Abstract
We used gene expression profiling to investigate whether the molecular effects induced by estrogens of different provenance are intrinsically similar. In this article we show that the physiologic estrogen 17β-estradiol, the phytoestrogen genistein, and the synthetic estrogen diethylstilbestrol alter the expression of the same 179 genes in the intact immature mouse uterus under conditions where each chemical has produced an equivalent gravimetric and histologic uterotrophic effect, using the standard 3-day assay protocol. Data are also presented indicating the limitations associated with comparison of gene expression profiles for different chemicals at times before the uterotrophic effects are fully realized. We conclude that the case has yet to be made for regarding synthetic estrogens as presenting a unique human hazard compared with phytoestrogens and physiologic estrogens.
Keywords: diethylstilbestrol, estrogen, gene expression, genistein, microarray, phytoestrogen, toxicogenomics, uterus
The question of whether phytoestrogens and synthetic estrogens are toxicologically similar, or intrinsically different, presents a challenge to all involved in human hazard and risk assessments. Although there is a general concern that exposure to nanogram or microgram amounts of environmental estrogens may be associated with adverse health effects, in the public mind there is a widespread belief that foods and dietary supplements containing milligram quantities of phytoestrogens confer only health benefits. An implicit distinction therefore seems to have been drawn between synthetic and plant-derived estrogens—a belief sustained in the public mind by the assumption that natural is good and synthetic is bad—but an untested and potentially misleading notion for those involved with science-based human hazard/risk assessments.
Phytoestrogens and synthetic estrogens are generally considered separately in the literature. For example, Howdeshell et al. (1999) suggested a possible association between the advance in first estrus observed in mice exposed in utero to 2.4 μg/kg of the synthetic environmental estrogen bisphenol A and reports of an increased incidence of hypospadias in boys (Paulozzi et al. 1997) and the earlier sexual maturation of girls (Herman-Giddens et al. 1997)—the implication being that synthetic estrogens present a greater hazard than the much higher levels of phytoestrogens being consumed by those same children. In contrast, there are reports of an increased incidence of hypospadias in boys born to vegetarians (North and Golding 2000), of alterations in the menstrual cycle (Cassidy et al. 1994), and of reduced breast cancer incidences (Messina 1999) among women eating diets rich in phytoestrogens. Support for these epidemiologic observations comes from experimental studies indicating that advances in sexual development in rodents can be induced by their exposure to phytoestrogens (Casanova et al. 1999; Cassidy and Faughnan 2000; Safe et al. 2002). In contrast to these separate lines of inquiry, Newbold and colleagues have evaluated potential similarities between natural and synthetic estrogens. In seminal studies, they demonstrated that neonatal exposure of female mice to equipotent uterotrophic doses of the phytoestrogen genistein (GEN; Figure 1) or the synthetic estrogen diethylstilbestrol (DES) leads to an identical incidence of uterine adenomas at 18 months of age (Newbold et al. 2001). However, in attempting to draw parallels, or distinctions, between phytoestrogens and synthetic estrogens, it is imperative to consider growing awareness of the complexity of estrogen signaling pathway and the pleuripotential biologic activities of most organic chemicals—irrespective of their origin.
Figure 1. Chemical structure of GEN, E2, and DES.
Estrogen signaling in mammalian cells is primarily mediated at the molecular level by two members of the nuclear receptor superfamily—estrogen receptors alpha (ER-α) and beta (ER-β). Ligand-activated ER-α and ER-β function as transcription factors, in conjunction with numerous coregulatory proteins, in order to activate or repress the transcription of ER-responsive genes (Hall et al. 2001; Moggs and Orphanides 2001). There is considerable variation in the binding affinity of ER-α and ER-β among different estrogens (Kuiper et al. 1998). In the case of the chemicals studied here, the physiologic estrogen 17β-estradiol (E2) and DES bind with a similar affinity to ER-α and ER-β, whereas GEN binds with approximately 20-fold higher affinity to ER-β than to ER-β (Kuiper et al. 1998). Concerning nonhormonal properties of the test chemicals (most of which have only be defined in vitro), GEN inhibits a range of enzymes, including tyrosine kinases (Akiyama et al. 1987), nitric oxide synthase (Duarte et al. 1997), and topoisomerase II (Okura et al. 1988), and also decreases calcium-channel activity (Potier and Rovira 1999), lipid peroxidation (Arora et al. 1998), and diacylglycerol synthesis (Dean et al. 1989). Likewise, DES is reported to induce aneuploidy in mammalian cells (Aardema et al. 1998) and to bind to rat liver DNA (Williams et al. 1993). More recently, some phytoestrogens were reported to inhibit the aromatase-mediated conversion of testosterone to E2 in vitro (Almstrup et al. 2002), and equol, the major circulating estrogenic metabolite associated with the dietary ingestion of phytoestrogens, is reported to selectively sequester dihydrotestosterone and thereby to act as a functional antiandrogen in vivo (Lund et al. 2004).
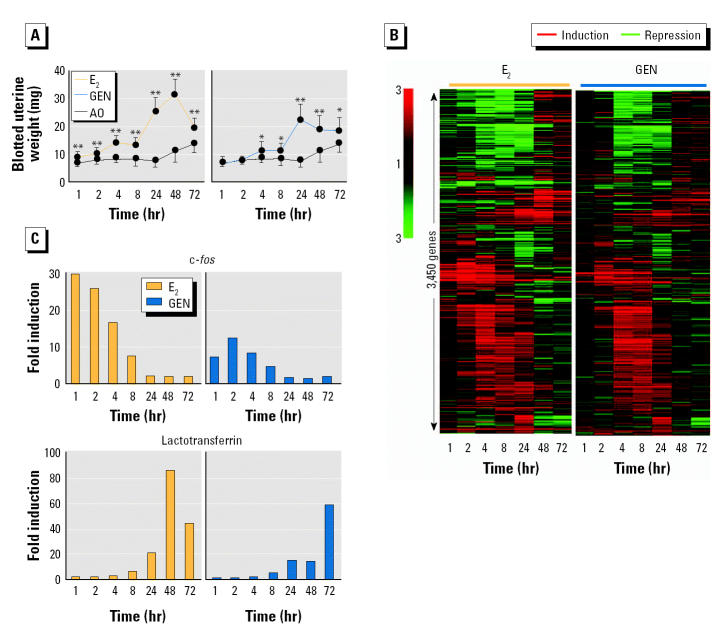
In order to advance understanding in this area, we decided to compare the genes expressed in the immature mouse uterus when it had grown in response to treatment with the estrogens E2, DES, and GEN. The immature mouse uterus was selected for our analysis because it is a major estrogen-responsive organ and forms the basis for a reference assay of estrogenic activity (Owens and Ashby 2002), including carcinogenesis (Newbold et al. 2001). Furthermore, it expresses both ER-α and ER-β (Weihua et al. 2000) and the androgen receptor (Frasor et al. 2003). We initially conducted a global analysis of gene expression in the mouse uterus at 1, 2, 4, 8, 24, 48, and 72 hr after exposure to a single high dose of either GEN (250 mg/kg) or E2 (400 μg/kg). These single high doses yielded a sustained uterotrophic response over 72 hr (Figure 2A) and were selected to avoid the complex transcriptional program that may result from the standard uterotrophic assay exposure regime in which each test compound is dosed by repeated administration on 3 consecutive days (Odum et al. 1997). Groups of 10 sexually immature mice [Alpk:APfCD-1; 19/20 days of age; maintained on RM1 diet (Special Diets Services Ltd., Witham, Essex, UK)] received a single subcutaneous injection of each compound or the test vehicle [arachis oil (AO); 5 mL/kg], and uterine RNA was isolated and pooled by group at each of the seven time points to determine gene expression levels among the 12,488 mouse genes represented on the Affymetrix MG-U74Av2 GeneChip (Affymetrix, High Wycombe, UK). Transcript profiling was performed using MG-U74Av2 GeneChip and Microarray Analysis Suite 5.0 (Affymetrix). Normalization and hierarchical clustering were performed with GeneSpring 6.0 (Silicon Genetics, Redwood City, CA, USA). MIAME (Minimum Information About a Microarray Experiment)-compliant microarray data are available as supplementary information and submitted to the Gene Expression Omnibus (GEO) database (GEO 2004). These data were analyzed using unsupervised hierarchical clustering and yielded temporal relationships between the expression profiles of 3,450 genes that were either up- or down-regulated (> 1.5-fold) by E2 and/or GEN (Figure 2B). Each chemical induced a similar, multistage transcriptional response (Figure 2B), although it is noteworthy that we observed variations in the magnitude and timing of both early (e.g., c-fos) and late (e.g., lactotransferrin) ER-responsive genes during the uterotrophic responses induced by E2 and GEN (Figure 2C).
Figure 2. Induction of very similar multistage transcriptional responses in the mouse uterus by E2 and GEN. (A) Blotted uterine weights (mean ± SD) of sexually immature mice (n = 10/group) at different times after a single subcutaneous dose of E2 (400 μg/kg), GEN (250 mg/kg), or AO (control; 5 mL/kg). See text for details of experiments. (B) Temporal expression profiles of 3,450 genes up-regulated or repressed (> 1.5-fold) by either E2 (400 μg/kg) or GEN (250 mg/kg) at one or more of seven different time points. The magnitude of altered gene expression (fold change vs. time-matched vehicle control) is indicated by color; genes are grouped according to similarity of their temporal expression profiles (Pearson correlation-based hierarchical clustering). (C) Northern blot analysis of temporal expression pattern of early (c-fos) and late (lactotransferrin) estrogen-responsive genes; the fold induction of gene expression relative to time-matched vehicle controls was calculated after data were normalized to the expression of the control gene RPB1 (accession number NM_009089). : *p < 0.05; : **p < 0.01; two-sided Student t-test.
A detailed description of the molecular functions of the genes affected, together with their association with physiologic changes during uterine growth, has been reported (Orphanides et al. 2003) and will be described in more detail in a future publication (Moggs et al., unpublished data).
These observations suggest that GEN does not induce “off-target” ER-independent transcriptional responses, that is, those associated with the properties of GEN other than estrogenicity. Furthermore, there was no evidence for the topoisomerase II–inhibiting properties of GEN in the bone marrow of the present mice despite demonstration of the sensitivity of that tissue to the potent micronucleus-inducing activity of the topoisomerase II inhibitor etoposide (data not shown). Together, these data led us to question whether a synthetic estrogen such as DES would also induce similar transcriptional responses in the immature mouse uterus.
In order to avoid temporal vagaries in gene expression (e.g., Figure 2C), we decided to anchor our transcript profiling data to the phenotype of the grown uterus by employing equipotent uterotrophic doses of E2, GEN, and DES. We compared the global gene expression profiles in the uteri of intact immature mice stimulated with three daily low doses of either GEN, DES, or E2, with an exposure regimen the same as that used in a standard 3-day uterotrophic assay (Odum et al. 1997). The route of administration and the doses of GEN and DES used were as described by Newbold et al. (2001) in their equivalent-outcome carcinogenicity bioassays of these two chemicals. Three independent replicates of four groups of sexually immature mice (Alpk:APfCD-1; 19/20 days of age; maintained on RM1 diet) received three daily subcutaneous injections of GEN (50 mg/kg), E2 (2.5 μg/kg), or DES (2 μg/kg). Control animals received the vehicle, AO (5 mL/kg). These doses elicited similar uterotrophic responses (72 hr after the initial dose; Figure 3A, Table 1) and identical histologic changes in the uteri of the treated animals (Table 1). Uterine RNA was isolated and pooled for each of the 12 groups and analyzed for changes in gene expression levels using the same Affymetrix microarray of 12,488 mouse genes. The data were analyzed using two independent statistical methods. First, unsupervised hierarchical clustering defined the global relationships (Euclidean distances) between the 12 gene expression profiles (Figure 3B). The three control groups clustered under one node, whereas the chemical treatment groups formed a separate node of compound-independent clusters, indicating equal similarity within and between the transcriptional responses induced by the three estrogens (Figure 3B). One-way analysis of variance (ANOVA), with Bonferroni (Holm 1979) correction (familywise error rate < 0.05) to minimize false positives, identified 179 genes where expression levels were altered by one or more chemical treatments (Figure 3C). Remarkably, Tukey post hoc testing revealed that all of these genes were affected in all nine compound treatment groups.
Figure 3. Equivalence of biologic responses induced in the mouse uterus by E2 (E), GEN (G), and DES (D). (A) Blotted uterine weights (mean ± SD) of three independent replicate groups (1–3) of sexually immature mice (n = 4/group) after three daily subcutaneous injections of either GEN (50 mg/kg), E2 (2.5 μg/kg), DES (2 μg/kg), or AO [control (C); 5 mL/kg]. (B) Unsupervised Euclidean-distance–based hierarchical clustering of 4,134 expressed genes. (C) Near-identical gene expression profiles induced by the three estrogens 72 hr after equipotent uterotrophic doses. Significant changes in gene expression induced by one or more of the three estrogens were identified by one-way ANOVA (parametric test, assuming equal variance). The magnitude of altered gene expression (fold change vs. vehicle control) is indicated by color.
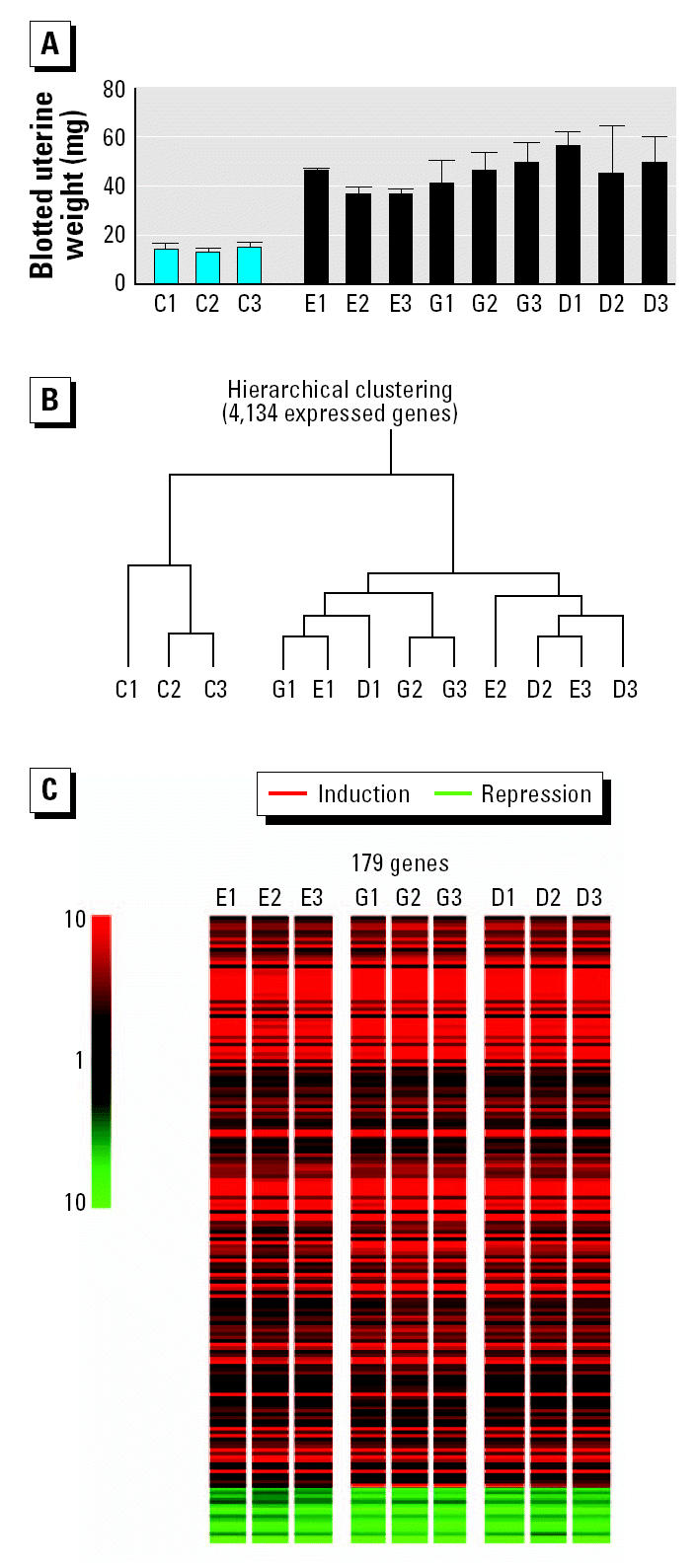
Table 1.
Blotted uterine weights and endometrial and epithelial cell heights (mean ± SD) after exposure to E2, GEN, or DES for 3 consecutive days.a
| Cell height (μm)
|
||||
|---|---|---|---|---|
| Compound | Dose (per kg) | Blotted uterine weight (mg) | Endometrium | Epithelium |
| AO | 5 mL | 13.0 ± 2.4 | 159.0 ± 23.1 (11) | 11.4 ± 1.1 |
| E2 | 2.5 μg | 45.3 ± 8.6* | 246.1 ± 52.4* (9) | 23.3 ± 1.4* |
| GEN | 50 mg | 39.8 ± 5.3* | 273.7 ± 63.3* (12) | 23.7 ± 3.1* |
| DES | 2.0 μg | 49.8 ± 13.0* | 273.2 ± 55.9* (10) | 22.6 ± 4.0* |
There were 12 animals/group, but not all of the histopathology samples were suitable for analyses; numbers in parentheses indicate the number of animals per group from which the histology data were generated.
Data were assessed for statistical significance using a two-sided Student t-test:
p < 0.01.
Table 2 highlights the high degree of similarity between the transcriptional responses to each of the three estrogens. These include established estrogen-responsive genes such as lactotransferrin, complement component 3, c-fos, small proline-rich protein 2A, and keratoepithelin (Hewitt et al. 2003; Naciff et al. 2003), together with many genes that have not previously been associated with estrogenicity (Table 2).
Table 2.
Quantitative data for 179 differentially expressed genes (from Figure 3C) regulated in the mouse uterus by all three estrogens (E2, GEN, and DES).a
| Fold change in expression (mean ± SD)
|
||||
|---|---|---|---|---|
| Gene name | GenBank accession no. | E2 | GEN | DES |
| Up-regulated genes | ||||
| Solute carrier family 9a3r1 | U74079 | 1.8 ± 0.01 | 2.0 ± 0.1 | 2.0 ± 0.2 |
| Keratin complex 2–8 | X15662 | 2.6 ± 0.2 | 3.1 ± 0.2 | 3.1 ± 0.3 |
| Laminin beta 3 | U43298 | 4.3 ± 0.1 | 5.5 ± 1.1 | 5.3 ± 0.7 |
| Claudin 7 | AF087825 | 4.5 ± 0.5 | 6.5 ± 1.0 | 5.8 ± 0.6 |
| bHLH-Zip transcription factor | U49507 | 2.6 ± 0.3 | 3.1 ± 0.3 | 2.9 ± 0.1 |
| RIKEN cDNA 1200008D14 | AW208938 | 3.0 ± 0.3 | 3.5 ± 0.1 | 3.3 ± 0.3 |
| Basic HLH-domain containing, class B2 | Y07836 | 5.9 ± 1.0 | 6.6 ± 0.9 | 6.6 ± 0.8 |
| RIKEN cDNA 9930104H07 | AW122310 | 3.0 ± 0.3 | 3.2 ± 0.4 | 3.3 ± 0.1 |
| Fucosyltransferase 2 | AF064792 | 27.5 ± 1.2 | 34.6 ± 8.5 | 36.7 ± 5.5 |
| Deleted in polyposis 1 | U28168 | 1.8 ± 0.1 | 2.0 ± 0.02 | 2.0 ± 0.1 |
| Microsomal glutathione S-transferase 3 | AI843448 | 2.9 ± 0.2 | 3.3 ± 0.6 | 3.3 ± 0.1 |
| Tumor-associated Ca signal transducer 2 | Y08830 | 4.0 ± 0.3 | 4.6 ± 0.9 | 4.6 ± 0.3 |
| Calpain 5 | Y10656 | 5.5 ± 0.4 | 6.3 ± 1.0 | 6.6 ± 0.6 |
| Mitochondrial creatine kinase | Z13969 | 9.7 ± 1.1 | 12.2 ± 2.1 | 13.1 ± 1.8 |
| ATPase 6v1a1 | AW123765 | 2.0 ± 0.1 | 2.1 ± 0.2 | 2.1 ± 0.2 |
| Tumor-associated Ca signal transducer 2 | AI563854 | 8.0 ± 0.4 | 9.2 ± 1.0 | 8.5 ± 0.4 |
| Lymphocyte antigen 6 complex, locus A | X04653 | 7.8 ± 0.9 | 8.8 ± 0.3 | 8.5 ± 0.4 |
| Chloride channel calcium-activated 3 | AV373378 | 26.4 ± 3.4 | 26.7 ± 1.0 | 26.2 ± 3.9 |
| Small proline-rich protein 2I | AJ005567 | 23.9 ± 1.5 | 24.7 ± 1.3 | 23.6 ± 1.6 |
| Oncoprotein induced transcript 1 | AA615075 | 19.0 ± 3.1 | 20.0 ± 1.2 | 18.9 ± 2.5 |
| Small proline-rich protein 2F | AJ005564 | 59.8 ± 8.4 | 65.8 ± 1.1 | 60.6 ± 2.6 |
| Small proline-rich protein 2E | AJ005563 | 12.0 ± 1.0 | 12.9 ± 0.8 | 12.1 ± 0.9 |
| Mucin 1 | M84683 | 8.3 ± 0.6 | 8.6 ± 0.3 | 8.5 ± 0.5 |
| Lipoocalin 2 | X81627 | 150.3 ± 15.0 | 175.7 ± 10.5 | 162.8 ± 6.5 |
| RIKEN cDNA 2210409B01 | AF109906 | 3.5 ± 0.6 | 4.0 ± 0.3 | 3.8 ± 0.8 |
| Interferon-activated gene 202A | M31418 | 7.9 ± 1.0 | 9.8 ± 2.5 | 8.8 ± 0.7 |
| Nuclear ankyrin-repeat protein | AA614971 | 3.7 ± 0.6 | 4.3 ± 0.7 | 4.1 ± 0.9 |
| RIKEN cDNA 5730469M10 | AI850090 | 22.0 ± 5.8 | 30.5 ± 9.1 | 27.0 ± 6.5 |
| RIKEN cDNA 1110034C02 | AI837104 | 1.5 ± 0.1 | 1.6 ± 0.1 | 1.6 ± 0.03 |
| IMAGE cDNA 4988271 | AV373294 | 8.0 ± 2.5 | 10.6 ± 1.6 | 9.2 ± 1.1 |
| RIKEN cDNA 5730493B19 | AW122413 | 12.7 ± 0.3 | 19.0 ± 4.1 | 15.7 ± 0.9 |
| Peptidoglycan recognition protein | AV092014 | 13.4 ± 1.5 | 18.3 ± 3.0 | 14.5 ± 2.1 |
| Inhibin beta-B | X69620 | 13.6 ± 3.1 | 19.4 ± 4.7 | 16.0 ± 1.4 |
| CEA-related cell adhesion molecule 2 | AF101164 | 11.9 ± 1.6 | 17.8 ± 5.3 | 14.2 ± 1.7 |
| Keratin complex 1–19 | M36120 | 4.4 ± 0.4 | 5.5 ± 1.1 | 4.8 ± 0.5 |
| CEA-related cell adhesion molecule 1 | M77196 | 15.9 ± 2.4 | 23.9 ± 5.9 | 19.0 ± 3.8 |
| SRC family-associated phosphoprotein 2 | AB014485 | 2.7 ± 0.04 | 3.2 ± 0.4 | 2.9 ± 0.3 |
| Peptidoglycan recognition protein | AF076482 | 7.7 ± 1.9 | 10.4 ± 2.7 | 9.0 ± 2.0 |
| CEA-related cell adhesion molecule 1 | M77196 | 19.5 ± 3.9 | 30.4 ± 8.8 | 22.2 ± 2.7 |
| CEA-related cell adhesion molecule 1 | X67279 | 6.4 ± 0.7 | 8.4 ± 1.1 | 7.1 ± 1.1 |
| Spermidine N1-acetyl transferase | L10244 | 8.3 ± 0.9 | 11.2 ± 0.8 | 9.3 ± 0.6 |
| RIKEN cDNA 0610007O07 | AI851762 | 2.7 ± 0.1 | 3.0 ± 0.3 | 2.8 ± 0.1 |
| Arginase 1 | U51805 | 79.4 ± 9.8 | 131.9 ± 20.0 | 99.6 ± 14.5 |
| Acetyl-coenzyme A synthetase 2 | AW125884 | 2.2 ± 0.2 | 2.0 ± 0.1 | 2.2 ± 0.2 |
| v-erb-b2 homolog 3 | AI006228 | 3.4 ± 0.4 | 3.1 ± 0.6 | 3.4 ± 0.4 |
| Phospholipase D3 | AF026124 | 2.6 ± 0.2 | 2.4 ± 0.2 | 2.6 ± 0.2 |
| RIKEN cDNA 0610031J06 | AW122935 | 1.9 ± 0.1 | 1.8 ± 0.1 | 1.8 ± 0.1 |
| Complement component 1q | X58861 | 2.1 ± 0.1 | 2.0 ± 0.1 | 2.0 ± 0.2 |
| Scotin | AW123754 | 2.0 ± 0.1 | 2.0 ± 0.2 | 2.0 ± 0.2 |
| CD24a antigen | M58661 | 3.2 ± 0.1 | 3.1 ± 0.1 | 3.3 ± 0.3 |
| Argininosuccinate synthetase 1 | M31690 | 2.7 ± 0.3 | 2.7 ± 0.3 | 2.8 ± 0.4 |
| ATPase 6v1a1 | U13837 | 2.1 ± 0.1 | 2.1 ± 0.2 | 2.2 ± 0.2 |
| Gelsolin-like actin-capping protein | X54511 | 3.6 ± 0.5 | 3.7 ± 0.5 | 3.7 ± 0.2 |
| Golgi phosphoprotein 2 | AW125446 | 4.5 ± 0.5 | 4.6 ± 0.5 | 4.7 ± 0.1 |
| Aldolase 1A | Y00516 | 2.3 ± 0.2 | 2.3 ± 0.1 | 2.4 ± 0.1 |
| Cathepsin L | X06086 | 6.4 ± 0.8 | 6.3 ± 1.1 | 6.9 ± 0.4 |
| CD14 antigen | X13333 | 3.0 ± 0.1 | 2.8 ± 0.1 | 3.1 ± 0.2 |
| Decay accelerating factor 2 | L41365 | 4.0 ± 0.1 | 3.8 ± 0.8 | 3.8 ± 0.2 |
| Actin-related protein 2/3 complex 1B | AW212775 | 2.1 ± 0.2 | 2.1 ± 0.1 | 2.1 ± 0.2 |
| Protective protein for β-galactosidase | J05261 | 2.0 ± 0.1 | 2.0± 0.1 | 2.0 ± 0.1 |
| Elastase 1 | M27347 | 2.7 ± 0.1 | 2.5 ± 0.2 | 2.6 ± 0.1 |
| Connexin 26 | M81445 | 10.6 ± 1.0 | 9.8 ± 0.5 | 10.4 ± 0.8 |
| Ceruloplasmin | U49430 | 15.1 ± 2.8 | 15.0 ± 5.5 | 14.5 ± 2.1 |
| Cathepsin H | U06119 | 3.0 ± 0.2 | 3.0 ± 0.3 | 3.0 ± 0.3 |
| Basigin | Y16258 | 1.6 ± 0.1 | 1.5 ± 0.1 | 1.7 ± 0.1 |
| Peptidylprolyl isomerase C–associated | X67809 | 2.2 ± 0.2 | 2.2 ± 0.1 | 2.4 ± 0.3 |
| Glutathione reductase 1 | AI851983 | 2.3 ± 0.2 | 2.3 ± 0.1 | 2.6 ± 0.3 |
| START domain–containing 3 | X82457 | 1.5 ± 0.1 | 1.4 ± 0.03 | 1.5 ± 0.01 |
| CD68 antigen | X68273 | 4.6 ± 0.6 | 4.2 ± 0.6 | 5.0 ± 0.7 |
| RIKEN cDNA E030027H19 | AW211760 | 2.7 ± 0.3 | 2.7 ± 0.2 | 2.9 ± 0.1 |
| cDNA sequence BC004044 | AI461767 | 3.1 ± 0.2 | 3.4 ± 0.1 | 3.8 ± 0.5 |
| E74-like factor 3 | AF016294 | 5.1 ± 0.8 | 5.9 ± 0.5 | 6.5 ± 0.5 |
| Glutathione S-transferase omega 1 | AI843119 | 5.0 ± 1.1 | 4.6 ± 0.6 | 4.1 ± 0.7 |
| Interferon-stimulated protein 20 | AW122677 | 4.2 ± 0.1 | 4.3 ± 0.7 | 3.4 ± 0.4 |
| Clusterin | D14077 | 3.6 ± 0.7 | 3.9 ± 0.9 | 3.4 ± 0.4 |
| Galectin 3 | X16834 | 7.4 ± 1.3 | 8.7 ± 0.7 | 6.9 ± 0.4 |
| Small proline-rich protein 2Ab | AJ005559 | 51.1 ± 4.0 | 78.3 ± 15.7 | 44.2 ± 5.2 |
| Complement component 3b | K02782 | 14.8 ± 1.8 | 18.8 ± 1.0 | 14.8 ± 0.8 |
| Small proline-rich protein 2C | AJ005561 | 220.3± 31.0 | 340.5 ± 37.1 | 214.1 ± 41.1 |
| Small proline-rich protein 2G | AJ005565 | 9.4 ± 0.9 | 11.0 ± 0.3 | 9.6 ± 0.6 |
| Prominin | AF039663 | 3.5 ± 0.5 | 3.6 ± 0.6 | 3.4 ± 0.3 |
| Lactotransferrinb | J03298 | 88.7 ± 18.4 | 99.2 ± 13.9 | 76.9 ± 21.8 |
| Carbonic anhydrase 2 | M25944 | 7.9 ± 0.5 | 8.2 ± 0.9 | 7.3 ± 0.4 |
| Complement component factor I | U47810 | 36.5 ± 4.4 | 38.4 ± 5.6 | 32.9 ± 4.2 |
| Mannosidase 2alphaB1 | U87240 | 2.0 ± 0.2 | 2.0 ± 0.1 | 1.9 ± 0.1 |
| Small proline-rich protein 2B | AJ005560 | 32.9 ± 3.7 | 39.2 ± 1.9 | 30.7 ± 5.0 |
| Small proline-rich protein 2Ab | AJ005559 | 269.8 ± 23.7 | 329.1 ± 42.9 | 59.1 ± 40.8 |
| RIKEN cDNA 5830413E08 | AI849939 | 3.3 ± 0.4 | 3.3 ± 0.5 | 3.0 ± 0.3 |
| RIKEN cDNA 1110029F20 | AW125508 | 4.1 ± 0.1 | 4.1 ± 0.4 | 3.7 ± 0.1 |
| Annexin A3 | AJ001633 | 2.7 ± 0.5 | 4.2 ± 0.7 | 3.2 ± 0.6 |
| Peptidase 4 | U51014 | 2.0 ± 0.1 | 2.9 ± 0.3 | 2.3 ± 0.2 |
| Laminin gamma 2 | U43327 | 6.3 ± 1.3 | 17.3 ± 6.8 | 10.2 ± 1.0 |
| Ubiquitin-like 3 | AW120725 | 1.5 ± 0.1 | 1.8 ± 0.1 | 1.7 ± 0.03 |
| Urate oxidase | M27695 | 23.8 ± 9.5 | 143.9 ± 62.9 | 43.8 ±17.7 |
| Amiloride binding protein 1 | AI197481 | 3.5 ± 1.0 | 10.1 ± 0.8 | 6.0 ± 1.2 |
| Keratin complex 1–19 | AU040563 | 4.5 ± 1.0 | 7.2 ± 1.0 | 5.6 ± 0.4 |
| Activated leukocyte cell adhesion molecule | L25274 | 3.6 ± 0.8 | 5.1 ± 0.9 | 4.4 ± 0.5 |
| CCAAT/enhancer binding protein β | M61007 | 2.3 ± 0.1 | 2.8 ± 0.3 | 2.6 ± 0.1 |
| Peptidyl arginine deiminase, type I | AB013848 | 8.6 ± 0.7 | 15.8 ± 3.1 | 12.0 ± 1.7 |
| Enolase 1 α | AI841389 | 2.5 ± 0.3 | 3.2 ± 0.5 | 3.0 ± 0.3 |
| p53 apoptosis effector related to Pmp22 | AI854029 | 2.9 ± 0.3 | 4.1 ± 0.8 | 3.7 ± 0.5 |
| β-Glucuronidase | M19279 | 1.9 ± 0.1 | 2.3 ± 0.2 | 2.2 ± 0.1 |
| Leucine-rich α-2-glycoprotein | AW230891 | 9.3 ± 1.1 | 17.6 ± 4.1 | 14.5 ± 2.6 |
| Quiescin Q6 | AW123556 | 3.7 ± 0.2 | 5.5 ± 1.2 | 4.8 ± 0.7 |
| GADD45a | U00937 | 1.9 ± 0.2 | 2.6 ± 0.3 | 2.3 ± 0.1 |
| Alkaline phosphatase 2 | J02980 | 9.2 ± 0.4 | 22.6 ± 6.2 | 15.8 ± 2.0 |
| Immediate early response 3 | X67644 | 5.5 ± 0.8 | 10.8 ± 2.2 | 8.9 ± 2.0 |
| Progressive ankylosis | AW049351 | 2.2 ± 0.1 | 2.9 ± 0.4 | 2.8 ± 0.4 |
| RAS p21 protein activator 4 | AA163960 | 6.8 ± 0.9 | 14.2 ± 2.5 | 12.1 ± 1.7 |
| Tumor-associated calcium signal transducer 1 | M76124 | 2.1 ± 0.2 | 2.7 ± 0.3 | 2.6 ± 0.2 |
| Hydroxysteroid (17-beta) dehydrogenase 11 | AA822174 | 1.9 ± 0.1 | 2.3 ± 0.3 | 2.4 ± 0.1 |
| Platelet-activating factor acetylhydrolase 1ba1 | U57746 | 1.9 ± 0.1 | 2.2 ± 0.2 | 2.3 ± 0.1 |
| Branched chain aminotransferase 1 | U42443 | 2.4 ± 0.2 | 3.4 ± 0.2 | 3.4 ± 0.01 |
| RIKEN cDNA 2400004E04 | AI846720 | 1.7 ± 0.1 | 2.4 ± 0.2 | 2.3 ± 0.2 |
| Myeloblastosis oncogene | M12848 | 2.8 ± 0.4 | 5.6 ± 1.1 | 5.0 ± 0.2 |
| K+ conductance calcium-activated channel N4 | AF042487 | 3.1 ± 0.2 | 4.2 ± 0.6 | 3.5 ± 0.8 |
| ATPase 6v1b2 | AI843029 | 1.7 ± 0.1 | 1.8 ± 0.1 | 1.8 ± 0.1 |
| Cystic fibrosis transmembrane regulator | M60493 | 3.4 ± 0.5 | 4.5 ± 0.8 | 3.9 ± 0.3 |
| RIKEN cDNA 1110008P14 | AI839839 | 4.3 ± 0.4 | 6.0 ± 1.0 | 5.1 ± 0.2 |
| Fused toes | Z67963 | 2.6 ± 0.2 | 3.2 ± 0.3 | 2.8 ± 0.1 |
| Solute carrier family 39a8 | AW124340 | 3.5 ± 0.5 | 4.5 ± 0.7 | 3.8 ± 0.3 |
| Cytochrome b-561 | AI846517 | 2.2 ± 0.2 | 2.5 ± 0.2 | 2.3 ± 0.2 |
| Secreted phosphoprotein 1 | X13986 | 30.2 ± 3.3 | 47.4 ± 7.4 | 31.1 ± 2.7 |
| Ion transport regulator Fxyd3 | X93038 | 5.3 ± 0.4 | 6.8 ± 1.1 | 5.5 ± 0.6 |
| Janus kinase 3 | L40172 | 2.1 ± 0.2 | 2.5 ± 0.2 | 2.2 ± 0.2 |
| Cytochrome b-245alpha | AW046124 | 2.9 ± 0.4 | 3.6 ± 0.4 | 2.9 ± 0.2 |
| RIKEN cDNA A430096B05 | AI465965 | 6.3 ± 1.0 | 8.6 ± 0.03 | 6.3 ± 0.6 |
| Small proline-rich protein 2J | AJ005568 | 8.6 ± 2.1 | 13.8 ± 3.2 | 8.5 ± 0.7 |
| Cathepsin B | M65270 | 2.2 ± 0.1 | 2.6 ± 0.2 | 2.2 ± 0.1 |
| RIKEN cDNA 1600025H15 | AI842734 | 2.2 ± 0.1 | 2.7 ± 0.3 | 2.2 ± 0.2 |
| c-fos oncogeneb | V00727 | 3.2 ± 0.4 | 4.7 ± 0.9 | 3.7 ± 0.8 |
| Guanine nucleotide binding protein γ5 | AI843937 | 1.6 ± 0.1 | 1.8 ± 0.1 | 1.6 ± 0.03 |
| Serine palmitoyltransferase lc2 | U27455 | 1.6 ± 0.1 | 2.0 ± 0.2 | 1.7 ± 0.1 |
| Cystatin B | U59807 | 1.5 ± 0.1 | 1.7 ± 0.1 | 1.5 ± 0.02 |
| Villin 2 | X60671 | 1.9 ± 0.2 | 2.4 ± 0.3 | 1.9 ± 01 |
| RIKEN cDNA 0610010O12 | AI849011 | 1.9 ± 0.1 | 2.5 ± 0.3 | 1.9 ± 0.03 |
| Matrix metalloproteinase 7 | L36244 | 47.8 ± 18.6 | 208.6 ± 83.3 | 48.2 ± 11.9 |
| RIKEN cDNA 4930422J18 | AV376312 | 2.0 ± 0.3 | 2.9 ± 0.3 | 2.0 ± 0.2 |
| RIKEN cDNA 1700017B05 | AW049360 | 1.6 ± 0.1 | 2.0 ± 0.1 | 1.6 ± 0.1 |
| Galactosidase beta 1 | M57734 | 1.8 ± 0.1 | 2.0 ± 0.1 | 1.7 ± 0.1 |
| Cathepsin C | U74683 | 2.6 ± 0.1 | 3.3 ± 0.2 | 2.3 ± 0.3 |
| Interferon-stimulated protein 15 | X56602 | 3.6 ± 0.6 | 2.0 ± 0.2 | 3.8 ± 0.7 |
| MAP kinase–interacting kinase 2 | Y11092 | 1.8 ± 0.1 | 1.4 ± 0.1 | 1.9 ± 0.1 |
| Glutathione S-transferase theta 2 | X98056 | 3.5 ± 0.4 | 2.9 ± 0.4 | 3.6 ± 0.2 |
| Gene name | accession no. | E2 | GEN | DES |
| Homeobox B6 | M18401 | 1.5 ± 0.02 | 1.5 ± 0.1 | 1.6 ± 0.03 |
| Procollagen VIalpha 3 | AF064749 | 2.1 ± 0.2 | 1.9 ± 0.2 | 2.1 ± 0.02 |
| Interferon regulatory factor 7 | U73037 | 11.7 ± 0.9 | 8.1 ± 0.7 | 12.6 ± 1.5 |
| Scavenger receptor class B2 | AB008553 | 2.7 ± 0.1 | 2.4 ± 0.3 | 2.6 ± 0.2 |
| Polyimmunoglobulin receptor | AB001489 | 8.1 ± 0.7 | 6.2 ± 0.8 | 7.9 ± 1.0 |
| Proteasome subunit β10 | Y10875 | 2.1 ± 0.04 | 1.9 ± 0.04 | 2.1 ± 0.1 |
| RIKEN cDNA 0610010E05 | AV312736 | 2.9 ± 0.3 | 2.5 ± 0.2 | 2.7 ± 0.4 |
| RIKEN cDNA 0610010E05 | AI854839 | 3.7 ± 0.5 | 3.0 ± 0.1 | 3.4 ± 0.3 |
| Xanthine dehydrogenase | X75129 | 12.2 ± 2.2 | 8.9 ± 1.2 | 10.1 ± 1.5 |
| Prominin | AF039663 | 3.5 ± 0.2 | 2.9 ± 0.2 | 3.1 ± 0.3 |
| Interferon-induced protein IFIT1 | U43084 | 17.5 ± 2.9 | 9.9 ± 1.5 | 14.0 ± 2.1 |
| Interferon-induced protein IFIT3 | U43086 | 8.1 ± 2.3 | 4.7 ± 0.2 | 6.7 ± 0.6 |
| Proteasome subunit β8 | U22033 | 2.0 ± 0.1 | 1.8 ± 0.2 | 2.1 ± 0.1 |
| RIKEN cDNA 1600023A02 | AW121336 | 1.9 ± 0.1 | 1.7 ± 0.1 | 2.0 ± 0.04 |
| Small proline-rich protein 1A | AF057156 | 11.1 ± 3.7 | 8.6 ± 0.8 | 14.7 ± 0.8 |
| MAP kinase-interacting kinase 2 | AI845732 | 2.0 ± 0.1 | 1.7 ± 0.1 | 2.0 ± 0.2 |
| Lymphocyte antigen 6 complex, locus E | U47737 | 2.0 ± 0.01 | 1.7 ± 0.1 | 2.0 ± 0.1 |
| Guanylate nucleotide binding protein 2 | AJ007970 | 3.0 ± 0.1 | 2.0 ± 0.2 | 2.6 ± 0.1 |
| Peptidyl arginine deiminase, type IIb | D16580 | 1.9 ± 0.3 | 9.1 ± 0.3 | 5.2 ± 1.5 |
| Down-regulated genes | ||||
| Solute carrier family 29a1 | AI838274 | 2.0 ± 0.2 | 2.9 ± 0.3 | 2.5 ± 0.1 |
| Lymphocyte specific 1 | D49691 | 1.6 ± 0.1 | 2.3 ± 0.2 | 1.8 ± 0.1 |
| Claudin 5 | U82758 | 2.0 ± 0.2 | 2.8 ± 0.1 | 2.7 ± 0.5 |
| Potassium channel td12 | AI842065 | 1.6 ± 0.04 | 2.0 ± 0.04 | 2.1 ± 0.1 |
| Zinc finger homeobox 1a | D76432 | 1.5 ± 0.1 | 1.7 ± 0.1 | 1.8 ± 0.01 |
| Monoamine oxidase A | AI848045 | 2.3 ± 0.2 | 2.7 ± 0.2 | 2.5 ± 0.4 |
| Histidine decarboxylase | X57437 | 4.8 ± 0.8 | 7.1 ± 0.6 | 5.4 ± 0.8 |
| α-2 Adrenergic receptor | M97516 | 3.0 ± 0.3 | 4.2 ± 1.1 | 3.6 ± 0.3 |
| Transcription factor 21 | AF035717 | 1.8 ± 0.2 | 2.2 ± 0.2 | 2.0 ± 0.1 |
| Homeobox D8 | X56561 | 2.2 ± 0.1 | 2.8 ± 0.03 | 2.5 ± 0.2 |
| Carboxypeptidase X2 | AF017639 | 4.1 ± 0.7 | 5.2 ± 1.0 | 4.2 ± 0.2 |
| RIKEN cDNA A230106A15 | AI848841 | 3.8 ± 0.2 | 4.7 ± 0.5 | 4.2 ± 0.7 |
| Reduced expression 3 | AA790008 | 3.1 ± 0.2 | 3.5 ± 0.3 | 3.2 ± 0.4 |
| TGF-β binding protein 4 | AA838868 | 1.8 ± 0.1 | 2.1 ± 0.1 | 1.8 ± 0.2 |
| Keratoepithelinb | L19932 | 11.5 ± 2.5 | 12.6 ± 0.9 | 9.7 ± 3.6 |
| GLI-Kruppel family member GLI | AB025922 | 11.6 ± 0.8 | 12.2 ± 2.6 | 8.2 ± 2.6 |
Abbreviations: CEA, carcinoembrionary antigen; SRC, steroid receptor coactivator; TGF, transforming growth factor.
Gene names (derived from the NetAffx database; Liu et al. 2003), GenBank accession numbers (GenBank 2004), and mean (± SD) fold induction/repression of gene expression are shown in the same order as the gene cluster in Figure 3C.
Genes mentioned in the text.
Although these three estrogens can alter the expression of some genes with different magnitudes [e.g., peptidyl arginine deiminase II is up-regulated to a lesser extent by E2 (1.86-fold ± 0.27) relative to GEN (9.11-fold ± 0.33) and DES (5.15-fold ± 1.53); Table 2], the present data show that the same genes are affected during equivalent uterotrophic responses. Previous studies have revealed both similarities and differences between transcriptional responses induced at a single time point after exposure to E2 and DES in the uteri of immature ovariectomized mice (Watanabe et al. 2003) and after exposure to either GEN, bisphenol A, or 17α-ethynyl estradiol in the reproductive tract of intact adult rats (Naciff et al. 2002). We suggest that these reported differences most probably arise from dose-dependent variations in the magnitude and kinetics of gene expression (Figure 2C), rather than from the operation of distinct mechanisms of estrogenic action.
Our data indicate that estrogens of differing provenance may have in common the potential for both beneficial and adverse health effects. This highlights the need for an holistic approach to hazard assessment wherein preconceptions are replaced by an objective assessment of the likely perturbations of physiologic functions caused by combined exposures to physiologic, synthetic, and plant-derived estrogens. This need is reinforced by data showing that plasma concentrations of isoflavones in infants fed soy formula are approximately 200 times higher than for those fed human milk (Setchell et al. 1997), by the estimated daily intake of approximately 29 mg of phytoestrogens for individuals taking dietary supplements (Committee on Toxicity of Chemicals in Food, Consumer Products and the Environment 2003), and by the demonstration that estrogens of different provenance can act additively in the rodent uterus (Tinwell and Ashby 2004).
References
- Aardema MJ, Albertini S, Arni P, Henderson LM, Kirsch-Volders M, Mackay JM, et al. Aneuploidy: a report of an ECETOC task force. Mutat Res. 1998;410:3–79. doi: 10.1016/s1383-5742(97)00029-x. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, et al. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- Almstrup K, Fernandez MF, Petersen JH, Olea N, Skakkebaek NE, Leffers H. Dual effects of phytoestrogens result in U-shaped dose–response curves. Environ Health Perspect. 2002;110:743–748. doi: 10.1289/ehp.02110743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora A, Nair MG, Strasburg GM. Antioxidant activities of isoflavones and their biological metabolites in a liposomal system. Arch Biochem Biophys. 1998;356:133–141. doi: 10.1006/abbi.1998.0783. [DOI] [PubMed] [Google Scholar]
- Casanova M, You L, Gaido KW, Archibeque-Engle S, Janszen DB, Heck HA. Developmental effects of dietary phytoestrogens in Sprague-Dawley rats and interactions of genistein and daidzein with rat estrogen receptors α and β in vitro. Toxicol Sci. 1999;51:236–244. doi: 10.1093/toxsci/51.2.236. [DOI] [PubMed] [Google Scholar]
- Cassidy A, Bingham S, Setchell KDR. Biological effects of a diet of soy protein rich in isoflavones on the menstrual cycle of premenopausal women. Am J Clin Nutr. 1994;60:333–340. doi: 10.1093/ajcn/60.3.333. [DOI] [PubMed] [Google Scholar]
- Cassidy A, Faughnan M. Phyto-oestrogens through the life cycle. Proc Nutr Soc. 2000;59:489–496. doi: 10.1017/s0029665100000719. [DOI] [PubMed] [Google Scholar]
- Committee on Toxicity of Chemicals in Food, Consumer Products and the Environment 2003. Phytoestrogens and Health. London:Food Standards Agency. Available: http://www.foodstandards.gov.uk/multimedia/pdfs/phytoreport0503 [accessed 2 June 2004].
- Dean NM, Kanemitsu M, Boynton AL. Effects of the tyrosine-kinase inhibitor genistein on DNA synthesis and phospholipid-derived second messenger generation in mouse 10T1/2 fibroblasts and rat liver T51B cells. Biochem Biophys Res Commun. 1989;165:795–801. doi: 10.1016/s0006-291x(89)80036-1. [DOI] [PubMed] [Google Scholar]
- Duarte J, Ocete MA, Perez-Vizcaino F, Zarzuelo A, Tamargo J. Effect of tyrosine kinase and tyrosine phosphatase inhibitors on aortic contraction and induction of nitric oxide synthase. Eur J Pharmacol. 1997;338:25–33. doi: 10.1016/s0014-2999(97)01311-3. [DOI] [PubMed] [Google Scholar]
- Frasor J, Barnett DH, Danes JM, Hess R, Parlow AF, Katzenellenbogen BS. Response-specific and ligand dose-dependent modulation of estrogen receptor (ER) α activity by ERβ in the uterus. Endocrinology. 2003;144:3159–3166. doi: 10.1210/en.2002-0143. [DOI] [PubMed] [Google Scholar]
- Genbank 2004. Bethesda, MD:National Center for Biotechnology Information, National Library of Medicine. Available: http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html [accessed 2 June 2004].
- GEO 2004. Gene Expression Omnibus Homepage. Bethesda, MD:National Center for Biotechnology Information, National Library of Medicine. Available: http://www.ncbi.nlm.nih.gov/geo/ [accessed 2 June 2004].
- Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- Herman-Giddens ME, Slora EJ, Wasserman RC, Bourdony CJ, Bhapkar MV, Koch GG, et al. Secondary sexual characteristics and menses in young girls seen in office practice: a study from the Pediatric Research in Office Settings network. Pediatrics. 1997;99:505–512. doi: 10.1542/peds.99.4.505. [DOI] [PubMed] [Google Scholar]
- Hewitt SC, Deroo BJ, Hansen K, Collins J, Grissom S, Afshari CA, et al. Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol Endocrinol. 2003;17:2070–2083. doi: 10.1210/me.2003-0146. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective Bonferroni test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Howdeshell KL, Hotchkiss AK, Thayer KA, Vandenbergh JG, vom Saal FS. Exposure to bisphenol A advances puberty. Nature. 1999;401:763–764. doi: 10.1038/44517. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, et al. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, et al. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund TD, Munson DJ, Haldy ME, Setchell KD, Lephart ED, Handa RJ. Equol is a novel anti-androgen that inhibits prostate growth and hormone feedback. Biol Reprod. 2004;70:1188–1195. doi: 10.1095/biolreprod.103.023713. [DOI] [PubMed] [Google Scholar]
- Messina MJ. Legumes and soybeans: overview of their nutritional profiles and health effects. Am J Clin Nutr. 1999;70:439S–450S. doi: 10.1093/ajcn/70.3.439s. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Orphanides G. Estrogen receptors: orchestrators of pleiotropic cellular responses. EMBO Rep. 2001;9:775–781. doi: 10.1093/embo-reports/kve185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naciff JM, Jump ML, Torontali SM, Carr GJ, Tiesman JP, Overmann GJ, et al. Gene expression profile induced by 17α-ethynyl estradiol, bisphenol A, and genistein in the developing female reproductive system of the rat. Toxicol Sci. 2002;68:184–199. doi: 10.1093/toxsci/68.1.184. [DOI] [PubMed] [Google Scholar]
- Naciff JM, Overmann GJ, Torontali SM, Carr GJ, Tiesman JP, Richardson BD, et al. Gene expression profile induced by 17α-ethynyl estradiol in the prepubertal female reproductive system of the rat. Toxicol Sci. 2003;72:314–330. doi: 10.1093/toxsci/kfg037. [DOI] [PubMed] [Google Scholar]
- Newbold RR, Banks EP, Bullock B, Jefferson WN. Uterine adenocarcinoma in mice treated neonatally with genistein. Cancer Res. 2001;61:4325–4328. [PubMed] [Google Scholar]
- North K, Golding J. A maternal vegetarian diet in pregnancy is associated with hypospadias. Br J Urol Int. 2000;85:107–113. doi: 10.1046/j.1464-410x.2000.00436.x. [DOI] [PubMed] [Google Scholar]
- Odum J, Lefevre PA, Tittensor S, Paton D, Routledge ED, Beresford NA, et al. The rodent uterotrophic assay: critical protocol features, studies with nonyl phenols, and comparison with a yeast estrogenicity assay. Regul Toxicol Pharmacol. 25:176–188. doi: 10.1006/rtph.1997.1100. [DOI] [PubMed] [Google Scholar]
- Okura A, Arakawa H, Oka H, Yoshinari T, Monden Y. Effect of genistein on topoisomerase activity and on the growth of [Val 12]Ha-ras-transformed NIH 3T3 cells. Biochem Biophys Res Commun. 1988;157:183–189. doi: 10.1016/s0006-291x(88)80030-5. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Moggs JG, Spurway T, Tinwell H, Ashby J, Kimber I. Use of gene expression profiling to understand the transcriptional programme associated with estrogen-induced uterine growth: implications for the use of surrogate molecular markers in toxicology [Abstract] Toxicol Sci. 2003;72:12. [Google Scholar]
- Owens JW, Ashby J. Critical review and evaluation of the uterotrophic bioassay for the identification of possible estrogen agonists and antagonists: in support of the validation of the OECD uterotrophic protocols for the laboratory rodent. Organisation for Economic Co-operation and Development. Crit Rev Toxicol. 2002;32:445–520. doi: 10.1080/20024091064291. [DOI] [PubMed] [Google Scholar]
- Paulozzi LJ, Erickson JD, Jackson RJ. Hypospadias: trends in two US surveillance systems. Pediatrics. 1997;100:831–834. doi: 10.1542/peds.100.5.831. [DOI] [PubMed] [Google Scholar]
- Potier B, Rovira C. Protein tyrosine kinase inhibitors reduce high-voltage activating calcium currents in CA1 pyramidal neurones from rat hippocampal slices. Brain Res. 1999;816:587–597. doi: 10.1016/s0006-8993(98)01241-4. [DOI] [PubMed] [Google Scholar]
- Safe SH, Pallaroni L, Yoon K, Gaido K, Ross S, McDonnell D. Problems for risk assessment of endocrine-active estrogenic compounds. Environ Health Perspect. 2002;110:925–929. doi: 10.1289/ehp.02110s6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setchell KDR, Zimmer-Nechemias L, Cai J, Heubi JE. Exposure of infants to phyto-oestrogens from soy-based infant formula. Lancet. 1997;350:23–27. doi: 10.1016/S0140-6736(96)09480-9. [DOI] [PubMed] [Google Scholar]
- Tinwell H, Ashby J. Sensitivity of the immature rat uterotrophic assay to mixtures of estrogens. Environ Health Perspect. 2004;112:575–582. doi: 10.1289/ehp.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Suzuki A, Kobayashi M, Lubahn DB, Handa H, Iguchi T. Similarities and differences in uterine gene expression patterns caused by treatment with physiological and non-physiological estrogens. J Mol Endocrinol. 2003;3:487–497. doi: 10.1677/jme.0.0310487. [DOI] [PubMed] [Google Scholar]
- Weihua Z, Saji S, Mäkinen S, Cheng G, Jensen EV, Warner M, et al. Estrogen receptor (ER) β, a modulator of ERα in the uterus. Proc Natl Acad Sci USA. 2000;97:5936–5941. doi: 10.1073/pnas.97.11.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GM, Iatropoulos M, Cheung R, Radi L, Wang CX. Diethylstilbestrol liver carcinogenicity and modification of DNA in rats. Cancer Lett. 1993;68:193–198. doi: 10.1016/0304-3835(93)90146-z. [DOI] [PubMed] [Google Scholar]