Abstract
Numerous bacteria secrete low molecular weight compounds termed siderophores that have a high affinity for iron ions. Siderophores have a well-documented role as iron-scavenging chemicals, chelating iron ions in the environment whereupon the ferrisiderophores reenter the bacterial cells by means of specific cell-surface receptors. The iron is then released for incorporation into bacterial proteins. Here we show that in addition to its role as an iron-scavenger, the siderophore pyoverdine that is secreted by Pseudomonas aeruginosa regulates the production of at least three virulence factors (exotoxin A, an endoprotease, and pyoverdine itself), which are major contributors to the ability of this bacterium to cause disease. Regulation occurs through a transmembrane signaling system that includes an outer membrane receptor for ferripyoverdine, a signal-transducing protein that is predicted to extend from the periplasm into the cytoplasm, and a sigma factor. Expression of genes that form part of the regulon is triggered by pyoverdine so that this siderophore acts as a signaling molecule to control the production of secreted products. Recognition that a siderophore acts as a signaling molecule has important implications for the understanding of interactions between bacterial cells.
Like other organisms, bacteria require iron as a cofactor for redox-dependent enzymes. In most environments the level of soluble iron is too low for sufficient iron to be acquired by passive diffusion of ions into the cell (1). Bacteria have evolved a number of different strategies to combat this problem and one of the most common of these is the secretion of iron-chelating compounds termed siderophores (2). These chelate ferric ions in the environment and the ferrisiderophore complexes are taken up by the bacteria through specific cell-surface receptor proteins. The iron is then released from the ferrisiderophore complex for incorporation into cellular proteins. About 500 siderophores have been identified, and most bacterial genera contain siderophore producers (3). The biological importance of siderophores has been demonstrated for a number of species, for example, siderophore-deficient mutants of pathogenic bacteria are invariably less virulent in disease models (4, 5). At least some bacteria are also able to use exogenous siderophores that are present in the environment, with the presence of an iron chelate inducing synthesis of the cognate receptor. In the best-characterized system, that of ferric-citrate uptake in Escherichia coli, binding of ferric citrate to outer membrane receptor protein FecA initiates a signal-transduction pathway (reviewed in refs. 6 and 7). FecA interacts with the periplasmic C terminus of a transmembrane protein, FecR, and this results in transduction of a signal to the cytoplasmic N-terminal part of FecR. FecR is an anti-sigma factor that controls the activity of a sigma factor protein, FecI, which directs transcription of genes required for transport of ferric citrate. Consequently, expression of these genes is up-regulated in response to the presence of environmental ferric citrate. A similar three-component system (Pup) in Pseudomonas putida WCS358 enables production of a ferrisiderophore receptor (PupB) in response to the presence of the cognate siderophores pseudobactin BN7 and pseudobactin BN8, which are produced by other strains of P. putida although not by strain WCS358 itself (reviewed in ref. 6). As well as up-regulating genes required for their transport, some siderophores have been reported to cause increased production of enzymes that are required for their synthesis (8–12).
Pseudomonas aeruginosa is an opportunistic pathogen that infects people with a range of predisposing conditions, such as cystic fibrosis or severe burns, or who are immunocompromised in some way (13). P. aeruginosa secretes two siderophores, pyoverdine and pyochelin, and mutants that are unable to synthesize pyoverdine have a greatly reduced ability to cause disease in animal models (14, 15). Expression of genes required for pyoverdine synthesis is directed by an alternative sigma factor, PvdS (16, 17). Expression of the pvdS gene is regulated by the iron-sensing repressor protein Fur so that PvdS (and hence pyoverdine) is only synthesized by the bacteria under conditions of iron limitation (18, 19).
P. aeruginosa also secretes a large number of proteins and many of these are virulence factors that play a crucial role in host–pathogen interactions (20). The production of virulence factors by P. aeruginosa and other species is controlled in part by cell-to-cell signaling that is mediated by quorum-sensing molecules, N-acyl-homoserine lactones (AHLs), which are released by the bacterial cells (21, 22). When the density of bacteria, and hence of the AHLs, is low they have minimal effect on gene expression. When the cell density increases and the concentration of AHLs reaches a threshold level, transcriptional activators trigger the expression of genes that influence the interactions of the bacteria with eukaryotic host organisms. Quorum-sensing molecules provide a means for intercellular communication between bacterial cells and within communities of bacteria.
Although AHLs are probably the most commonly occurring molecules known to mediate intercellular bacterial signaling, other signaling molecules have been identified (23). Here we show that pyoverdine, secreted by P. aeruginosa, controls production of at least three virulence factors—an exotoxin, an endoprotease, and pyoverdine itself—required by the bacteria to cause disease.
Materials and Methods
Growth of Bacteria.
Bacteria were grown at 37°C by using L-agar and L-broth or King's B agar and broth (24) for analysis of pyoverdine production, or at 32°C in D-TSB broth (25) for analysis of expression of the toxA and prpL genes. Antibiotics and the iron-chelating compound ethylenediamine-di(o-hydroxy-phenylacetic acid) (EDDA) were added at the same concentrations as described (18, 25). Pyoverdine, purified from P. aeruginosa PAO as described (26), was added as required to a final concentration of 45 μM unless otherwise stated.
DNA Analysis and Manipulations.
Database searches were conducted at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) with blast algorithms (27). The predicted sequence of FpvR (PA2388) was obtained from the Pseudomonas Genome Project web site (www.pseudomonas.com) (28). It was aligned with those of FecR and PupR with eclustalw (29) and was analyzed for hydrophobicity and likely membrane-spanning regions with tmhmm and tmpred (http://au.expasy.org/tools/#transmem). Levels of similarity between proteins were calculated with gap (30).
Chromosomal DNA was isolated from P. aeruginosa PAO1 by using the method of Chen and Kuo (31). DNA manipulations were performed by using standard methods (32) and as recommended by manufacturers of enzymes and kits. The fpvR gene and DNA fragments spanning the promoters of the pvdS, prpL, and toxA genes were amplified from P. aeruginosa PAO1 genomic DNA by PCR with appropriate primers; details are available on request. The fpvR gene was cloned into pUCP22 (33) to generate plasmid pUCP22∷fpvR. The 590-bp pvdS promoter fragment was cloned into plasmid pMP190 (34) to give pMP190∷PpvdS, which contains a transcriptional pvdS-lacZ fusion. The 524-bp toxA promoter fragment was cloned into the promoter probe vector pPZ20 (35), giving pPZ-toxA that contains an in-frame translational fusion of codon 8 of the toxA gene to the promoterless lacZ gene. Plasmid pPZ-prpL contains a 284-bp DNA fragment cloned into pPZTC, which is a transcriptional fusion version of pPZ30 (35) and carries a transcriptional fusion of the prpL promoter to the lacZ gene. All plasmid constructs were confirmed by DNA sequencing. P. aeruginosa strains were genetically transformed with plasmid constructs by using the magnesium chloride/heat-shock method as described (25).
Construction of fpvR, fpvA, and fpvR fpvA Mutant Strains.
A 1.4-kb kanamycin-resistance cassette in pNRE1 (36) was cloned into the unique BglII site within the fpvR gene. The resulting fpvR∷kan construct was cloned into pSUP202 (37) and this suicide plasmid was introduced into P. aeruginosa PAO1 by conjugation from E. coli strain S17–1 (37). Transconjugants in which the chromosomal fpvR gene had been replaced by the fpvR∷kan allele were identified as described (18). The mutant strains grew at similar rates to P. aeruginosa PAO1.
Plasmid pEXGm∷fpvA was constructed by cloning an internal 828-bp EcoRI/SalI fragment of fpvA into pEX18Gm (38). pEXGm∷fpvA was transferred into P. aeruginosa by conjugation, resulting in insertional inactivation of fpvA. The presence of the intended mutations in all engineered strains was confirmed by Southern blotting, and in some cases by PCR or Western blotting.
Promoter Activity Assays.
Activities of the pvdE and pvdS promoters were determined through β-galactosidase production with the reporter plasmids pMP190∷PpvdE (39) and pMP190∷PpvdS. Overnight cultures (18-h incubations) were diluted in King's B broth to an approximate OD600 of 0.1, and pyoverdine (60 μM) was added if required. Bacteria were then incubated for 6.5 h until the OD600 was ≈2.0, and β-galactosidase assays were performed according to the method of Miller (40).
Activities of the toxA and prpL promoters were determined similarly with pPZ-toxA and pPZ-prpL except that bacteria were grown in D-TSB for 6 h at 32°C, pyoverdine was added as required, and the cultures were incubated for a further 18 h before β-galactosidase assays were carried out.
Measurement of Secreted Products.
The amount of pyoverdine produced by strains of P. aeruginosa was determined as described (41) except that bacteria were incubated in King's B broth for 6.5 h (OD600 of ≈2.0). The amounts of exotoxin A in culture supernatants were estimated by carrying out serial dilutions of supernatants followed by Western blotting with an antiexotoxin antibody (25).
Results
Pyoverdine Regulates Its Own Production and Production of Exotoxin A and PrpL Protease.
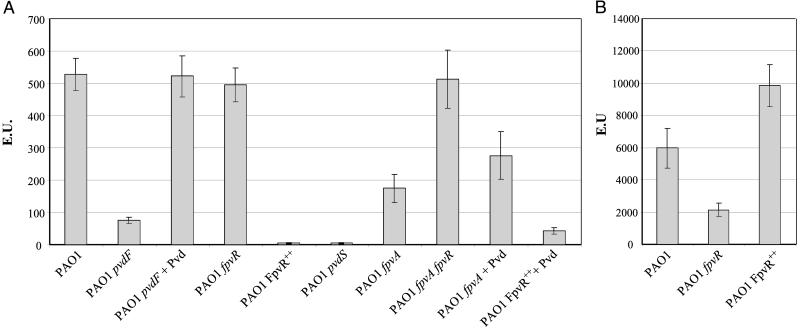
We first addressed the question, does pyoverdine regulate the expression of genes encoding pyoverdine-synthesis enzymes? This question was examined with a P. aeruginosa pvdF mutant strain that makes no detectable pyoverdine (42) in conjunction with the promoter reporter construct pMP190∷PpvdE in which expression of lacZ depends on transcription initiated from the upstream pvdE promoter (18, 39). pvdE promoter activity was much lower in the pvdF mutant than in wild-type bacteria (Fig. 1A). Addition of pyoverdine to the pvdF mutant increased lacZ expression to a level similar to that obtained with wild type. Similar results were obtained with PpvdA-lacZ and PpvdD-lacZ fusion constructs (data not shown), where PpvdA and PpvdD were the promoters of the pvdA and pvdD pyoverdine-synthesis genes. In each case, promoter activity was lower in the pvdF mutant than in wild-type bacteria and was restored to wild-type levels by the addition of pyoverdine. These data show that pyoverdine acts as a signal molecule resulting in increased transcription of pyoverdine-synthesis genes.
Figure 1.
Activities of the pvdE and pvdS promoters. The P. aeruginosa strains shown were grown in King's B medium, and β-galactosidase was assayed. (A) pvdE promoter activity was assayed with plasmid pMP190∷PpvdE. (B) pvdS promoter activity was assayed with pMP190∷PpvdS. + Pvd, pyoverdine was added to a concentration of 60 μM.
The concentration of pyoverdine required to induce gene expression was measured (Fig. 2). Even at the lowest amount examined (0.6 μM pyoverdine), expression from the pvdE promoter was significantly higher than that observed with uninduced cells. Maximal expression was obtained with 12 μM pyoverdine.
Figure 2.
Expression from the pvdE promoter in response to varying concentrations of pyoverdine. P. aeruginosa PAO1pvdF containing pMP190∷PpvdE was grown in King's B medium containing pyoverdine at the concentrations shown, and β-galactosidase was assayed. EU, β-galactosidase enzyme units.
Expression of the pvdA, pvdD, and pvdE genes requires the alternative sigma factor PvdS (18, 19). PvdS is also required for production of exotoxin A, a potent virulence factor of P. aeruginosa (25). In addition, PvdS is required for production of PrpL, an extracellular endoproteinase with elastolytic activity that contributes to infection in a rat lung model (41). As pyoverdine is required for expression from PvdS-dependent pvd promoters, we hypothesized that pyoverdine is also required for PvdS-dependent expression of the genes toxA and prpL that encode exotoxin A and PrpL protease.
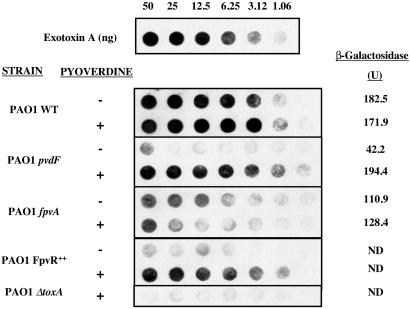
This hypothesis was tested for exotoxin A (Fig. 3). The absence of pyoverdine (pvdF mutant strain) resulted in levels of exotoxin A that were very greatly reduced relative to wild-type P. aeruginosa. The addition of pyoverdine restored production of exotoxin A. Expression of the toxA gene was examined through the use of the promoter-lacZ fusion construct pPZ-toxA. Promoter activity was greatly reduced in the pvdF mutant and was restored to wild-type levels by the addition of pyoverdine (Fig. 3).
Figure 3.
Production of exotoxin A in response to the signaling pathway. Culture supernatants for the strains shown, which had been grown with or without the addition of pyoverdine, were serially diluted. and the presence of exotoxin A was determined with an anti-exotoxin antibody. toxA promoter activity was determined by measuring the amounts of β-galactosidase produced by strains carrying the toxA-lacZ reporter fusion plasmid construct pPZ-toxA. All SDs were less than 12%. ND, not determined.
A specific assay for PrpL protease is not available, but expression of prpL was examined with a prpL-lacZ fusion construct (Table 1). Expression of prpL was lower in the pvdF mutant than in wild-type bacteria, and expression was increased to wild-type levels by the addition of pyoverdine.
Table 1.
Expression of a prpL-lacZ fusion
| Strain | β-Galactosidase activity (EU)*
|
|
|---|---|---|
| −Pyoverdine | +Pyoverdine | |
| PAO1 (pPZ-prpL) | 8.6 (100) | 9.4 (109) |
| PAO1 pvdF (pPZ-prpL) | 1.9 (22) | 9.8 (113) |
Values are the average of at least three different experiments; SDs were less than 12% in each case. Values in parentheses are the % of that obtained with PAO1 without added pyoverdine.
The amounts of β-galactosidase made by strains PAO1 and PAO1pvdF containing plasmid pPZ-prpL were assayed. EU, β-galactosidase enzyme units.
These data show that pyoverdine up-regulates expression of genes required for synthesis of pyoverdine, exotoxin A, and PrpL protease.
Identification of the Anti-Sigma Factor FpvR.
The Fec and Pup systems control expression of iron-transport genes in response to the presence of the relevant iron chelate in E. coli and P. putida, respectively (6). In each case, the iron chelate binds to an outer membrane protein (FecA or PupB), causing transmission of a signal through a membrane-spanning anti-sigma factor (FecR or PupR) to the corresponding sigma factor (FecI or PupI) with a consequent increase in gene expression. PvdS has sequence similarity to FecI and PupI, and FpvA, the outer membrane receptor for ferripyoverdine, has sequence similarity to FecA and PupB. We therefore carried out a blast search of the P. aeruginosa PAO1 genome sequence with the FecR protein sequence to identify homologues that may regulate the activity of PvdS. Thirteen homologues were identified. One of these (PA2388) was linked to pyoverdine-synthesis genes pvdA (PA2386), pvdD (PA2399), pvdE (PA2397), and pvdF (PA2396), as well as the fpvA gene (PA2398) that encodes FpvA. The PA2388 ORF, designated fpvR, potentially encodes a protein of 331 amino acid residues (molecular weight, 36,956).
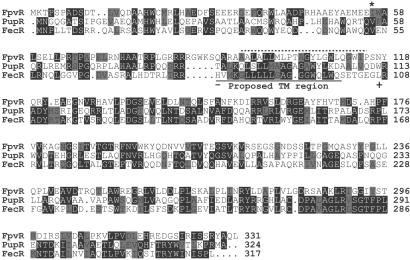
An alignment of the FpvR protein sequence with those of FecR and the related PupR protein is shown in Fig. 4. The FecR protein is thought to have a single transmembrane region on the basis of hydropathy profile and evidence from hybrid FecR-β-lactamase proteins (43). The programs tmhmm and tmpred predicted a single transmembrane helix spanning residues 93–115 of FpvR with the N-terminal part of the protein being in the cytoplasm (Fig. 4). These predictions are consistent with the proposed membrane-spanning topology of FecR (Fig. 4).
Figure 4.
Alignment of the sequences of FpvR, PupR, and FecR. The sequence of FpvR (PA2388 in the P. aeruginosa PAO1 genomic sequence) was aligned with those of PupR and FecR. Positions where identical or similar residues are present in at least two of the sequences are highlighted with identical residues shaded black and similar residues (A and G; D and E; F, W and Y; I, L, M, and V; N and Q; S and T; R and K) shaded gray. FpvR has 27.4% identity (49.0% similarity) with PupR and 35.1% identity (43.0% similarity) with FecR. Sites where fusion of β-lactamase with the N-terminal portion of FecR resulted in sensitivity (−) (cytoplasmic or intramembrane) or resistance (+) (periplasmic) to ampicillin (43) are indicated, and the predicted membrane-spanning region of FecR is underlined. A broken line indicates the region of FpvR that is predicted with TMHMM and TMPRED to constitute a transmembrane helix. The asterisk (*) corresponds to the site where a mutation was introduced into the gene encoding FpvR (see text).
To test whether FpvR is an anti-sigma factor for PvdS, pUCP22∷fpvR, in which fpvR is expressed from the plac promoter, was transformed into P. aeruginosa PAO1 to give strain PAO1FpvR++. This strain was unable to grow under iron-limiting conditions and did not produce any detectable pyoverdine (Table 2). This phenotype was identical to that of a pvdS mutant strain (18) (Table 2). An fpvR mutant of P. aeruginosa PAO1 was constructed by homologous recombination, with the mutation being confirmed by Southern blotting and PCR (data not shown). Growth of this mutant was not restricted under iron-limiting conditions, and the bacteria produced amounts of pyoverdine that were comparable to wild type (Table 2). These results indicate that FpvR is a negative regulator of pyoverdine synthesis and may be an anti-sigma factor for PvdS.
Table 2.
Growth and pyoverdine production of strains of P. aeruginosa
| Strain | Sensitivity to EDDA*
|
Pyoverdine, μM† | |
|---|---|---|---|
| +EDDA | −EDDA | ||
| PAO | +++ | +++ | 103.4 (12.1) |
| PAOfpvR | +++ | +++ | 81.9 (8.0) |
| PAO1FpvR++ | − | +++ | <1.0 |
| PAO1pvdS | − | +++ | <1.0 |
| PAO1pvdF | − | +++ | <1.0 |
| PAO1fpvA | − | +++ | 31.7 (3.1) |
| PAO1fpvAfpvR | + | +++ | 85.9 (5.6) |
Bacterial strains were grown on King's B agar with (+EDDA) or without (−EDDA) the iron-chelating compound EDDA. +++, wild-type levels of growth; +, slight growth; −, no growth; EDDA, ethylenediamine-di(O-hydroxy-phenylacetic acid).
The amounts of pyoverdine present in the supernatants of cultures grown without EDDA were determined as described in Materials and Methods. Values shown are the averages of at least three experiments with SDs shown in parentheses.
The pMP190∷PpvdE promoter construct was used to test this hypothesis further. Expression of lacZ was similar in the wild-type and fpvR mutant strains (Fig. 1A). However, overexpression of fpvR had a profound effect on pvdE promoter activity with expression in P. aeruginosa PAO1FpvR++ being similar to expression in a pvdS mutant strain (Fig. 1A). These results are consistent with the amounts of pyoverdine produced by the fpvR and FpvR++ strains (Table 2) and with the hypothesis that FpvR is an anti-sigma factor for PvdS. The addition of pyoverdine to P. aeruginosa PAO1FpvR++ resulted in a significant increase in transcription from the pvdE promoter although even with pyoverdine added, expression was only 8% of that seen with wild-type bacteria.
An alternative hypothesis was that instead of being an anti-sigma factor FpvR is a transcriptional repressor that regulates transcription of the pvdS gene. This hypothesis was tested by using plasmid pMP190∷PpvdS. Expression from the pvdS promoter was less in the fpvR mutant than in wild-type P. aeruginosa (Fig. 1B). Overexpression of FpvR caused a 65% increase in pvdS expression. These results show that FpvR is not a repressor of pvdS transcription, and indeed increased levels of FpvR resulted in increased PvdS expression. Expression from the pvdS promoter is regulated by the iron-responsive Fur repressor protein (19), and the increased activity of the pvdS promoter in the FpvR++ strain (and decreased activity in the fpvR mutant) may be the result of altered amounts of intracellular iron as a consequence of changes in the amount of FpvR protein.
Overexpression of FpvR also very greatly reduced the amount of exotoxin A that was produced and this was reflected in reduced toxA expression (Fig. 3). Addition of pyoverdine increased the amount of exotoxin A in culture supernatant to levels comparable to those obtained with wild-type bacteria. Collectively, these data are consistent with the hypothesis that FpvR is an anti-sigma factor that inhibits PvdS-mediated transcription.
FpvA Is Part of the Pyoverdine Regulatory Pathway.
By analogy with the Fec and Pup systems, the activity of FpvR and hence of PvdS was likely to be regulated by pyoverdine in concert with the FpvA ferripyoverdine receptor protein. To test this hypothesis, fpvA and fpvA fpvR mutant strains of P. aeruginosa were constructed with the mutations being confirmed by Southern analysis and Western blotting for the FpvA protein (data not shown). The fpvA mutant produced lower amounts of pyoverdine than wild type (Table 2), as has been observed by others (K. Poole, personal communication), showing that the FpvA receptor protein has a role in regulating pyoverdine production. The effect of the fpvA mutation on pyoverdine production was alleviated in the fpvR background, with the fpvA fpvR double mutant making similar amounts of pyoverdine to the wild-type and fpvR strains.
The involvement of FpvA in pvd gene expression was also examined. The level of pvdE promoter activity in the fpvA mutant was 33% of that obtained with the wild type whereas in the fpvA fpvR double mutant, expression was comparable with that in the wild-type strain (Fig. 1A). These data are consistent with the amounts of pyoverdine produced by these strains (Table 2). The addition of pyoverdine to the fpvA mutant resulted in a slight increase in pvdE promoter activity, but levels of expression were lower than those observed for wild-type bacteria or after addition of pyoverdine to the pvdF mutant strain.
The P. aeruginosa mutant lacking FpvA made detectable exotoxin A but this was reduced relative to the wild-type, and addition of pyoverdine had no effect on exotoxin production (Fig. 3). Expression of toxA as judged by using the toxA-lacZ fusion was intermediate between that of the wild-type and the pvdF mutant and was not affected by the addition of pyoverdine, consistent with the effect of this mutation on exotoxin production.
These data indicate that the ferripyoverdine receptor protein FpvA is required for transmission of the pyoverdine signal and are consistent with a model in which FpvA modulates the activity of FpvR, which in turn affects the activity of PvdS and hence transcription of the toxA and pyoverdine-synthesis genes.
Discussion
The research described here shows that a siderophore, pyoverdine, regulates its own production and also controls production of at least two other virulence factors, exotoxin A and PrpL protease, by P. aeruginosa. Regulation takes place through a signaling pathway involving the FpvA, FpvR, and PvdS proteins (Fig. 5). Signaling is initiated by the presence of (ferri)pyoverdine that interacts with the cell-surface FpvA receptor protein. FpvA is predicted to interact with part of the FpvR protein that is located in the periplasm, and this interaction results in transmission of a signal through the cytoplasmic membrane to the cytoplasmic domain of FpvR. This event enables PvdS to become active, interacting with RNA polymerase to cause transcription of the genes required for production of pyoverdine, exotoxin A, and PrpL protease. It remains to be determined whether production of other secreted products is also regulated by this system. It also remains to be determined whether apopyoverdine or ferripyoverdine, both of which can bind to the FpvA receptor (44), is the active signaling molecule. The system is responsive to pyoverdine at a concentration of 0.6 μM (the lowest amount tested) (Fig. 2). This is comparable to the concentration of pyoverdine in sputum from patients with cystic fibrosis (0.48–1.55 μM) (45), indicating that this signaling system is likely to be functioning in P. aeruginosa in these patients.
Figure 5.
Model of the FpvA/FpvR/PvdS signaling pathway. (Ferri)pyoverdine complexes bind the FpvA receptor protein, transmitting a signal to the FpvR protein that otherwise suppresses the activity of PvdS. PvdS then binds to RNA polymerase, causing expression of genes required for synthesis of pyoverdine and PrpL protease (prpL) and of the ptxR gene that encodes a transcriptional activator of the toxA gene.
This signaling system has clear similarities to the Fec and Pup signaling systems that control production of ferrisiderophore receptor proteins in E. coli and P. putida WCS358, respectively (reviewed in ref. 6). It has been shown that in the Fec system, FecA and FecI both interact physically with FecR (46). Interactions between the protein components of the FpvA/FpvR/PvdS pathway have not yet been demonstrated. However, the N-terminal part of FpvA has been shown to be located in the periplasm (P.A.B. and I.L.L., unpublished data), and the presence of a likely membrane-spanning domain in FpvR is consistent with the suggestion that this protein spans the cytoplasmic membrane and thus could interact with both FpvA and PvdS. It will be of interest to compare protein–protein interactions in the Fec system with those in the Fpv/PvdS system.
Biologically, the pyoverdine-mediated signaling pathway described here differs from the Fec and Pup systems in two important respects. First, unlike the Fec and Pup systems, pyoverdine regulates its own production and controls production of at least two secreted proteins. Second, the Fec and Pup signaling pathways both respond to exogenous iron chelates that are not produced by the organisms in which these systems have been studied. By contrast, P. aeruginosa PAO both produces pyoverdine and responds to its presence. In this respect, the system has conceptual parallels with quorum-sensing. In P. aeruginosa, quorum-sensing occurs through AHLs that are secreted by bacterial cells and as the cell density increases so does the concentration of AHL, resulting in increased expression of a range of genes including those that give rise to the AHLs (21, 22). In principle, pyoverdine-mediated gene expression works in the same way; when the cell density is low, the concentration of pyoverdine is low, whereas at high cell density the higher concentration of pyoverdine will cause increased expression of genes for synthesis of exotoxin, PrpL protease, and pyoverdine itself. There are obvious differences between the two systems. Although there is some evidence that AHLs can act directly on mammalian tissues (22), their primary role is as signaling molecules and they interact directly with transcriptional regulators in the cytoplasm. By contrast, pyoverdine is a siderophore as well as a signaling molecule and exerts its effect through a signaling pathway rather than as a transcriptional coregulator. Chemically, AHLs and pyoverdine are also very different. Nonetheless the pyoverdine-mediated signaling system potentially represents a further mechanism by which cells of P. aeruginosa can respond to increased cell density. There is evidence that expression of pyoverdine-synthesis genes is responsive to AHLs (47, 48), indicating an interaction between the quorum-sensing and pyoverdine systems, and it will be of interest to determine the molecular nature of the interactions between the two systems.
What are the advantages to P. aeruginosa of having pyoverdine as a signaling molecule? It has been proposed that AHL-mediated quorum-sensing provides pathogenic bacteria with a mechanism to delay production of virulence factors, and detection by the host immune system, until bacterial cell numbers have reached a sufficient level to enable a productive infection (22, 49). The same logic can be applied to pyoverdine-mediated signaling as the concentration of pyoverdine will be low in the initial stages of infection but will increase as the number of bacterial cells increases. However, P. aeruginosa can exist in a variety of habitats, including fresh water and soil, as well as cause infections. Simplistically, it would seem inefficient for the bacteria to secrete pyoverdine if there is a low probability of it being recaptured as ferripyoverdine and taken up by the cells (this may be the case in an aquatic environment). However, if there is a reasonable chance of secreted pyoverdine being taken up by the bacteria as ferripyoverdine—for example, when a large number of bacteria are present in a limited space, such as in a biofilm—then this becomes an efficient mechanism of iron uptake. From this point of view, it makes biological sense for pyoverdine to be synthesized under conditions where successful uptake can occur, and for synthesis to be repressed if there is a low probability of recapture of secreted pyoverdine.
How widespread is this mechanism of gene regulation? A survey of genome databases revealed over 70 uncharacterized homologues of FpvR in a wide range of bacteria including human pathogens (Bordetella species and Burkholderia pseudomallei), a plant pathogen (Pseudomonas syringae), soil bacteria (P. putida and Nitrosomonas europaea), and an aquatic bacterium (Caulobacter crescentus) (I.L.L. and P.A.B., unpublished observations). These may well be components of siderophore-responsive transmembrane signaling systems analogous to the FpvA/FpvR/PvdS pathway described here and to the Fec and Pup systems. In many cases, the genes that are up-regulated in response to signaling may be limited to those required for siderophore transport, as is the case for the Fec and Pup systems. However, given the large number of uncharacterized FpvR-like genes, it would be surprising if the pyoverdine system was the only example in which a siderophore acts as a signaling molecule to regulate production of secreted proteins. More broadly, over 500 siderophores have been identified. It would seem likely that many of these may regulate their own production, although the mechanisms may be different from those of the Fpv system. Autoregulation has been demonstrated for production of pyochelin by P. aeruginosa (10) and alcaligin by Bordetella species (12), both of which use an AraC-type regulator. Other siderophores may also regulate synthesis of additional extracellular products.
In summary, the results presented here demonstrate that pyoverdine acts as a signaling molecule to regulate its own production and to control production of secreted proteins. This finding has important implications for our understanding of the molecular processes involved in infections caused by P. aeruginosa. Given the widespread production of siderophores by bacteria, it is likely that siderophores will also act as signaling molecules to control gene expression in other species.
Acknowledgments
We are grateful to Daphne Raj for construction and preliminary characterization of pMP190∷PpvdS, to the Pseudomonas genome sequencing consortium for making data available before publication, and to Clive Ronson for helpful comments on an earlier version of this manuscript. This research was supported by a grant from the University of Otago Research Committee (to I.L.L.), by the Cystic Fibrosis Foundation, and by National Institutes of Allergy and Infectious Diseases Grant AI15940 (to M.L.V.).
Abbreviation
- AHL
N-acyl-homoserine lactone
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Griffiths E. In: Iron and Infection. Bullen J J, Griffiths E, editors. Chichester, U.K.: Wiley; 1987. pp. 1–25. [Google Scholar]
- 2.Guerinot M L. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 3.Drechsel H, Winkelmann G. In: Transition Metals in Microbial Metabolism. Winkelmann G, Carrano C J, editors. Amsterdam: Harwood Academic; 1997. pp. pp.1–49. [Google Scholar]
- 4.Ratledge C, Dover L G. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 5.Expert D. Annu Rev Phytopathol. 1999;37:307–334. doi: 10.1146/annurev.phyto.37.1.307. [DOI] [PubMed] [Google Scholar]
- 6.Crosa J H. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun V, Killmann H. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 8.Venturi V, Weisbeek P, Koster M. Mol Microbiol. 1995;17:603–610. doi: 10.1111/j.1365-2958.1995.mmi_17040603.x. [DOI] [PubMed] [Google Scholar]
- 9.Callanan M, Sexton R, Dowling D N, O'Gara F. FEMS Microbiol Lett. 1996;144:61–66. doi: 10.1111/j.1574-6968.1996.tb08509.x. [DOI] [PubMed] [Google Scholar]
- 10.Reimmann C, Serino L, Beyeler M, Haas D. Microbiology. 1998;144:3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 11.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brickman T J, Kang H Y, Armstrong S K. J Bacteriol. 2001;183:483–489. doi: 10.1128/JB.183.2.483-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fick R B., Jr . Pseudomonas aeruginosa the Opportunist: Pathogenesis and Disease. Boca Raton, FL: CRC; 1993. pp. 1–5. [Google Scholar]
- 14.Meyer J-M, Neely A, Stintzi A, Georges C, Holder I A. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takase H, Nitanai H, Hoshino K, Otani T. Infect Immun. 2000;68:1834–1839. doi: 10.1128/iai.68.4.1834-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leoni L, Orsi N, de Lorenzo V, Visca P. J Bacteriol. 2000;182:1481–1491. doi: 10.1128/jb.182.6.1481-1491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson M J, Lamont I L. Biochem Biophys Res Commun. 2000;273:578–583. doi: 10.1006/bbrc.2000.2996. [DOI] [PubMed] [Google Scholar]
- 18.Cunliffe H E, Merriman T R, Lamont I L. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leoni L, Ciervo A, Orsi N, Visca P. J Bacteriol. 1996;178:2299–2313. doi: 10.1128/jb.178.8.2299-2313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyczak J B, Cannon C L, Pier G B. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 21.Parsek M R, Greenberg E P. Proc Natl Acad Sci USA. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitehead N A, Barnard A M L, Slater H, Simpson N J L, Salmond G P C. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro J A. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 24.King E O, Ward M K, Raney D E. J Lab Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 25.Ochsner U A, Johnson Z, Lamont I L, Cunliffe H E, Vasil M L. Mol Microbiol. 1996;21:1019–1028. doi: 10.1046/j.1365-2958.1996.481425.x. [DOI] [PubMed] [Google Scholar]
- 26.Meyer J-M, Stintzi A, Vos D D, Cornelis P, Tappe R, Taraz K, Budzikiewicz H. Microbiology. 1997;143:35–43. doi: 10.1099/00221287-143-1-35. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, et al. Nature (London) 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Needleman S B, Wunsch C D. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 31.Chen W, Kuo T. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Russell D W, Irwin N. Molecular Cloning: A Laboratory Manual. 3rd. Ed. Plainview, New York: Cold Spring Harbor Lab. Press; 2000. [Google Scholar]
- 33.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Gene. 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 34.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 35.Schweizer H P. Gene. 1991;103:97–92. doi: 10.1016/0378-1119(91)90396-s. [DOI] [PubMed] [Google Scholar]
- 36.Berryman T A. The Application of Gene Recombinant DNA Technology in the Analysis of Candida albicans. Dunedin, New Zealand: University of Otago; 1994. p. 15. [Google Scholar]
- 37.Simon R, O'Connell M, Labes M, Puhler A. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 38.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 39.Wilson M J, McMorran B J, Lamont I L. J Bacteriol. 2001;183:2151–2155. doi: 10.1128/JB.183.6.2151-2155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller J H. Experiments in Molecular Genetics. Plainview, New York: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 41.Wilderman P J, Vasil A I, Johnson Z, Wilson M J, Cunliffe H E, Lamont I L, Vasil M I. Infect Immun. 2001;69:5385–5394. doi: 10.1128/IAI.69.9.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMorran B J, Kumara H M C S, Sullivan K, Lamont I L. Microbiology. 2001;147:1517–1524. doi: 10.1099/00221287-147-6-1517. [DOI] [PubMed] [Google Scholar]
- 43.Welz D, Braun V. J Bacteriol. 1998;180:2387–2394. doi: 10.1128/jb.180.9.2387-2394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schalk I J, Hennard C, Dugave C, Poole K, Abdallah M A, Pattus F. Mol Microbiol. 2001;39:351–360. doi: 10.1046/j.1365-2958.2001.02207.x. [DOI] [PubMed] [Google Scholar]
- 45.Haas B, Kraut J, Marks J, Zanker S C, Castignetti D. Infect Immun. 1991;59:3997–4000. doi: 10.1128/iai.59.11.3997-4000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enz S, Mahren S, Stroeher U H, Braun V. J Bacteriol. 2000;182:637–646. doi: 10.1128/jb.182.3.637-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stintzi A, Evans K, Meyer J-M, Poole K. FEMS Microbiol Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 48.Whiteley M, Lee K M, Greenberg E P. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]