hu Y J, Ulmer J, Ward M D, et al. Cancer Res. 2001;61:6002–6007. [PubMed] [Google Scholar]
- 32.Aravind L, Landsman D. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.