Abstract
Invasive fungal infections have emerged as a major cause of morbidity and mortality in immunocompromised patients. Conventional identification of pathogenic fungi in clinical microbiology laboratories is time-consuming and, therefore, often imperfect for the early initiation of an adequate antifungal therapy. We developed a diagnostic microarray for the rapid and simultaneous identification of the 12 most common pathogenic Candida and Aspergillus species. Oligonucleotide probes were designed by exploiting the sequence variations of the internal transcribed spacer (ITS) regions of the rRNA gene cassette to identify Candida albicans, Candida dubliniensis, Candida krusei, Candida glabrata, Candida tropicalis, Candida parapsilosis, Candida guilliermondii, Candida lusitaniae, Aspergillus fumigatus, Aspergillus flavus, Aspergillus niger, and Aspergillus terreus. By using universal fungal primers (ITS1 and ITS4) directed toward conserved regions of the 18S and 28S rRNA genes, respectively, the fungal ITS target regions could be simultaneously amplified and fluorescently labeled. To establish the system, 12 precharacterized fungal strains were analyzed; and the method was validated by using 21 clinical isolates as blinded samples. As the microarray was able to detect and clearly identify the fungal pathogens within 4 h after DNA extraction, this system offers an interesting potential for clinical microbiology laboratories.
Invasive fungal infections have emerged as major causes of morbidity and mortality in immunocompromised patients. Candidiasis and aspergillosis are the most common invasive fungal infections (IFIs) in patients receiving immunosuppressive treatment for cancer or organ transplantation. An IFI incidence of up to 23% has been reported among high-risk patients, such as patients with prolonged neutropenia following chemotherapy for acute leukemia or allogeneic transplant recipients, especially following nonmyeloablative transplantation (16, 17, 47). In addition to the increasing incidence of IFIs, the number of fungal species which must be considered as potential fungal pathogens has also increased during the last few decades. Application of antifungal agents against common fungal pathogens has led to the emergence of resistant species, e.g., Candida krusei, Aspergillus terreus, and members of the class Zygomycetes (26, 36, 54). The conventional identification of pathogenic fungi in clinical microbiology laboratories based on phenotypic features and physiological tests is time-consuming and, therefore, often imperfect for the early initiation of an antifungal therapy. Several human fungal pathogens are characterized by high rates of intrinsic resistance. Therefore, identification of a fungal pathogen to the species level rather than antifungal drug susceptibility testing is presently the most important step in the selection of the adequate antifungal agent (39).
Until now, a variety of molecular methods have been applied for the detection and identification of fungal pathogens, including species or group discrimination with specific primers (35), identification of PCR products by gel or capillary electrophoresis (4, 8, 44), restriction fragment length polymorphism analysis (53), single-strand conformational polymorphism (48), and Southern or slot blot hybridization assays with oligonucleotide probes (10, 12, 21, 25, 40). Furthermore, a number of PCR-enzyme immunoassays (EIAs) were developed (11, 13, 30, 32, 41). One study showed the applicability of PCR-EIA for the resolution of discrepancies in phenotype-based identification between different institutions (6). This PCR-EIA (11) discriminates the different fungal species by applying the variability of internal transcribed spacer (ITS) region 2 (ITS2). More recently, real-time PCR assays have been described for the quantitative detection of either Candida or Aspergillus species (19) in serum (3, 7, 50) or other specimen types (51). Diaz and Fell applied the Luminex technology to the detection of pathogenic yeasts of the genus Trichosporon (9). These previously described methods feature only limited multiplexing capability, resulting in high costs if all relevant species must be considered. An economically more efficient approach would be the application of a protocol which is capable of identifying a panel of relevant species in a highly parallel fashion.
It was the aim of the present study to develop a species identification array by making use of the high multiplexing capacity of DNA microarrays in the field of molecular detection for identification of human fungal pathogens. As targets, 12 Candida and Aspergillus species representing the pathogens that are the most frequently isolated from invasive fungal infections as well as species with known intrinsic resistance to antifungal agents were chosen.
MATERIALS AND METHODS
Microorganisms.
Ten reference strains, obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ; Candida albicans DSM 1386, Candida dubliniensis DSM 13268, Candida krusei DSM 3433, Candida parapsilosis DSM 5784, Aspergillus fumigatus DSM 819, and Aspergillus niger DSM 1988) and the American Type Culture Collection (ATCC; Torulopsis glabrata ATCC 90030, Candida tropicalis ATCC 750, Candida guillermondii ATCC 6260, and Aspergillus flavus ATCC 20043), as well as two clinical isolates from the Institute of Medical Microbiology and Hygiene, University of Tübingen (Candida lusitaniae VA 13910 and Aspergillus terreus H15), were used as precharacterized strains. Additionally, 21 clinical isolates, including 15 Candida strains belonging to 11 species, 4 Aspergillus strains belonging to 3 species, and 1 strain each of Saccharomyces cerevisiae and Pseudallescheria boydii from the Institute of Medical Microbiology and Hygiene, University of Tübingen, were included in this study (see Table 2) and were used as blinded samples. All clinical isolates were identified by standard laboratory procedures, including macro- and micromorphologies, germ tube formation, growth characteristics at different temperatures, and assimilation tests (ID 32C; Biomerieux).
TABLE 2.
Evaluation of the array with blinded samples
| No. | Isolateb | Isolate by conventional identification method | Array result | Sequencing resulta |
|---|---|---|---|---|
| 1 | C. albicans | ST 3477-03 | C. albicans | + |
| 2 | C. albicans | VA 115470-03 | C. albicans | + |
| 3 | C. albicans | VA 115839-03 | C. albicans | + |
| 4 | C. dubliniensis | ST 2792-03 | C. dubliniensis | + |
| 5 | C. dubliniensis | VA 103469-04 | C. dubliniensis | + |
| 6 | C. glabrata | VA 104009-04 | C. glabrata | + |
| 7 | C. guillermondii | UR 9406-03 | C. guillermondii | + |
| 8 | C. krusei | ST 3382-03 | C. krusei | + |
| 9 | C. lusitaniae | ST 3324-03 | C. lusitaniae | + |
| 10 | C. lusitaniae | VA 115231-03 | C. lusitaniae | + |
| 11 | C. parapsilosis | VA 115230-03 | C. parapsilosis | + |
| 12 | C. tropicalis | UR 9344-03 | C. tropicalis | + |
| 13 | Candida kefyr | VA 116042-03 | Nontarget species | Candida kefyr |
| 14 | Candida norvegensis | ST 3481-03 | C. krusei | Candida norvegensis |
| 15 | Candida pelliculosa | ST 3352/2-03 | Nontarget species | Candida pelliculosa |
| 16 | A. fumigatus | VA 104001-04 | A. fumigatus | + |
| 17 | A. niger | ST 717-04 | A. niger | + |
| 18 | A. niger | VA 3590-04 | A. niger | + |
| 19 | Aspergillus spp.c | VA 103936-04 | A. flavus | + |
| 20 | Pseudallescheria boydii | VA 103543-04 | Nontarget species | Pseudallescheria boydii |
| 21 | Saccharomyces cerevisiae | ST 3352/1-03 | Nontarget species | Saccharomyces cerevisiae |
+, concordance with the array result.
Clinical isolates were identified by standard laboratory procedures.
The content of this sample could not be identified to the species level by the described methods.
DNA extraction.
Prior to DNA extraction, the fungal strains were cultured on Sabouraud dextrose agar at 30°C for 42 h (yeasts) or 72 h (molds). Candida and other yeast colonies were inoculated in sterile saline. Mold cultures were washed with 10 ml of sterile saline (0.9%) to obtain conidia. The fungal suspensions were centrifuged at 3,000 × g, and then the pellets were incubated with 1 ml of leukocyte lysis buffer consisting of 10 mM Tris (pH 7.6), 10 mM EDTA (pH 8.0), 50 mM NaCl, 0.2% sodium dodecyl sulfate, and 200 μg of proteinase K (Roche Diagnostics, Penzberg, Germany) per ml at 65°C overnight. The suspensions were again centrifuged (3,000 × g), and the pellets were exposed to three cycles of freezing in liquid nitrogen for 30 s and heating at 95°C for 4 min to disrupt the fungal cells. Thereafter, the pellets were resuspended and incubated in 1,000 μl recombinant lyticase (L4276; Sigma) at 37°C for 45 min. After centrifugation at 1,600 × g, the DNA was extracted by using a QIAmp DNA mini kit (QIAGEN, Hilden, Germany).
Primers.
Oligonucleotide primers were obtained from Sigma-Genosys, Steinheim, Germany. Amplification of the ITS1 and ITS2 regions was performed with universal fungal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATG-3′), as described by White et al. (52). The sequence of ITS1 is complementary to a conserved region at the end of the 18S rRNA gene, and ITS4 binds to a conserved region at the beginning of the 28S rRNA gene, leading to amplification of the ITS regions and the 5.8S rRNA gene, which is located between the noncoding ITS regions. ITS1- and ITS4-specific primers were also used for sequencing of the purified amplicon. Additionally, the primers ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) and ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) were used for sequencing, leading to overlapping fragments.
DNA sequencing.
For DNA sequencing, the full ITS region was amplified by PCR in a final reaction volume of 50 μl. Each reaction mixture contained approximately 100 ng template DNA; 0.4 pmol (each) forward (ITS1) and reverse (ITS4) primers; 100 μM (each) dATP, dCTP, dGTP, and dTTP; 1× Expand High Fidelity reaction buffer containing 1.5 mM MgCl2 (Roche); 2.5% dimethyl sulfoxide (Fluka Chemie, Buchs, Switzerland); and 2.6 U of Expand High Fidelity DNA polymerase (Roche). The amplification was performed in a Mastercycler gradient (Eppendorf, Hamburg, Germany). An initial denaturation step (94°C for 5 min) was followed by 35 cycles (with each cycle consisting of DNA denaturation at 94°C for 30 s, primer annealing at 55°C for 30 s, and elongation at 72°C for 1 min) and a final extension step at 72°C for 7 min. A no-template negative control was included in each PCR run. The PCR product was purified with a QIAquick Spin PCR purification kit (QIAGEN), according to the protocol of the manufacturer. The DNA was eluted in 30 μl of double-distilled H2O (ddH2O). The purified PCR products were sequenced with the primers ITS1, ITS2, ITS3, and ITS4 and the BigDye Terminator cycle sequencing kit (Applied Biosystems, Darmstadt, Germany). Sequencing was performed on an ABI Prism 377 DNA sequencer (Applied Biosystems). All fungal strains were sequenced in both directions with two primers each, resulting in four overlapping fragments. The sequence data were assembled by using SeqMan II software (version 5.00; DNAStar, Madison, Wis.). The sequences obtained have been submitted to GenBank.
Capture probe design.
All species- or genus-specific oligonucleotide probes and process controls were designed from ITS sequences available in the GenBank database (EMBL and DDBJ databases). Multiple-sequence alignments were carried out by using ClustalX software (42) or SeqMan II software (version 5.00; DNAStar). By comparison of the sequences of the ITS1 and ITS2 regions of the target species, regions with interspecies variations could be identified and were used to develop species-specific probes. Conserved regions served either as targets for probes which are able to discriminate between Candida and Aspergillus (genus-specific probes) or as process controls. In addition to the position of the probe within the target region, thermodynamic parameters were considered in order to get a set of probes with at least similar hybridization efficiencies. Oligonucleotide probes of various lengths (16 to 24 bases) but within a very narrow range of melting temperatures (Tms; ±3.5°C) were designed. The Tms of the probes (Table 1) and the stabilities of possible secondary structures were calculated with the software Oligo (version 6.65; Molecular Biology Insights, Cascade, Colo.) by using the following parameters and thresholds: 50 mM univalent ions, 1.5 mM free Mg2+, and 250 pM DNA for Tm calculation; desired stability of hairpins, ΔG > 0 kcal/mol (where G is Gibb’s free energy); and desired stability of duplexes, ΔG > −4 kcal/mol. All probes were designed as sense and antisense probes. To check the specificities of the probes, additional database searches were performed by using the BLAST and FASTA programs to compare the probe sequences with the sequences in the EMBL and GenBank DNA databases. The sequences of the probes developed are shown in Table 1.
TABLE 1.
Oligonucleotide probe sequencesa
| Melting temp (°C)b | Length | Namec | Positiond | Probe sequence (5′-3′)e |
|---|---|---|---|---|
| 52.7 | 24 | Calb1S | ITS1 | AACTTGTCACACCAGATTATTACT |
| 52.5 | 21 | Calb2S | ITS2 | CTTGAAAGACGGTAGTGGTAA |
| 51.7 | 23 | Cdub1S | ITS1 | AACTTGTCACGAGATTATTTTTA |
| 54.2 | 18 | Cdub2S | ITS2 | CTTGAAAGACGGTAGTGGTAA |
| 52.0 | 22 | Cpar1S | ITS1 | AACTTGTCACGAGATTATTTTTA |
| 51.2 | 24 | Cgla1S | ITS1 | TTGCTAAGGCGGTCTCTG |
| 53.6 | 21 | Ckru1S | ITS1 | GCGGAGTATAAACTAATGGATA |
| 50.8 | 23 | Ctro1S | ITS1 | TTACTACACACAGTGGAGTTTACT |
| 53.5 | 23 | Cgui1S | ITS1 | CGAAAACAACAACACCTAAAA |
| 52.3 | 18 | Clus1S | ITS2 | AAACCAAACTTTTTATTTACAGT |
| 54.1 | 20 | Afla1S | ITS1 | GGAGACACCACGAACTCTGT |
| 51.5 | 18 | Afla2S | ITS2 | CGAACGCAAATCAATCTT |
| 53.6 | 18 | Anig1S | ITS1 | ACCCCAACACGAACACTG |
| 54.3 | 20 | Anig2S | ITS1 | CCAACACGAACACTGTCTGA |
| 51.6 | 21 | Ater1S | ITS1 | AGTCTGAGTGTGATTCTTTGC |
| 52.1 | 21 | Ater2S | ITS1 | GTCTGAGTTGTGATTCTTTGC |
| 52.5 | 19 | Afum1S | ITS2 | CCGACACCCAACTTTATTT |
| 52.3 | 17 | Can1S | 5.8S rRNA gene | TCTTGGTTCTCGCATCG |
| 51.3 | 17 | Can2S | 5.8S rRNA gene | ATGCCTGTTTGAGCGTC |
| 54.1 | 16 | Asp1S | 5.8S rRNA gene | ATGCCTGTCCGAGCGT |
| 53.0 | 16 | Asp2S | ITS1 | CGGCTTGTGTGTTGGG |
| 53.8 | 17 | PC1S | 18S rRNA gene | GTGAACCTGCGGAAGGA |
| 51.8 | 17 | PC2S | 5.8S rRNA gene | GCATCGATGAAGAACGC |
| 51.0 | 17 | PC3S | 18S rRNA gene | TCCGTAGGTGAACCTGC |
| 48.9 | 18 | Negative hybridization control | TCTAGACAGCCACTCATA | |
| 61.7 | 19 | Positive hybridization control | GATTGGACGAGTCAGGAGC | |
| 48.9 | 18 | Spotting control | TCTAGACAGCCACTCATA-Cy3 |
For each species- or genus-specific probe or process control, only the sense version of the probe is shown.
Calculated with Oligo (version 6.65; Molecular Biology Insights) and the parameters described.
S, sense; the name of the corresponding “antisense”probe uses AS (e.g., Calb1AS).
Position within the amplified target region.
The sequence of the 14-thymidine spacer at the 5′ end is not shown.
Oligonucleotide array fabrication.
Oligonucleotide arrays were constructed with 51 different oligonucleotide capture and control probes. The oligonucleotides were purchased from Metabion (Planegg-Martinsried, Germany) in desalted purity and quality controlled by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Each species- or genus-specific capture probe had a 14-thymidine spacer and an amino modification at the 5′ end and was spotted onto the microarray in triplicate. The array production process and the subsequent washing steps were carried out as described previously (15), with modifications. In brief, by using a MicroGrid II microarrayer with two MicroSpot 2500 pins (BioRobotics, Cambridge, United Kingdom), the oligonucleotide probes, which were dissolved in spotting buffer (160 mM Na2SO4, 130 mM Na2HPO4) to a final concentration of 20 μM (prelabeled spotting control; 10 μM), were spotted onto epoxy-coated glass slides (Elipsa, Berlin, Germany). Covalent immobilization was achieved by incubating the oligonucleotide array at 60°C for 30 min in a drying compartment (Memmert, Schwabach, Germany). After the probes were spotted, the slides were rinsed for 5 min in 0.1% (vol/vol) Triton X-100 in ddH2O, 4 min in 0.5 μl of concentrated HCl per ml of ddH2O, and 10 min in a 100 mM KCl solution with constant stirring. Subsequently, the slides were incubated in blocking solution (25% [vol/vol] ethylene glycol, 0.5 μl of concentrated HCl per ml of ddH2O), with the spotted side facing upwards, at 50°C in a heating compartment (OV5; Biometra, Göttingen, Germany); rinsed in ddH2O for 1 min; and finally, dried under a flow of nitrogen. The spot size and the spot-to-spot distance were estimated to be 160 μm and 320 μm, respectively. The processed slides were stored dry for a maximum of 20 days at room temperature in the dark until further use.
Controls.
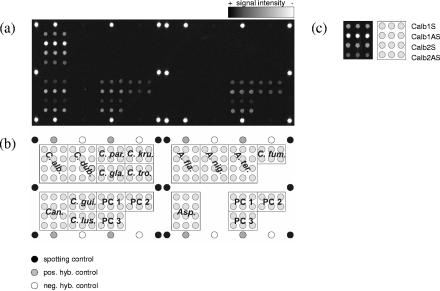
In addition to the species- and genus-specific capture probes, each array also included several controls: a prelabeled spotting control (5′-TTTTTTTTTTTTTTCTAGACAGCCACTCATA-cyanine3 [Cy3]-3′); a positive hybridization control (5′-TTTTTTTTTTTTTGATTGGACGAGTCAGGAGC-3′) complementary to a labeled oligonucleotide target (5′-Cy3-GCTCCTGACTCGTCCAATC-3′), which was spiked during hybridization; and a negative hybridization control (5′-TTTTTTTTTTTTTTCTAGACAGCCACTCATA-3′). All these control sequences are unrelated to sequences found in fungi. Three process controls (PC1 to PC3; Table 1) were designed by using universal fungal sequences. Additionally, the process controls were designed as sense and antisense probes. The spotting controls were set on the side positions of each subgrid spotted with the same pin. The positive and negative hybridization controls were spotted alternately at the upper and lower edges of each subarray. The process controls were spotted within both subgrids (Fig. 1a).
FIG. 1.
(a) Fluorescence image after hybridization with 0.5 pmol of C. albicans isolate VA 115839-03 target DNA for 1 h at 53°C. The signal intensity is encoded in the 65,636 gray scales of the 16-bit TIF image. (b) Layout of the capture probes on the array. All species- and genus-specific probes and the process controls were spotted in triplicate. The other control probes are positioned at the corners of each subarray. Each subarray contains a set of process controls. pos. hyb., positive hybridization, neg. hyb., negative hybridization. (c) The enlarged results for the C. albicans probes describe the arrangement of different probes for the same species or genus within the indicated fields of the array.
Amplification, labeling, and purification of target DNA.
The target DNA used for hybridization on the oligonucleotide arrays was synthesized by PCR. For amplification and labeling, 100 ng of template DNA was supplemented with 0.4 pM (each) forward (ITS1) and reverse (ITS4) primers; 100 μM (each) dATP, dGTP, and dTTP; 50 μM dCTP; 50 μM Cy3-dCTP (ratio of unlabeled dCTP/labeled dCTP, 1:1; Amersham Biosciences, Freiburg, Germany); 1× Expand High Fidelity reaction buffer containing 1.5 mM MgCl2 (Roche); 2.5% dimethyl sulfoxide (Fluka Chemie); and 2.6 U of Expand High Fidelity DNA polymerase (Eppendorf, Hamburg, Germany) in a total volume of 50 μl. The amplification was performed in a Mastercycler gradient (Eppendorf). The reaction profile was as follows: 5 min of initial denaturation at 94°C; 35 cycles of DNA denaturation at 94°C for 30 s, primer annealing at 55°C for 30 s, and elongation at 72°C for 1 min; and a final extension step at 72°C for 4 min. A no-template negative control was included in each PCR run. After purification of the PCR product with a QIAquick Spin PCR purification kit (QIAGEN), the rate of incorporation of Cy3-dCTP, expressed as number of nucleotides/number of incorporated fluorescent dyes (NT/F), was determined by measurement of the optical density (ND-1000 spectrophotometer; NanoDrop Technologies, Rockland, Maine).
Hybridization.
The purified target DNA (0.5 pmol) together with control DNA (Cy3-GCTCCTGACTCGTCCAATC; 0.05 pmol) was hybridized within a gene frame (15 by 16 mm) closed with a coverslip (Abgene House, Hamburg, Germany) in 70 μl of 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]). Prior to hybridization, the hybridization mixture was incubated for 10 min at 95°C, stored for 1 min on ice, and immediately used. For hybridization with amplified target DNA from the 12 precharacterized strains, the DNA solution on the glass slide, within the gene frame, was incubated in a thermomixer with an exchangeable thermoblock for slides (Eppendorf) for 2 h at 50°C and at an agitation speed of 1,200 rpm. In order to improve the hybridization conditions, hybridizations with target DNA from a subset of target species were also carried out for 2 h at 53°C, 55°C, and 57°C. During the evaluation process, target DNA amplified from the 21 blinded samples was hybridized for 1 h at 53°C with agitation at 1,200 rpm. The reduction of the assay time was due to previous optimization experiments (data not shown). After hybridization, the slides were washed with 2× SSC (1× is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% sodium dodecyl sulfate, then 2× SSC, and finally, 0.2× SSC for 10 min each time. The washing procedure was performed at room temperature with agitation in a glass container. The slides were subsequently dried with N2.
Data acquisition and processing.
After the hybridization reaction, the data from the oligonucleotide arrays were read by acquisition of the fluorescence signals by using a 418 Array Scanner (Affymetrix, Santa Clara, Calif.). The laser power and gain were adjusted to 100%. Image processing and calculation of signal intensities were performed with ImaGene (version 3.0; Biodiscovery, Los Angeles, Calif.). For calculation of the individual net signal intensities, the local background was subtracted from the raw spot intensity value. Further data processing was performed by using Microsoft Excel software (Microsoft, Richmond, Wash.). Only net signal intensities which were significantly higher than the background were considered. Therefore, a cutoff 1 of 300 intensity units (three times the detection limit of the array scanner) was applied. By definition, all values less than 300 were set equal to 0. For normalization, the net signal intensity of each spot was divided by the mean net signal intensity of the three replicates of the antisense version of process control 3 (referred to as “signal intensity” [I] of PC3 antisense) and multiplied by 100. All other process control probes were found to be unsuitable for data processing and were not used. The mean of the three normalized net signal intensities (referred to as the “relative signal intensity” [RI] of one probe) and the standard deviation of those values were calculated. Accordingly, the probe PC3 antisense always had an RI value of 100%. To discriminate between specific and unspecific hybridization signals, a cutoff 2 was defined. Only RI values which were significantly greater than 10% (RI > 10 + three times the standard deviation of the normalized net signal intensities of one probe) of the signal intensity of the process control 3 antisense were considered specific signals.
Nucleotide sequence accession numbers.
The accession numbers of the sequences deposited in GenBank are AY939782 (A. flavus ATCC 20043), AY939783 (A. niger isolate ST 717-04), AY939784 (A. niger isolate VA 3590-04), AY939785 (A. flavus isolate VA 103936-04), AY939786 (C. albicans ATCC 10231), AY939787 (A. niger ATCC 16404), AY939788 (A. terreus isolate H15), AY939789 (C. albicans isolate ST 3477-03), AY939790 (A. fumigatus ATCC 9197), AY939791 (C. albicans isolate VA 115470-03), AY939792 (C. guilliermondii ATCC 6260), AY939793 (C. glabrata ATCC 90030), AY939794 (C. glabrata isolate VA 104009-04), AY939795 (C. guilliermondii isolate UR 9406-03), AY939796 (C. krusei isolate ST 3382-03), AY939797 (C. lusitaniae isolate VA 115231-03), AY939798 (C. parapsilosis ATCC 22019), AY939799 (C. norvegensis isolate ST 3481-03), AY939800 (C. pelliculosa isolate ST 3352/2-03), AY939801 (C. tropicalis isolate UR 9344-03), AY939802 (P. boydii isolate VA 103543-04), AY939803 (C. parapsilosis isolate VA 115230-03), AY939804 (A. fumigatus VA 104001-04), AY939805 (C. dubliniensis isolate ST 2792-03), AY939806 (C. kefyr isolate VA 116042-03), AY939807 (C. albicans isolate VA 115839-03), AY939808 (C. krusei ATCC 24210), AY939809 (C. dubliniensis isolate R2), AY939810 (C. tropicalis ATCC 750), AY939811 (C. lusitaniae VA 13910-03), AY939812 (C. lusitaniae isolate ST 3324-03), AY939813 (C. dubliniensis isolate VA 103469-04), AY939814 (S. cerevisiae isolate ST 3352/1-03), and DQ105856 (C. dubliniensis DSM 13268).
RESULTS
We constructed an oligonucleotide microarray for the rapid and simultaneous identification of the most common pathogenic Candida and Aspergillus species. By using the internal transcribed spacer regions of the rRNA gene complex as the diagnostic region, probes were designed against either the ITS1 region or the ITS2 region, or both (Table 1). By exploiting the full sequence of the amplicon obtained with primers ITS1 and ITS4, the sequence of the central 5.8S rRNA gene was used to design either genus-specific probes or process controls. The conserved region at the end of the 18S rRNA gene was used for the design of process controls. The size of the PCR product generated with those primers, using extracted genomic DNA as the template, varied, depending on the species, from 382 bp (C. lusitaniae) to 882 bp (C. glabrata). The ITS regions of A. fumigatus, A. niger, and A. terreus were amplified by adding 2.5% of dimethyl sulfoxide to resolve the secondary structures. The fluorescent label (Cy3) was incorporated during PCR. The NT/F ratios varied from 80 to 220, depending on the quality of the template DNA and on the species.
Setting up the system and proof of concept.
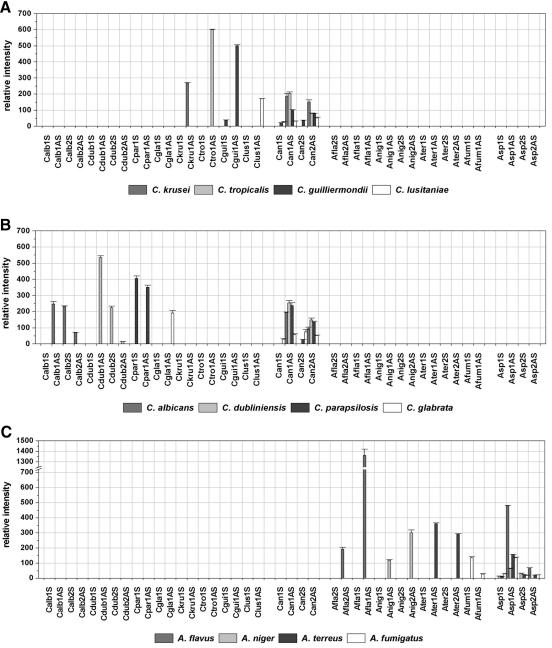
To examine the specificities and hybridization efficiencies of the capture probes that were designed, to optimize hybridization conditions, and to establish the quantification and subsequent data processing, the array was tested by using 12 precharacterized strains, 1 strain of each target species. By hybridization for 2 h at 50°C, different hybridization patterns for the different species could clearly be seen on the microarray. Figure 1 shows the fluorescence intensities of the probes after hybridization with the C. albicans DSM 1386 target DNA. The hybridization efficiencies of the sense and antisense versions of one probe often showed big differences. At least one of the two versions showed a high hybridization efficiency, with fluorescent net signal intensities ranging from 21,000 to 55,400 intensity units (with species-specific probes) or 6,600 to 37,500 intensity units (with genus-specific probes). By using epoxy-coated glass slides in combination with the washing procedures described above, backgrounds which allowed the use of a low cutoff, 1 of 300 intensity units, could be achieved. Weak cross hybridizations (∼1,000) were observed only between the A. niger-specific probe Anig1 antisense and the A. fumigatus or A. terreus target DNA. This was expected, as there is no sequence difference between A. fumigatus and A. terreus at the position within the ITS1 region to which the probe Anig1 antisense binds. The second A. niger-specific probe, Anig2 sense, showed weak but significant signals (I, ∼400) only with A. fumigatus. Furthermore, we included in the array layout three process controls which were designed to be specific for universal fungal sequences with the aim of using one as an internal standard. The assumption was that the intensity of a process control would be similarly dependent on the parameters of the experiment (function of the labeling PCR, hybridization conditions), as was the case for the other probes on the array. To rate the suitability of the three different process controls as an internal standard, the relative signal intensities of certain probes calculated with different process controls on different slides were compared (data not shown). Due to the small deviations of the relative signal intensities on different slides with probe PC3 antisense, for calculation of the RI values it fulfilled the requirements for a normalization procedure and was chosen as the internal standard. Figure 2 shows the quantified data sets of all probes after hybridization with the labeled target DNA of the different precharacterized strains by using PC3 antisense for normalization and cutoff 2. This cutoff was chosen as a consequence of the weak cross hybridizations observed. By using this threshold, every target species could be unequivocally identified.
FIG. 2.
Relative intensities after hybridization with labeled target DNA from the 12 target species. Cutoff 1, I = 300; cutoff 2, RI = 10. The mean RIs and their standard deviations are calculated for the triplicate spots on one slide (n = 3).
Optimization of hybridization conditions.
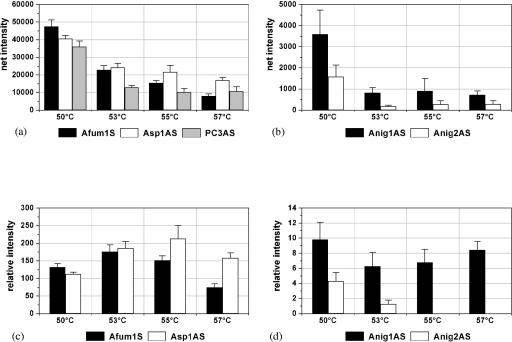
To optimize the assay system, the performance of the probes during hybridization was examined under more stringent hybridization conditions. This was achieved by raising the hybridization temperature. The aim of these experiments was the optimization of the discrimination of target species by not loosing too much net signal intensity (and, therefore, sensitivity). A. fumigatus was representative in both aspects (the sole species which showed cross hybridization to other species-specific probes and which had relative low net signal intensities) and was chosen as the model target species. The increase in the temperature gradually resulted in a decrease of the net signal intensities of the specific signals (Fig. 3a) as well as of the unspecific signals (Fig. 3b). The most significant decrease was observed by raising the temperature from 50 to 53°C. The relative signal intensities of the species-specific probe Afum1 sense and the genus-specific probe Asp1 antisense had maxima at 53°C and 55°C, respectively (Fig. 3c). The highest unspecific signal had a minimum at 53°C (Fig. 3d). The concordance of the optimum of the specific signal of Afum1S and the minimum of the unspecific signal of Anig1AS at 53°C indicated that the assay would have the strongest discrimination power at this hybridization temperature. Consequently, all following hybridizations were performed at 53°C. Further experiments with A. fumigatus, C. lusitaniae, and C. krusei DNA showed the assay time could be reduced by reducing the hybridization duration from 2 to 1 h (data not shown).
FIG. 3.
Behaviors of I and RI of the species-specific probe Afum1 sense, the genus-specific probe Asp1 antisense, the process control PC3 antisense (a and c), and the A. niger-specific probes Anig1 antisense and 2 antisense (b and d) after hybridization with 0.5 pmol labeled A. fumigatus DSM 819 target DNA for 2 h at different hybridization temperatures. The columns depict the mean net signal intensities or the mean relative signal intensities and their standard deviations over three slides of the same experiment (n = 9).
Testing of clinical isolates.
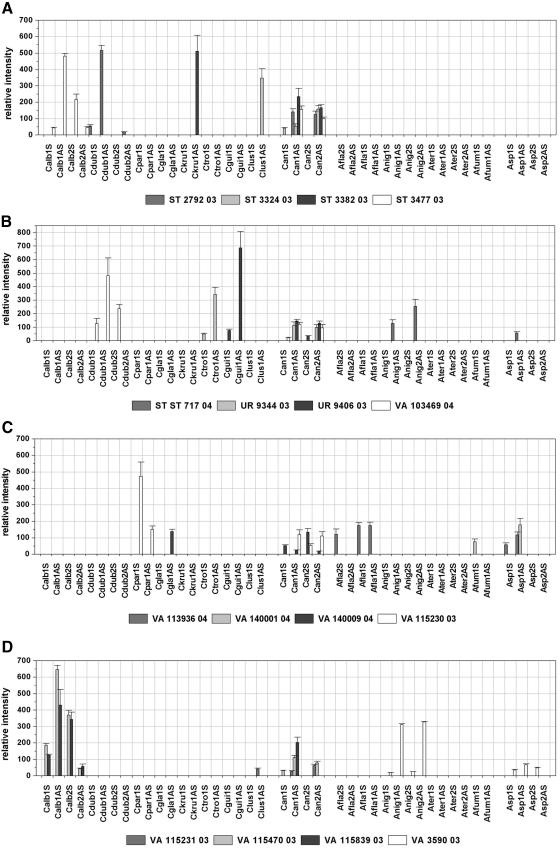
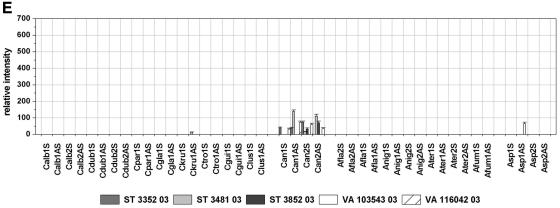
To examine the specificity of the designed probes and to assess their applicability for clinical isolates, the performance of the array was validated by using 21 blinded samples containing genomic DNA of clinical isolates of either the target species or fungal nontarget species. The application of nontarget species served as a cross-reactivity test. The array result was compared in each case to the result of the direct sequencing of the PCR product amplified out of the same sample in a separate PCR by using the same primers used for the labeling PCR. The identities of the sequences obtained were determined by performing searches of the sequences with the sequences in the GenBank DNA database by using the BLAST program. Additionally, the sequences were analyzed by comparison to the sequences used for probe design. By comparing the array results and the results of the sequencing to the previous identification obtained by standard methods, the array was able to unequivocally identify the contents of 16 samples (Table 2). Due to the absence of species-specific signals and the presence of genus-specific and control signals, four other isolates were identified as nontarget species by the array. In those cases the sequencing led to the identification of the fungal species and confirmed the array result. In the case of sample ST 3481-03, the species-specific probe for C. krusei, probe Ckru1AS, showed a weak hybridization signal with an RI value of 14 (Fig. 4e). Although this probe showed much higher RI values when with the C. krusei DSM 3433 strain (RI = 270; Fig. 2b) or isolate ST 3382-03 (RI = 511; Fig. 4a) was hybridized, we interpreted the result as specific, according to the global cutoff 2 described above. Sequencing identified the fungal content as C. norvegensis, which was concordant with the conventional identification results (Table 2). Accordingly, the sequence analysis revealed a 15-bp match between the C. norvegensis sequence and C. krusei-specific probe Ckru1AS. Regarding the full ITS amplicon, the sequences of the C. krusei DSM 3433 strain and the C. norvegensis isolate ST 3481-03 showed 81% homology, with high homology within the central 5.8S rRNA gene and sequence variations concentrating within the ITS regions. Other isolates also showed minor differences in sequence compared with those of the precharacterized strains either within regions not used for probe design (isolates ST 3382-03 and VA 104001-04) or within the diagnostic position (isolate VA 115231-03). Nevertheless, the correct identification of those isolates was possible, proving the robustness of the array (Fig. 4a, c, and d; Table 2). The fluorescence intensities of the probes quantified after hybridization with the labeled target region of the different isolates are shown in Fig. 4.
FIG. 4.
Relative intensities after hybridization with labeled target DNA from blinded samples of clinical isolates. Cutoffs 1 and 2 are as described in the legend to Fig. 2. The columns depict mean relative signal intensities and their standard deviations over three slides of the same experiment (n = 9).
DISCUSSION
With the development of more aggressive therapeutic regimens, especially for the treatment of hematological malignancies, the incidence of invasive opportunistic yeast and mold infections has increased. The early initiation of antifungal therapy is critical in reducing the high mortality rate in patients with IFIs. Early and accurate identification of the fungal pathogen is the most important and critical step in providing adequate antifungal treatment in time. The conventional method of identification of pathogenic fungi used in clinical microbiology is based on phenotypic features and physiological tests and is therefore time-consuming. Instead, molecular genotyping methods could provide a rapid and specific means of identification of fungal pathogens. In view of the increasing number of fungal species isolated from clinical samples as potential pathogens, a broad-spectrum detection system as it is represented by the microarray technology would be desirable. Until today, diagnostic DNA microarrays were applied for the identification of viruses (5, 27-29, 56), bacteria (14, 24, 45, 46, 49), and mechanisms of resistance to certain antibiotics (15, 18, 55). To our knowledge, this study describes for the first time the application of the DNA microarray technology in combination with a quantification and data-processing method for the rapid and reliable detection of fungal pathogens. To achieve this, oligonucleotide capture probes were designed by exploiting the variations in the sequences of the ITS regions in a way similar to that described by other research groups (20, 23). The ITS regions were chosen as the target for several reasons: (i) as part of the rRNA gene complex, the ITS regions are present in numerous copies in the fungal genome; (ii) universal fungal primers based on the conserved regions of the rRNA genes are available; and (iii) the level of variability of the ITS regions of the target species is higher than that of the rRNA genes and is high enough for species resolution.
A frequent challenge in DNA microarray-based species detection studies is the discrimination between specific and unspecific signals, especially in the case of mixture analysis. Cross hybridization was observed in several identification microarray studies (1, 46), which made it necessary to design more than one probe per species (2). For this reason, we introduced a simple normalization procedure based on process controls. While the first cutoff in the study presented here was basically related to the background intensity, cutoff 2 was determined by the intensity value normalized on the basis of the process control for the most severe cross hybridization. The use of a global and very stringent cutoff indicates the possibility of misidentifications, as was found in the case of C. krusei and C. norvegenis. On the other hand, this low threshold left a lot of space between the cutoff 2 and the relative signals for the most efficient species-specific probes (well above 100) and for the genus-specific probes (from 50 to 250). For this reason, the use of an individual cutoff seems more appropriate for future studies, as this option would offer the possibility to take the sequence similarities of closely related species into account. Naturally, such an approach would require the analysis of larger numbers of samples in order to better reflect sequence variations in the clinical routine. On the other hand, the stringent cutoff offers the advantage of a higher resolution for samples with mixtures of microorganisms. Future studies will show at which ratios different species can be detected by using the present cutoff definition or if a modification will be necessary.
Two aspects of the specificity of the detection system can be discussed. First, the misidentification of sample ST 3481-03 as C. krusei due to sequence similarities between C. krusei and C. norvergensis suggests the need for the design of an additional C. krusei-specific probe which would be able to distinguish between these two species. Second, the big difference between the RI value of the perfect match and the cross hybridization would allow an individual cutoff 2 higher than 10 for C. krusei-specific probe Ckru1AS instead of a universal one. Nevertheless, a considerable robustness was demonstrated for an isolate which showed a point mutation within the diagnostic position (C. lusitaniae VA 115231-03).
With multiplexing capacity as one of the key features of DNA microarrays, the method developed in the present study should particularly be compared to methods which are able to detect more than one parameter simultaneously. Using two multiplex liquid hybridization mixtures, Hendolin and colleagues needed 2 days for the detection of eight Candida and Aspergillus species from tissue samples (22). A line probe assay was designed for the simultaneous detection of 11 different Candida and Aspergillus species and Cryptococcus neoformans in spiked blood samples within 1 working day (38). However, the capacity of this format is nearly used up with this number of species (57). In contrast, microarrays have the potential to discriminate between thousands of targets, and principally, the design of specific probes which are able to discriminate between a set of species by using a particular target or targets is the only limiting factor. In this study a set of 12 target species was chosen, and the procedure could be performed within 4 h after DNA extraction. Additional time must be counted for DNA extraction, but optimized protocols for extraction in 4 h have been reported previously (31, 34). The opportunity to extend the amount of detectable species in combination with the fast assay procedure makes this array-based method an interesting tool for the rapid and parallel detection of fungal pathogens. The most time-saving effect from the introduction of molecular methods for the diagnosis of fungal infections could be expected if this microarray technique were successfully applied directly to clinical samples that are microscopically positive for fungal elements rather than fungal cultures. The quality of such a culture-independent diagnostic method is mainly dependent on the sensitivity of the DNA extraction method and the PCR protocol, as well as on the specificity of the target identification at the end of the detection process. All three steps of this diagnostic process are part of an ongoing study.
To have a diagnostic tool which can prevail in the routine clinical laboratory, the spectrum of detectable species should be expanded. In addition to IFIs due to non-C. albicans Candida species (43) and non-A. fumigatus Aspergillus species, the incidence of invasive mold infections caused by Zygomycetes (e.g., Rhizopus, Mucor, Rhizomucor, or Absidia spp.) and Fusarium spp. increased significantly in the late 1990s (37). It will be the aim of future studies to develop a more comprehensive array that includes these fungi as targets to improve the diagnosis of emerging yeast and mold infections. Also, yeasts like Cryptococcus neoformans, which particularly play a role in the care of AIDS patients, and representatives of the groups Trichosporon and Malassezia should be included as target species. As more and more molecular targets which are responsible for resistance or pathogenicity are currently identified (33), DNA microarrays also present a promising platform for the parallel species identification and the detection of pathogenicity factors or genes that confer resistance (18). The combined information about the species and its resistance would then lead to more efficient therapies while reducing the unwanted selection pressure of antifungal agents at the same time.
Although today only a limited number of laboratories that specialize in molecular biology are prepared with suitable technical equipment, a lot of work is under way, especially for the application of other, simpler readout systems for microarray slides. As the number of promising applications for microarrays targeting bacterial, parasitic, and fungal pathogens increases, it could be expected that commercial microarray systems will also be offered in the future at a reasonable price to a broader spectrum of customers.
Conclusions.
The results presented in this paper showed the feasibility of the DNA microarray-based method for the detection and identification of 12 of the most common fungal pathogens belonging to the genus Candida or Aspergillus. The assay was demonstrated with isolated genomic DNA originating from precharacterized fungal strains and blinded clinical isolates. While mixture analysis and its application to primary specimens must be performed in the future, the microarray method can already bring great improvements to the laborious standard identification procedures and be a rapid tool for the identification of isolated fungal pathogens.
REFERENCES
- 1.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, T., C. Koob, M. Schedl, A. Mehlen, H. Meier, D. Knopp, E. Frahm, U. Obst, K. H. Schleifer, R. Niessner, and W. Ludwig. 2000. A nested array of rRNA targeted probes for the detection and identification of enterococci by reverse hybridization. Syst. Appl. Microbiol. 23:563-572. [DOI] [PubMed] [Google Scholar]
- 3.Challier, S., S. Boyer, E. Abachin, and P. Berche. 2004. Development of a serum-based TaqMan real-time PCR assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 42:844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coignard, C., S. F. Hurst, L. E. Benjamin, M. E. Brandt, D. W. Warnock, and C. J. Morrison. 2004. Resolution of discrepant results for Candida species identification by using DNA probes. J. Clin. Microbiol. 42:858-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa, C., D. Vidaud, M. Olivi, E. Bart-Delabesse, M. Vidaud, and S. Bretagne. 2001. Development of two real-time quantitative TaqMan PCR assays to detect circulating Aspergillus fumigatus DNA in serum. J. Microbiol. Methods 44:263-269. [DOI] [PubMed] [Google Scholar]
- 8.De Baere, T., G. Claeys, D. Swinne, G. Verschraegen, A. Muylaert, C. Massonet, and M. Vaneechoutte. 2002. Identification of cultured isolates of clinically important yeast species using fluorescent fragment length analysis of the amplified internally transcribed rRNA spacer 2 region (ITS2). BMC Microbiol. 2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz, M. R., and J. W. Fell. 2004. High-throughput detection of pathogenic yeasts of the genus trichosporon. J. Clin. Microbiol. 42:3696-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flahaut, M., D. Sanglard, M. Monod, J. Bille, and M. Rossier. 1998. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J. Clin. Microbiol. 36:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita, S., B. A. Lasker, T. J. Lott, E. Reiss, and C. J. Morrison. 1995. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J. Clin. Microbiol. 33:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukushima, M., K. Kakinuma, H. Hayashi, H. Nagai, K. Ito, and R. Kawaguchi. 2003. Detection and identification of Mycobacterium species isolates by DNA microarray. J. Clin. Microbiol. 41:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm, V., S. Ezaki, M. Susa, C. Knabbe, R. D. Schmid, and T. T. Bachmann. 2004. Use of DNA microarrays for rapid genotyping of TEM beta-lactamases that confer resistance. J. Clin. Microbiol. 42:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groll, A. H., P. M. Shah, C. Mentzel, M. Schneider, G. Just-Nuebling, and K. Huebner. 1996. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J. Infect. 33:23-32. [DOI] [PubMed] [Google Scholar]
- 17.Hagen, E. A., H. Stern, D. Porter, K. Duffy, K. Foley, S. Luger, S. J. Schuster, E. A. Stadtmauer, and M. G. Schuster. 2003. High rate of invasive fungal infections following nonmyeloablative allogeneic transplantation. Clin. Infect. Dis. 36:9-15. [DOI] [PubMed] [Google Scholar]
- 18.Hamels, S., J. L. Gala, S. Dufour, P. Vannuffel, N. Zammatteo, and J. Remacle. 2001. Consensus PCR and microarray for diagnosis of the genus Staphylococcus, species, and methicillin resistance. BioTechniques 31:1364-1366, 1368, 1370-1372. [DOI] [PubMed] [Google Scholar]
- 19.Haugland, R. A., M. Varma, L. J. Wymer, and S. J. Vesper. 2004. Quantitative PCR analysis of selected Aspergillus, Penicillium and Paecilomyces species. Syst. Appl. Microbiol. 27:198-210. [DOI] [PubMed] [Google Scholar]
- 20.Healy, M., K. Reece, D. Walton, J. Huong, K. Shah, and D. P. Kontoyiannis. 2004. Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 42:4016-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebart, H., J. Loffler, C. Meisner, F. Serey, D. Schmidt, A. Bohme, H. Martin, A. Engel, D. Bunje, W. V. Kern, U. Schumacher, L. Kanz, and H. Einsele. 2000. Early detection of aspergillus infection after allogeneic stem cell transplantation by polymerase chain reaction screening. J. Infect. Dis. 181:1713-1719. [DOI] [PubMed] [Google Scholar]
- 22.Hendolin, P. H., L. Paulin, P. Koukila-Kahkola, V. J. Anttila, H. Malmberg, M. Richardson, and J. Ylikoski. 2000. Panfungal PCR and multiplex liquid hybridization for detection of fungi in tissue specimens. J. Clin. Microbiol. 38:4186-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry, T., P. C. Iwen, and S. H. Hinrichs. 2000. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J. Clin. Microbiol. 38:1510-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakinuma, K., M. Fukushima, and R. Kawaguchi. 2003. Detection and identification of Escherichia coli, Shigella, and Salmonella by microarrays using the gyrB gene. Biotechnol. Bioeng. 83:721-728. [DOI] [PubMed] [Google Scholar]
- 25.Kan, V. L. 1993. Polymerase chain reaction for the diagnosis of candidemia. J. Infect. Dis. 168:779-783. [DOI] [PubMed] [Google Scholar]
- 26.Kauffman, C. A. 2004. Zygomycosis: reemergence of an old pathogen. Clin. Infect. Dis. 39:588-590. [DOI] [PubMed] [Google Scholar]
- 27.Kim, C. J., J. K. Jeong, M. Park, T. S. Park, T. C. Park, S. E. Namkoong, and J. S. Park. 2003. HPV oligonucleotide microarray-based detection of HPV genotypes in cervical neoplastic lesions. Gynecol. Oncol. 89:210-217. [DOI] [PubMed] [Google Scholar]
- 28.Lapa, S., M. Mikheev, S. Shchelkunov, V. Mikhailovich, A. Sobolev, V. Blinov, I. Babkin, A. Guskov, E. Sokunova, A. Zasedatelev, L. Sandakhchiev, and A. Mirzabekov. 2002. Species-level identification of orthopoxviruses with an oligonucleotide microchip. J. Clin. Microbiol. 40:753-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsley, M. D., S. F. Hurst, N. J. Iqbal, and C. J. Morrison. 2001. Rapid identification of dimorphic and yeast-like fungal pathogens using specific DNA probes. J. Clin. Microbiol. 39:3505-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loeffler, J., H. Hebart, U. Schumacher, H. Reitze, and H. Einsele. 1997. Comparison of different methods for extraction of DNA of fungal pathogens from cultures and blood. J. Clin. Microbiol. 35:3311-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loeffler, J., H. Hebart, S. Sepe, U. Schumacher, T. Klingebiel, and H. Einsele. 1998. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med. Mycol. 36:275-279. [DOI] [PubMed] [Google Scholar]
- 33.Loeffler, J., S. L. Kelly, H. Hebart, U. Schumacher, C. Lass-Florl, and H. Einsele. 1997. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol. Lett. 151:263-268. [DOI] [PubMed] [Google Scholar]
- 34.Loeffler, J., K. Schmidt, H. Hebart, U. Schumacher, and H. Einsele. 2002. Automated extraction of genomic DNA from medically important yeast species and filamentous fungi by using the MagNA Pure LC system. J. Clin. Microbiol. 40:2240-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannarelli, B. M., and C. P. Kurtzman. 1998. Rapid identification of Candida albicans and other human pathogenic yeasts by using short oligonucleotides in a PCR. J. Clin. Microbiol. 36:1634-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marr, K. A. 2004. Invasive Candida infections: the changing epidemiology. Oncology (Huntington) 18:9-14. [PubMed] [Google Scholar]
- 37.Marr, K. A., R. A. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34:909-917. [DOI] [PubMed] [Google Scholar]
- 38.Martin, C., D. Roberts, W. M. van Der, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 38:3735-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rex, J. H., and M. A. Pfaller. 2002. Has antifungal susceptibility testing come of age? Clin. Infect. Dis. 35:982-989. [DOI] [PubMed] [Google Scholar]
- 40.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 33:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin, J. H., F. S. Nolte, and C. J. Morrison. 1997. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J. Clin. Microbiol. 35:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627-630. [DOI] [PubMed] [Google Scholar]
- 44.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volokhov, D., V. Chizhikov, K. Chumakov, and A. Rasooly. 2003. Microarray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 41:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volokhov, D., A. Rasooly, K. Chumakov, and V. Chizhikov. 2002. Identification of Listeria species by microarray-based assay. J. Clin. Microbiol. 40:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wald, A., W. Leisenring, J. A. vanBurik, and R. A. Bowden. 1997. Epidemiology of Aspergillus infections in a large cohort of patients undergoing bone marrow transplantation. J. Infect. Dis. 175:1459-1466. [DOI] [PubMed] [Google Scholar]
- 48.Walsh, T. J., A. Francesconi, M. Kasai, and S. J. Chanock. 1995. PCR and single-strand conformational polymorphism for recognition of medically important opportunistic fungi. J. Clin. Microbiol. 33:3216-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, R. F., M. L. Beggs, L. H. Robertson, and C. E. Cerniglia. 2002. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples. FEMS Microbiol. Lett. 213:175-182. [DOI] [PubMed] [Google Scholar]
- 50.White, P. L., A. Shetty, and R. A. Barnes. 2003. Detection of seven Candida species using the Light-Cycler system. J. Med. Microbiol. 52:229-238. [DOI] [PubMed] [Google Scholar]
- 51.White, P. L., D. W. Williams, T. Kuriyama, S. A. Samad, M. A. Lewis, and R. A. Barnes. 2004. Detection of Candida in concentrated oral rinse cultures by real-time PCR. J. Clin. Microbiol. 42:2101-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. Innis, D. Gelfand, J. Sninsky, and T. White (ed.), PCR protocols: a guide to methods and application. Academic Press, Inc., San Diego, Calif.
- 53.Williams, D. W., W. A. Coulter, M. J. Wilson, A. J. Potts, and M. A. Lewis. 2001. Identification of Candida dubliniensis, based on ribosomal DNA sequence analysis. Br. J. Biomed. Sci. 58:11-16. [PubMed] [Google Scholar]
- 54.Wingard, J. R. 2005. The changing face of invasive fungal infections in hematopoietic cell transplant recipients. Curr. Opin. Oncol. 17:89-92. [DOI] [PubMed] [Google Scholar]
- 55.Yu, X., M. Susa, C. Knabbe, R. D. Schmid, and T. T. Bachmann. 2004. Development and validation of a diagnostic DNA microarray to detect quinolone-resistant Escherichia coli among clinical isolates. J. Clin. Microbiol. 42:4083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, W., W. Liu, Q. Liu, L. Zhang, Z. Zhou, X. Liu, and H. Zhang. 2002. Genotyping of hepatitis C virus by hepatitis gene diagnosis microarray. Zhonghua Yi. Xue. Za Zhi. 82:1249-1253. [PubMed] [Google Scholar]
- 57.Zwart, G., E. J. van Hannen, M. P. Kamst-van Agterveld, K. Van der Gucht, E. S. Lindstrom, J. Van Wichelen, T. Lauridsen, B. C. Crump, S. K. Han, and S. Declerck. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]