Abstract
Moloney murine leukemia virus (M-MuLV) IN-IN protein interactions important for catalysis of strand transfer and unimolecular and bimolecular disintegration reactions were investigated by using a panel of chemically modified M-MuLV IN proteins. Functional complementation of an HHCC-deleted protein (NΔ105) by an independent HHCC domain (CΔ232) was severely compromised by NEM modification of either subunit. Productive NΔ105 IN-DNA interactions with a disintegration substrate lacking a long terminal repeat 5′-single-stranded tail also required complementation by a functional HHCC domain. Virus encoding the C209A M-MuLV IN mutation exhibited delayed virion production and replication kinetics.
Productive retroviral infections require the stable insertion of the reverse-transcribed viral genome into the host chromosome, a step that is mediated by the virus-encoded IN protein. Following reverse transcription of the retrovirus genome, the IN protein processes the termini of the viral long terminal repeats (LTRs) by endonucleolytic cleavage of the 3′-terminal dinucleotides. This 3′-processing reaction exposes a 5′-CA-3′ dinucleotide that is subterminally embedded in the LTRs and is conserved among known retroelements (3, 27, 28, 31, 41, 45). The 3′ processing of the viral DNA termini leaves behind a 5′ overhang or single-stranded (ss) tail which may impart IN-DNA complex stability in vitro (16, 48). Integration of the viral DNA copy occurs by a staggered transesterification of the recessed viral 3′ ends and the phosphodiester backbone of the target DNA (19). Repair of the resultant single strand gaps creates the hallmark duplication of target DNA sequence flanking the retroviral integration site (4, 28, 45).
Biochemical and genetic studies suggest that IN functions as a multimer which coordinates the integration of both viral DNA termini (18, 23, 24, 26, 28, 35, 37, 44, 46). The retroviral IN protein contains three functionally important domains: an N-terminal HHCC domain, a central catalytic core, and a C-terminal domain implicated in nonspecific DNA binding (2, 34, 47, 49, 50). The results of chemical cross-linking, equilibrium sedimentation, gel filtration, and yeast two-hybrid studies suggest that regions throughout IN are mediators of homomeric IN-IN interactions (1, 7, 23, 26, 46). The crystal and solution structures of the N-terminal HHCC (containing zinc), central catalytic core, and C-terminal subdomains of IN have substantiated the dimeric nature of these subdomains independent of DNA (5, 7, 15, 33). Furthermore, recent findings indicate that metal-dependent IN-IN (17, 52) and IN-DNA interactions (38) are involved in functional, higher-order multimerization of the IN protein.
Various assays for IN function have been developed including integration (3′ processing and strand transfer) (12, 29), disintegration (10), and coordinated disintegration (8, 13, 35). For disintegration, the substrate is unimolecular; strand transfer and coordinated disintegration are dependent on bimolecular assembly of the substrate mediated by multimeric IN-DNA interactions. The domains of IN which are important for these interactions vary among IN proteins and are implicated in such interactions through mutational analysis (6, 24, 30, 43, 46, 47), analysis of modified substrates (9, 14, 42), and the ability of defective IN mutants to complement each other in trans (8, 18, 24, 35, 44).
In previous studies, the C terminus of M-MuLV IN could only be minimally truncated (28 amino acids) in vitro and in vivo (24, 39). The M-MuLV HHCC domain was shown to be essential for 3′ processing (24). M-MuLV IN lacking the HHCC region could catalyze unimolecular disintegration (24) and single and double disintegration on a tailed crossbone substrate (13). Under less-saturating enzyme conditions, double-end disintegration was strictly dependent on trans-subunit IN interactions promoted by the HHCC domain (13). With the crossbone substrate, the intramolecular “foldback” reaction is mechanistically similar to 3′ processing (13). The foldback reaction was found to be dependent on both the LTR 5′-ss tail and N-terminal HHCC domain of M-MuLV IN, whereas either determinant sufficed to promote intermolecular activity (13). Collectively, these data suggested differing roles of the HHCC domain in the formation of the initial IN-DNA complexes and coordinated IN-IN interactions.
Prior studies had indicated that the M-MuLV IN was sensitive to N-ethylmaleimide (NEM) modification (25). Complementation studies performed with human immunodeficiency virus type 1 IN have identified an NEM-sensitive site (Cys 56) which is required in trans to the HHCC domain (17). In the present work, multimeric M-MuLV IN-IN interactions important for strand transfer and unimolecular and coordinated (bimolecular) disintegration reactions were studied by complementation analysis of NEM-modified M-MuLV IN subunits. A central cysteine within the catalytic core domain, C209, and the HHCC domain were identified as NEM-sensitive sites, both of which were necessary to mediate functional complementation. Furthermore, a critical requirement of the C209 NEM-sensitive site for efficient viral replication was demonstrated, suggesting a novel model for multimeric IN function.
NEM modification of wild-type (WT) and mutant M-MuLV IN proteins.
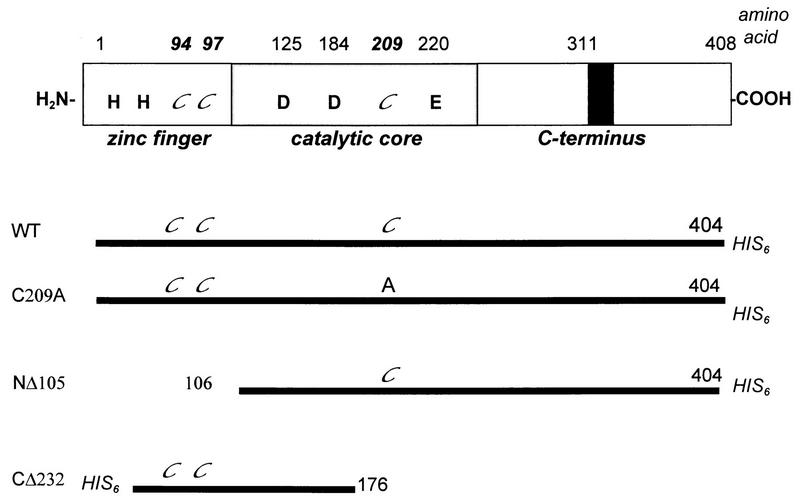
Several M-MuLV IN mutants, which have been previously characterized (13, 24), were used to probe the NEM sensitivity of the IN-IN and IN-DNA interactions (Fig. 1). CΔ232 is an inactive C-terminal truncation that contains the HHCC domain and terminates just beyond the first aspartate residue of the active site (Fig. 2 and 3, lanes 3 and 4) (24). A point mutant that eliminates the sole non-HHCC cysteine residue located within the central catalytic core domain of M-MuLV IN, C209A, is active for integration and disintegration reactions (Fig. 2 and 3, lanes 5 and 6) (24). NΔ105 lacks the N-terminal HHCC domain and is inactive for 3′ processing but is active for unimolecular disintegration (Fig. 2B, lanes 7 and 8) and coordinated disintegration of a tailed crossbone substrate (Fig. 3B, lane 3). In strand transfer assays, however, NΔ105 retains a limited capacity to mediate integration into a single target site (Fig. 2A, lanes 7 and 8) (24). Full integration activity of NΔ105 can be restored by complementation with CΔ232 (Fig. 2A, MOCK, lane 11) or by increasing the reducing conditions and protein levels in the assay (24).
FIG. 1.
M-MuLV IN mutants used for NEM alkylation studies. The conserved domains and residues of IN are depicted, with the cysteines highlighted in italics (top); the black box represents a C-terminal insertion of 36 amino acids unique to M-MuLV IN. Cysteine targets of NEM modification in the WT, C209A, NΔ105, and CΔ232 proteins are highlighted in italics (bottom). Residues at which NΔ105 and CΔ232 initiate or terminate are also indicated.
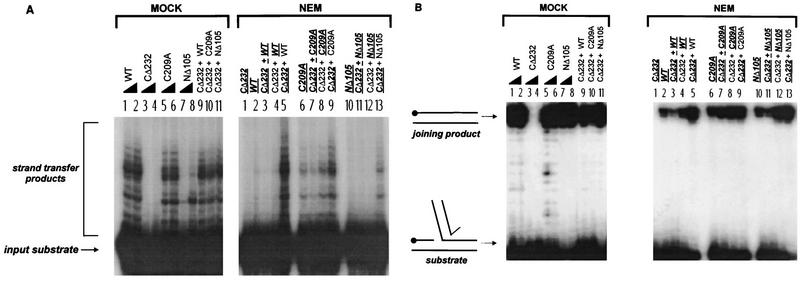
FIG. 2.
Activity of NEM-alkylated M-MuLV IN proteins in strand transfer and unimolecular disintegration reactions. (MOCK) Lanes 1 to 8, odd and even numbered lanes are reactions that respectively contained 10 and 20 pmol of the mock-treated proteins; lanes 9 to 11, complementation reactions which contained 10 pmol each of the indicated mock-treated proteins. (NEM) Assays were performed with 10 pmol of each of the indicated proteins. M-MuLV IN proteins that were subjected to NEM alkylation are indicated by bold underlined characters. Non-underlined proteins are untreated IN. Lane 1, NEM-alkylated CΔ232 negative-control reaction; lanes 2 to 13, reactions that contained individual or mixed pairs of treated and untreated proteins. (A) Strand transfer reactions with substrate 2784/2785 (13, 24). (B) Unimolecular disintegration reactions with the standard Y substrate (Y3154ds [13, 14]). The position of the 5′ 32P-labeled C strand of oligonucleotide 3152 (14) is indicated (•).
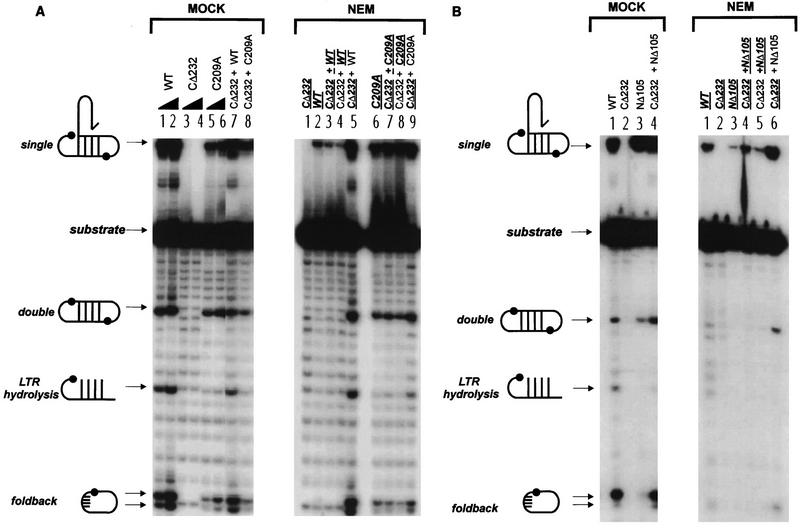
FIG. 3.
Activity of NEM-alkylated M-MuLV IN proteins in coordinated bimolecular disintegration reactions. Reaction products of coordinated disintegration are indicated at the left of each panel. The input crossbone oligonucletide substrate consists of tailed half-crossbones 4166-4167 (13). M-MuLV IN proteins that were subjected to NEM alkylation are indicated by bold underlined characters. Non-underlined proteins are untreated IN. (A) Tailed coordinated disintegration reactions mediated by WT and C209A M-MuLV IN. (MOCK) Lanes 1 to 6, odd and even numbered lanes are reactions that respectively contained 10 and 20 pmol of the mock-treated proteins; lanes 7 and 8, complementation reactions which contained 10 pmol each of the indicated mock-treated proteins. (NEM) Assays were performed with 10 pmol of each of the indicated proteins. Lane 1, NEM-alkylated CΔ232 negative-control reaction; lanes 2 to 9, reactions that contained individual or mixed pairs of treated and untreated proteins. (B) Tailed coordinated disintegration reactions contrasting WT and NΔ105 M-MuLV IN. The activity of complementation mixtures containing 10 pmol of each indicated protein pair (MOCK, lane 4, and NEM, lanes 4 to 6) is compared to the activity of reaction mixtures containing 10 pmol of the individual proteins (lanes 1 to 3).
To investigate the nature of the IN-IN interactions important for functional complementation, each of the M-MuLV IN proteins was subjected to NEM alkylation followed by quenching with excess dithiothreitol (DTT). Strand transfer and unimolecular and bimolecular disintegration assays were performed in parallel. A total of 120 pmol of each protein to be treated was separately diluted to a final concentration of 2 to 2.5 pmol/μl with ice-cold diluent (20 mM HEPES [pH 7.4]–20% glycerol) and equilibrated on ice for 5 min. As a control, mock reactions were performed in which each of the M-MuLV IN proteins were preincubated on ice with 40 mM DTT for 20 min, followed by incubation with 10 mM NEM (freshly prepared; Sigma Chemical Corp) for a further 20 min. For the NEM reactions, proteins were first incubated on ice in 10 mM NEM for 20 min, followed by a 20-min quench with 40 mM DTT on ice. The NEM and mock-treated M-MuLV IN proteins were then assayed individually and for functional complementation. A final DTT concentration of 40 mM was present in both mock and NEM reactions. In complementation assays where only one of the IN protein pairs was alkylated, DTT was present at a 20 mM concentration. Reactions were incubated at 37°C for 1.5 h and terminated. Similar results were obtained when the nonalkylated protein in the mixture was either untreated, mock treated (indicated in the figure legends), or DTT treated (data not shown).
Effects of NEM modification on strand transfer and unimolecular disintegration reactions.
The results of the NEM treatment and complementation of the M-MuLV IN proteins for strand transfer and disintegration are shown in Fig. 2A and B, respectively. Compared to the control mock reactions (Fig. 2A, MOCK, lanes 1 and 2), NEM-treated WT protein was inactive for strand transfer (Fig. 2A, NEM, lane 2). Similar to WT IN, the strand transfer activity of NΔ105 protein was severely compromised by alkylation (Fig. 2A, NEM, lane 10 versus Mock, lane 7). NΔ105 (at 20 pmol) remained sensitive to NEM (data not shown). Since NΔ105 contains only a single cysteine, the loss of NΔ105 strand transfer activity indicates that residue C209 in the central catalytic core is NEM sensitive. These results indicate that modification of C209 blocks the catalytic functions of IN. C209 per se is not essential for catalysis in vitro, since the mutant C209A is capable of mediating single-end strand transfer (Fig. 2A, lanes 5 and 6) and unimolecular (Fig. 2B, lanes 5 and 6) and bimolecular (Fig. 3A, lanes 5 and 6) disintegration reactions. This is supported by NEM treatment of C209A (Fig. 2A, NEM, lane 6), which cannot be modified in the catalytic core and maintained activity, albeit yielding reduced levels of strand transfer, double disintegration, and foldback product. The partial diminution of activity implies that the HHCC domain may serve as an additional target for NEM modification.
Complementation analysis allowed us to directly assess the sensitivity of the HHCC domain versus that of the central catalytic core’s C209 site to NEM modification since these domains are present as separate proteins. Previous analysis of NΔ105-C209A, a double mutant that both lacks the HHCC domain and contains an alanine substitution for residue C209, indicated that although it retains limited integration and disintegration activity it cannot be functionally complemented by CΔ232 (24). This implied that residue C209 and the HHCC domain may be in close proximity. It was therefore tested whether the CΔ232 protein could protect residue C209 from alkylation with NEM. For premixed complementation reactions, 60 pmol of each protein pair was mixed together and then diluted immediately to a total protein concentration of 2 pmol/μl. Premixing CΔ232 with WT IN prior to NEM treatment did not protect WT or NΔ105 IN from alkylation (Fig. 2A, NEM, lanes 3 and 11) and caused a slight reduction in the strand transfer activity of C209A (Fig. 2A, NEM, lane 7).
The effect of NEM treatment on multimeric IN-IN interactions could therefore be addressed in complementation reactions in which only one of the pairs of M-MuLV IN proteins was alkylated. Untreated CΔ232 was unable to complement NEM-treated WT, C209A, and NΔ105 M-MuLV IN for the strand transfer reactions (Fig. 2A, NEM, lanes 4, 8, and 12, respectively). When the HHCC domain of CΔ232 was alkylated and tested for complementation, little effect on the strand transfer activity of untreated WT and C209A IN proteins was noted (Fig. 2A, NEM, lanes 5 and 9, respectively). A low level of strand transfer activity was seen for complementation of NΔ105 by NEM-treated CΔ232 (Fig. 2A, NEM, lane 13); however, this level of complementation was below that of the MOCK-treated sample (Fig. 2A, MOCK, lane 11). This is a result of functional complementation by a limited number of CΔ232 molecules that were resistant to alkylation (51).
In the unimolecular disintegration reactions, NEM modification of the individual M-MuLV IN proteins (WT, C209A, and NΔ105) only resulted in a partial loss of activity (Fig. 2B, NEM, lanes 2, 6, and 10, respectively). For the unimolecular disintegration reactions, mixing of CΔ232 with WT, C209A, or NΔ105 prior to NEM treatment did not protect the NEM-sensitive subunits against alkylation (Fig. 2B, NEM, lane 3, 7, or 11, respectively). Premixing CΔ232 with WT or NΔ105, in fact, decreased the yield of joining product. Under these conditions, the HHCC domain may assist in exposing the cysteines for NEM modification. For complementation, the untreated CΔ232 protein stimulated the disintegration activity of NEM-treated WT and C209A by 1.5- to 2-fold and that of NΔ105 by 4-fold (Fig. 2B, NEM, lanes 4, 8, and 12, respectively). The alkylated CΔ232 HHCC domain did not interfere with productive unimolecular IN-DNA interactions mediated by the WT, C209A, and NΔ105 proteins (Fig. 2B, NEM, lanes 5, 9, and 13, respectively).
NEM alkylation and multimeric IN function: effects on bimolecular coordinated disintegration reactions.
The NEM-treated M-MuLV IN proteins were assayed for coordinated disintegration to more directly address the effect of alkylation on the productive assembly of bimolecular IN-DNA complexes. In the bimolecular disintegration reactions, four major products have been identified by using tailed crossbone substrates: intermolecular single disintegration and circular double disintegration products, a foldback product formed by an intramolecular attack by the target DNA 3′-OH, and an LTR hydrolysis product (Fig. 3) (13). The catalytic properties of WT IN, NΔ105, C209A, and CΔ232 on this substrate have previously been characterized (13).
NEM treatment of WT (Fig. 3A, NEM, lane 2 and Fig. 3B, NEM, lane 1) and NΔ105 (Fig. 3B, NEM, lane 3) clearly compromised activity. This point is illustrated by the severe reduction of single disintegration and by the total ablation of double disintegration and foldback reactions compared to the control WT (Fig. 3A, NEM, lanes 1 and 2) and NΔ105 (Fig. 3B, NEM, lane 3). In contrast, C209A was only moderately affected by NEM alkylation, with a slight decrease in the level of double disintegration and foldback products (Fig. 3A, NEM, lane 6). C209A has been previously characterized as catalyzing intramolecular foldback reactions less efficiently than WT (13). To identify targets of NEM important for coordinated disintegration and to test if the loss of IN function could be rescued, different combinations of treated and untreated proteins were assayed for complementation. Mixing of CΔ232 with WT, C209A (Fig. 3A and B, NEM, lanes 3 and 7), or NΔ105 (Fig. 3B, NEM, lane 4) prior to NEM treatment did not protect the NEM-sensitive subunits against alkylation. The activity of NEM-treated C209A was not changed by complementation with untreated CΔ232 (Fig. 3A, NEM, lane 8). Significantly, the coordinated disintegration activity mediated by NEM-alkylated WT (Fig. 3A, NEM, lanes 4) and NΔ105 proteins (Fig. 3B, NEM, lane 5) could not be restored by complementation with an untreated CΔ232 HHCC domain. The NEM-alkylated CΔ232 HHCC domain did not alter the activity of untreated WT (Fig. 3A, NEM, lane 5) or C209A (Fig. 3A, NEM, lane 9), indicating that the loss of complementation functions was not due to incomplete quenching of unreacted NEM. Importantly, NEM-treated CΔ232 was unable to complement untreated NΔ105 (Fig. 3B, NEM, lane 6). The untreated CΔ232 protein did function as a multimer with NΔ105, as indicated by the restoration of double disintegration and foldback products to WT levels in the control reactions (Fig. 3B, MOCK, lane 4, compared to NΔ105, lane 3 and WT IN, lane 1).
Thus, when either residue C209 or the HHCC domain was alkylated by NEM treatment, functional complementation could not be achieved. These data collectively suggest that residue C209 of M-MuLV IN, in conjunction with the HHCC domain, participates in the multimeric assembly of bimolecular IN-DNA complexes and coordinated IN-IN interactions.
The HHCC domain of M-MuLV IN is required for productive interactions with an untailed unimolecular disintegration substrate.
The function of the HHCC in retroviral integration remains obscure. Its proposed role in IN-IN interactions, LTR positioning, or in the maintenance of stable IN-LTR DNA complexes (13, 16, 17, 24, 25) has been inferred from biochemical analysis of mutant proteins through multiple assay systems. The importance of the HHCC domain in the absence of the LTR 5′-ss tail has been documented for coordinated disintegration reactions (13). The requirements for bimolecular substrate assembly in coordinated disintegration, however, is distinct from that of a unimolecular disintegration reaction. In the context of unimolecular disintegration, the HHCC domain is not critical when an LTR 5′-ss tail is present on the Y substrate (24). To further define the function of the HHCC domain, conditions were identified which strictly require the HHCC for unimolecular disintegration (Fig. 4A).
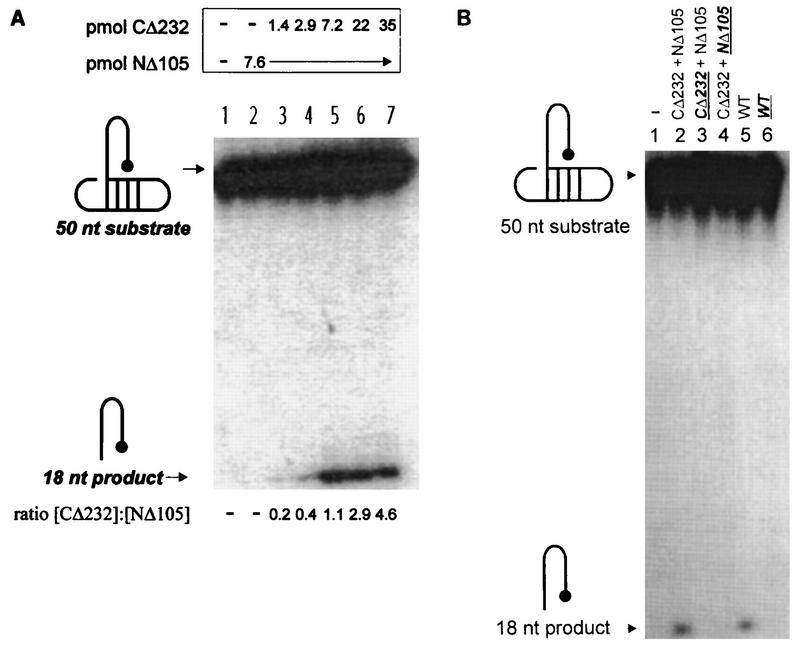
FIG. 4.
Influence of the HHCC domain, residue C209, and the LTR 5′-ss tail on NΔ105. The structures of the 5′-32P-labeled untailed dumbbell substrate (7440, 5′-TGAAAGCGTAAGCTTTCAACCTGCGTAAGCAGGTAGACCGCAAGGTCT-3′) and the reaction product of unimolecular disintegration are indicated on the left of each panel. (A) Requirement of the HHCC domain in trans to NΔ105 for unimolecular disintegration of an untailed dumbbell disintegration substrate. Lane 1 contains a control buffer reaction. The molar quantities of the proteins and their ratios in each reaction are respectively indicated above and below each lane. (B) Effect of NEM alkylation of residue C209 or the HHCC domain on productive interactions with an untailed dumbbell substrate. Individual M-MuLV IN proteins and complementation protein pairs are indicated as in Fig. 2 and 3. NEM-treated samples are indicated in bold and are underlined.
In the absence of both the HHCC domain and the LTR 5′-ss tail, no disintegration activity of the NΔ105 protein was detected by using an untailed dumbbell substrate (Fig. 4A, lane 2). The critical requirement of the HHCC domain for productive interactions with the untailed disintegration substrate was demonstrated by functional complementation with the CΔ232 protein (Fig. 4A, lanes 3 to 7), with maximal disintegration occurring at a 1:1 molar ratio of CΔ232 to NΔ105 protein. This suggests that in the absence of the 5′-ss tail, productive unimolecular IN-DNA interactions are dependent on a functional IN multimer containing an HHCC domain in stoichiometric levels with the catalytic core and C terminus of M-MuLV IN. In this simplified assay, the HHCC domain is required directly for LTR stabilization and not bimolecular substrate assembly.
This necessity of the HHCC domain for productive IN-DNA interactions in the absence of the LTR 5′-ss tail was further investigated by NEM alkylation of the NΔ105 and CΔ232 complementation pairs (Fig. 4B). In comparison to the complementation of NΔ105 by CΔ232 (Fig. 4B, lane 2), NEM treatment of the CΔ232 protein resulted in complete loss of functional complementation (Fig. 4B, lane 3). When the C209 site of NΔ105 was alkylated, complementation by untreated CΔ232 was also inhibited (Fig. 4B, lane 4). These data indicate that both the HHCC domain of CΔ232 and residue C209 of NΔ105 represent NEM-sensitive sites when present as independent domains. In the context of WT M-MuLV IN (Fig. 4B, lane 5), NEM alkylation also caused a complete loss of function with the untailed dumbbell substrate (Fig. 4B, lane 6).
Residue C209 of M-MuLV IN influences the retrovirus life cycle in vivo.
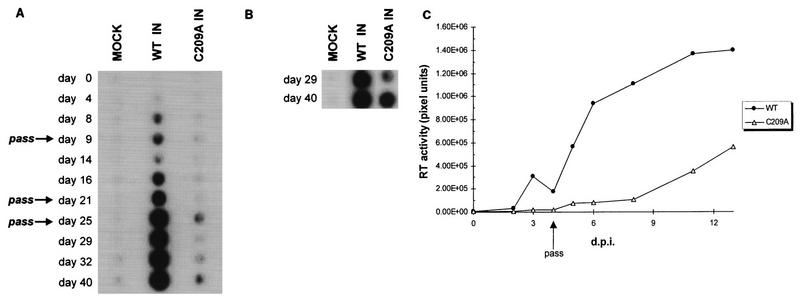
The results of NEM alkylation on multimeric M-MuLV IN function in vitro prompted an investigation of its importance to viral replication in vivo. M-MuLV proviral DNA clones (pNCA-C [11, 20]) that encoded either WT or C209A IN proteins were transiently transfected into Rat-1 cells by the DEAE-dextran method (36). Rat-1 cells were maintained in 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine calf serum (HyClone), 2 mM glutamine, 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 100 μg of gentamycin sulfate/ml. Viral propagation was measured over time by assaying supernatant medium for reverse transcriptase (RT) activity (21). In comparison to WT M-MuLV, C209A IN virion production was delayed 2 to 3 weeks (Fig. 5A, WT and C209A). RT activity levels of the C209A IN producer cells remained low throughout the time course. This implied that the C209A mutant virus was infecting cells extremely poorly and was hindered by dilution of the low level of infected cells during passage. To test this possibility, plates from passage on day 21 were maintained through day 40 with changes of the medium every 1.5 weeks and assayed for viral spread. Under these conditions, the C209A IN-infected cells showed increasing RT activity levels on days 29 and 40 (Fig. 5B). No virus was detected in the control cells (Fig. 5A and B, MOCK), and the WT maintained the high level of viral release under these conditions.
FIG. 5.
In vivo analysis of the effects of the C209A IN mutation on viral replication. (A) Time course of viral RT release into medium by Rat-1 producer cells. Days posttransfection are indicated on the left. Passages of the transfected producer cell line are indicated by arrows. (B) Accumulation of C209A viral RT in medium of plates maintained from passage on day 21 through day 40. (C) Time course of viral spread in Rat-1 cells. WT and C209A (pNCA-c/C209A IN) viruses collected at 40 days posttransfection were normalized for RT activity and used to infect fresh Rat-1 cells. RT activity was quantitated on a Molecular Dynamics PhosphorImager (Sunnyvale, Calif.). The pixel densities for each time point during the infection represent relative RT activity and are indicated on the y axis. The drop in RT activity at day 4 was due to passage of the infected cell lines. d.p.i., days postinfection.
The delayed spread of the viral C209A IN mutant suggested a defect in replication kinetics affected by the C209A IN mutation. Supernatants from the WT and C209A IN producer cells at 40 days postinfection were normalized with respect to RT activity levels and used to infect fresh Rat-1 cells to address this possibility. The kinetics of C209A IN viral spread showed a 1.5- to 2-week delay compared to WT M-MuLV (Fig. 5C). To confirm that the phenotype of the C209A IN virus was not due to reversion or second-site mutations, low-molecular-weight viral DNA was isolated from infected cells (22), and the full IN coding region was amplified by PCR (data not shown). Sequencing of the 1.2-kb IN coding region confirmed that the C209A mutation was retained in the producer cell virions at 40 days postinfection (data not shown). Though a single nucleotide change was found, this was in a codon wobble position which did not affect the amino acid sequence of the IN protein (ACT→ACG, Thr112).
Immunoprecipitation of C209A M-MuLV IN viral particles produced in the RT time course (Fig. 5B) indicated that normal levels of proteins (i.e., CA, MA, TM, SU, RT, and IN) are incorporated into the C209A M-MuLV IN virions (data not shown). Similarly, Southern blot analysis of unintegrated DNA indicated the levels of linear viral DNA from C209A were similar to those of the WT virus at days 2 and 4 (data not shown). PCR amplification of C209A and WT M-MuLV regions of pol occurred with equivalent levels of input Hirt DNA. These results indicate that the replication defect probably does not reflect aberrant levels of viral protein synthesis, protein incorporation into mature viral particles, or subsequent viral DNA synthesis (32, 40). Taken together with the in vitro data, the replication-defective phenotype of the C209A IN virus suggests that residue C209 may affect in vivo multimeric IN functions that are important for coordinated integration (37).
Model for assembly of IN multimers.
The in vitro integration and unimolecular and bimolecular disintegration reactions mediated by M-MuLV IN have been shown to be subtly influenced by substrate determinants and domains of the IN protein (13, 14, 24, 25). Thus, the assembly of different productive IN-DNA complexes may be achieved through complicated and distinct multimeric IN-IN and IN-DNA interactions. The combined approach of complementation and chemical modification of M-MuLV IN mutants in each of the in vitro reactions was used to investigate these different states of IN function.
A model for the assembly of functional multimers for each of the integration and disintegration substrates is presented in Fig. 6. Two symmetric dimers of M-MuLV IN bind independent LTRs in a fashion that is analogous to how one holds a pen: the thumb, index finger, and middle finger each contribute to the mechanism and stability of the grasp. In the working model, each of these stabilizing factors are represented by the HHCC domain of IN, the CA dinucleotide of the LTR, and the LTR 5′-ss tail. Elimination of one or more of these factors would cripple the stability of LTR binding to various degrees. This is exemplified in Fig. 4, where unimolecular disintegration of a 5′-untailed substrate is dependent on the HHCC domain.
FIG. 6.
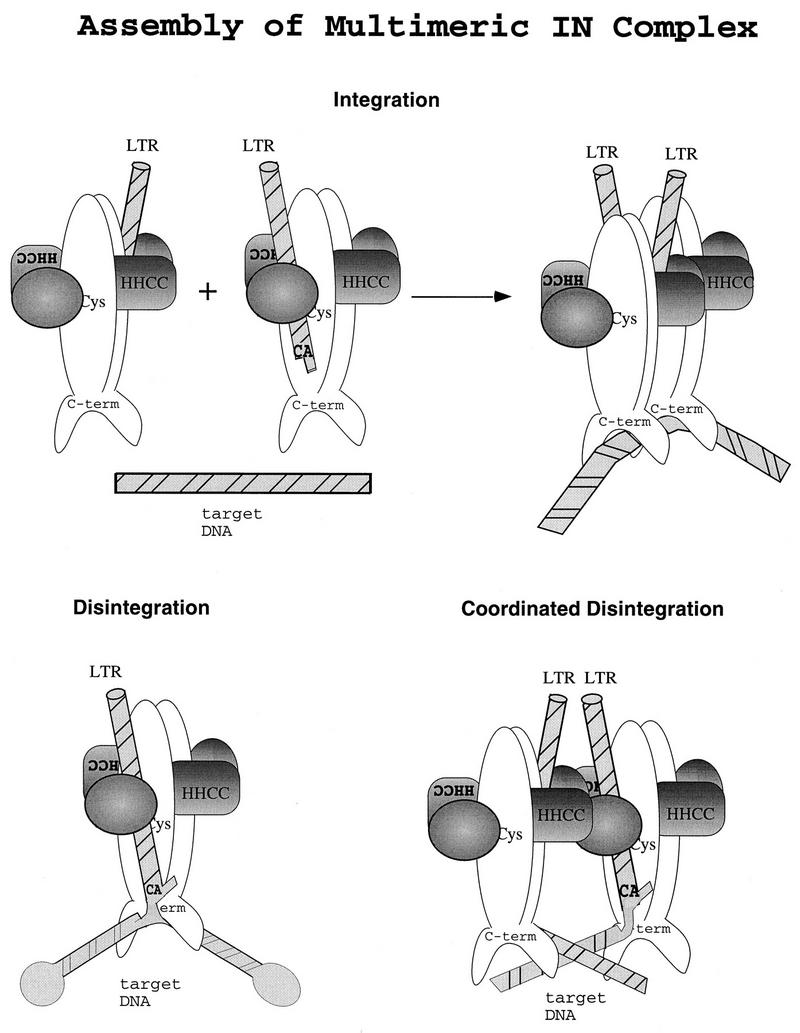
Model of multimeric M-MuLV IN functions in vitro and in vivo. An illustration of M-MuLV IN domains and multimeric subunit interactions is shown. A dimeric interface is formed by parallel association of M-MuLV IN monomers, reflecting the structure studies of the catalytic core and C terminus of human immunodeficiency virus type 1 and avian sarcoma virus IN (5, 15, 33). DNA is depicted as striped bars. The N terminus of M-MuLV IN through the HHCC region is shaded. (Top) LTR coordination and multimeric assembly of subunits for integration. (Bottom) Substrate coordination and subunit interactions for unimolecular disintegration (left) and bimolecular coordinated disintegration (right).
We have previously demonstrated that under nonsaturating conditions, coordinated disintegration mediated by the HHCC-deleted NΔ105 protein was strongly dependent on trans-subunit interactions promoted by an independent HHCC domain (13). The model presented here proposes that coordinated IN activity is arbitrated by an interaction of the HHCC domains at the multimerization interface, in the vicinity of the C209. Two LTR strand transfer and coordinated disintegration would be dependent on this assembly, whereas the unimolecular disintegration substrate is essentially “preassembled,” with the attacking nucleophile position maintained by the substrate structure, and may be supported by a minimal protomeric subunit.
The sensitivity of the C209 and the HHCC domain to NEM modification can be visualized in this model. Both the C209 and the HHCC region participate in the productive assembly of multimeric IN-IN complexes. For unimolecular disintegration, the structural arrangement of the attacking target 3′-OH and scissile bond diminishes any steric constraints that could be imposed by NEM alkylation. These would only be visible under conditions of suboptimal LTR recognition, such as the absence of the 5′-ss tail. C209 is not essential for catalysis; the C209A full-length protein as well as the C209A-NΔ105 truncated protein can catalyze single LTR strand transfer (24). The addition of the large maleimide group onto the C209 blocks IN-DNA and/or IN-IN interactions required for the strand transfer and coordinated disintegration reactions.
While NEM treatment of the core C209 site greatly reduced catalytic activities of the M-MuLV IN proteins, variation was detected in the sensitivity of the HHCC domain to NEM. The N-terminal HHCC domain may represent multiple functional domains. CΔ232 consists of the N-terminal 176 amino acids of M-MuLV IN, of which the HHCC structure per se only accounts for 41 amino acids. For M-MuLV, the N terminus of IN contains an additional 56 amino acids which are not conserved among related retroviral IN proteins. For HIV-1, the nuclear magnetic resonance solution structure (7) identified the dimer interface within the HHCC domain. For M-MuLV, an HHCC construct smaller than CΔ232 is a dimer as well (51). The IN constructs which are catalytically active were not renatured in the presence of exogenously added zinc. Preliminary data (32a) indicate an inverse relationship between NEM sensitivity and zinc coordination. The absence of zinc may also explain the inability of the HHCC domain to complement LTR hydrolysis in the coordinated disintegration reactions. It is possible that NEM may inhibit one function of the HHCC, such as LTR positioning, while maintaining a second function, such as dimerization. Additional studies to separate these functions are under way.
Acknowledgments
This work was supported by American Cancer Society grant RPG-95-056-03-VM and NSF-International Program NSF-INT-9408501/Fundacion ANDES (travel grant). G.A.D. was supported by NIH training grant 5T32AI07043.
REFERENCES
- 1.Andrake M D, Skalka A M. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 2.Andrake M D, Skalka A M. Retroviral intregrase, putting the pieces together. J Biol Chem. 1996;271:19633–19636. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 3.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J Mol Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 6.Bushman F D. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronengorn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 8.Chow S A, Brown P O. Juxtaposition of two viral DNA ends in a bimolecular disintegration reaction mediated by multimers of human immunodeficiency virus type 1 or murine leukemia virus integrase. J Virol. 1994;68:7869–7878. doi: 10.1128/jvi.68.12.7869-7878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow S A, Brown P O. Substrate features important for recognition and catalysis by human immunodeficiency virus type 1 integrase identified by using novel DNA substrates. J Virol. 1994;68:3896–3907. doi: 10.1128/jvi.68.6.3896-3907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 11.Colicelli J, Goff S P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985;42:573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- 12.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 13.Donzella G A, Jonsson C B, Roth M J. Coordinated disintegration reactions mediated by Moloney murine leukemia virus integrase. J Virol. 1996;70:3909–3921. doi: 10.1128/jvi.70.6.3909-3921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donzella G A, Jonsson C B, Roth M J. Influence of substrate structure on disintegration activity of Moloney murine leukemia virus integrase. J Virol. 1993;67:7077–7087. doi: 10.1128/jvi.67.12.7077-7087.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 16.Ellison V, Brown P O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellison V, Gerton J, Vincent K A, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 18.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 20.Felkner R H, Roth M J. Mutational analysis of N-linked glycosylation sites of the SU protein of Moloney murine leukemia virus. J Virol. 1992;66:4258–4264. doi: 10.1128/jvi.66.7.4258-4264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goff S P, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants; use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–371. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 23.Jones K S, Coleman J, Merkel G W, Laue T M, Skalka A M. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992;287:16037–16040. [PubMed] [Google Scholar]
- 24.Jonsson C B, Donzella G A, Gaucan E, Smith C M, Roth M J. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J Virol. 1996;70:4585–4597. doi: 10.1128/jvi.70.7.4585-4597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsson C B, Roth M J. Role of the His-Cys finger of Moloney murine leukemia virus integrase protein in integration and disintegration. J Virol. 1993;67:5562–5571. doi: 10.1128/jvi.67.9.5562-5571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalpana G V, Goff S P. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 28.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 29.Katzman M, Katz R, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulkosky J, Skalka A M. HIV DNA integration: observations and inferences. J Acquired Immune Defic Syndr. 1990;3:839–851. [PubMed] [Google Scholar]
- 32.Leavitt A D, Robles G, Alesandro N, Varmus H E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Leon, Oscar. Unpublished data.
- 33.Lodi P L, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Grononborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 34.Lutzke R A, Vink C, Plasterk R H A. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazumder A, Engelman A, Craigie R, Fesen M, Pommier Y. Intermolecular disintegration and intramolecular strand transfer activities of wild-type and mutant HIV-1 integrase. Nucleic Acids Res. 1994;22:1037–1043. doi: 10.1093/nar/22.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCutchan J H, Pagano J S. Enhancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968;41:351–357. [PubMed] [Google Scholar]
- 37.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pemberton I K, Buckle M, Buc H. The metal ion-induced cooperative binding of HIV-1 integrase to DNA exhibits a marked preference for Mn(II) rather than Mg(II) J Biol Chem. 1996;271:1498–1506. doi: 10.1074/jbc.271.3.1498. [DOI] [PubMed] [Google Scholar]
- 39.Roth M J. Mutational analysis of the carboxyl terminus of the Moloney murine leukemia virus integration protein. J Virol. 1991;65:2141–2145. doi: 10.1128/jvi.65.4.2141-2145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth M J, Schwartzberg P, Tanese N, Goff S P. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J Virol. 1990;64:4709–4717. doi: 10.1128/jvi.64.10.4709-4717.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 42.VanDenEnt F M I, Vink C, Plasterk R H A. DNA substrate requirements for different activities of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:7825–7832. doi: 10.1128/jvi.68.12.7825-7832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.vanGent D C, Groeneger A A M O, Plasterk R H A. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.vanGent D C, Vink C, Groeneger A A M O, Plasterk R H A. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varmus H E, Brown P O. Retroviruses. In: Howe M, Berg D, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 53–108. [Google Scholar]
- 46.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vink C, Groeneger A A M O, Plasterk R H A. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type 1 integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vink C, Lutzke R A, Plasterk R H A. Formation of a stable complex between the human immunodeficiency virus integrase protein and viral DNA ends. Nucleic Acids Res. 1994;22:4103–4110. doi: 10.1093/nar/22.20.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woerner A M, Klutch M, Levin J G, Marcus-Sekura C J. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992;8:297–304. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 50.Woerner A M, Marcus-Sekura C J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993;21:3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, F., O. Leon, and M. J. Roth. Unpublished data.
- 52.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]