Abstract
There is evidence that hypoxia-inducible factor-1α (HIF-1α) interacts with the tumor suppressor p53. To characterize the putative interaction, we mapped the binding of the core domain of p53 (p53c) to an array of immobilized HIF-1α-derived peptides and found two peptide-sequence motifs that bound to p53c with micromolar affinity in solution. One sequence was adjacent to and the other coincided with the two proline residues of the oxygen-dependent degradation domain (P402 and P564) that act as switches for the oxygen-dependent regulation of HIF-1α. The binding affinity was independent of the hydroxylation state of P564. We found from NMR spectroscopy that these sequence motifs bind to the DNA-binding site of p53c. Because the two sequences are homologous and separated by 120 residues, and one is in a largely unstructured transactivation domain, we speculate that each sequence motif in HIF-1α binds to a different subunit of the p53 tetramer, leading to very tight binding. The binding data support the proposal that p53 provides a route for the degradation in hypoxic tumor cells of HIF-1α that is not hydroxylated at the two proline residues.
Keywords: NMR, peptide, array, hypoxia, VHL
The two cancer-related transcription factors p53 and hypoxia-inducible factor (HIF)-1α are reported to interact either directly (1) or in a complex with the coactivator protein p300 (2). The interaction has been suggested to mediate hypoxia-induced apoptosis (3) and constitute a pathway of HIF-1α degradation (4) but is controversial (5). The interface between the proteins has not been investigated yet on the molecular level.
The active transcription factor HIF-1 is a heterodimer consisting of HIF-1α and HIF-1β [aryl hydrocarbon receptor nuclear translocator (ARNT); ref. 6]. In adaptation to oxygen deficiency, the dimer activates genes involved in angiogenesis, anaerobic metabolism, and iron homeostasis (7). HIF-1β is ubiquitous in the cell, whereas HIF-1α is tightly regulated. A class of hydroxylases acts as oxygen sensors and hydroxylates two proline residues of HIF-1α (P402 and P564) at normal levels of oxygen (8, 9). The von Hippel Lindau (VHL)–E3 complex recognizes the hydroxyproline residues, ubiquitinates HIF-1α, and drives it to degradation in the proteasome (10, 11). The transcription factor is not hydroxylated during hypoxia and, therefore, is not degraded. HIF-1 belongs to the Per-ARNT-Sim (PAS) transcription factor family with an N-terminal DNA-binding domain (basic helix–loop–helix) followed by two folded PAS domains contributing to the dimerization (Fig. 1; ref. 6). The C-terminal part of HIF-1α holds the ODDD and two transactivation domains, one of which overlaps with the ODDD (12). The two proline residues that are hydroxylated both reside in the ODDD (Fig. 1).
Fig 1.
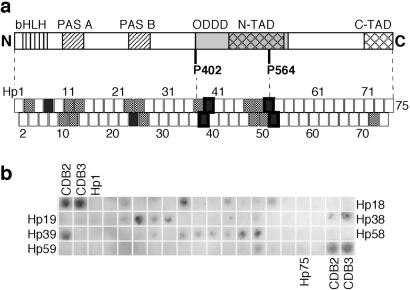
Binding of p53c to an immobilized peptide array screening the HIF-1α sequence. (a) Map of how the array peptides correlated to the HIF-1α structure. The peptides have a sequence overlap with their neighbors and are numbered Hp1–75. HIF-1α consists of the following domains: DNA-binding domain basic helix–loop–helix (bHLH), dimerization domains PAS A and PAS B, oxygen-dependent degradation domain (ODDD), and two transactivation domains (N-TAD and C-TAD). P402 and P564 act as switches at oxygen-regulatory hydroxylation. HIF-1α-derived peptides with strong binding signals are indicated by filled boxes, and weakly binding peptides are indicated by dotted boxes. Peptide boxes with a thick frame have been analyzed in soluble form. (b) Average of three immunoblot experiments of p53c bound to HIF-1α peptide array. The 9-mer peptides CDB2 and CDB3 (15) were used as positive controls.
The tumor suppressor p53 is a homotetramer that is induced by DNA damage and cellular stress. When active, it may induce either cell-cycle arrest or apoptosis. The core domain of p53 (p53c) is the major folded unit of the protein. It harbors a sequence-specific DNA-binding site and frequently is mutated in human cancers (13). The level of p53 in the cell is tightly regulated in a similar fashion to that of HIF-1α. The protein Mdm2, an E3 ligase, is induced by p53 and acts as a feedback inhibitor (14). It binds to the N-terminal transactivation domain of p53 and mediates ubiquitination and degradation of p53.
In this study we probed the interaction between HIF-1α and p53 at the molecular level and localized the binding sites. An array of immobilized peptides derived from the HIF-1α sequence was used to map p53c-binding sites in HIF-1α. The putative sequence motifs and their derivatives then were synthesized for solution studies to determine their binding affinities by biophysical methods and to identify their binding sites in p53c by using NMR spectroscopy.
Materials and Methods
Peptide Array.
We designed an array of 18-mer peptides corresponding to the human HIF-1α sequence with a seven-residue overlap. The array of peptides, immobilized on cellulose, was produced by Jerini Bio Tools GmbH (Berlin). The peptides were acetylated at their N termini and attached to cellulose via their C termini by an amide bond. As controls, peptides CDB2 and CDB3, which are known to bind to p53c (15), were used as the two first and two last peptides in the array.
Screening of p53c Binding to Peptide Array.
The human p53c (residues 94–312) and p53 core+tet (residues 94–360) were expressed and purified (16). The analysis of binding to cellulose-bound peptides (17) was modified slightly. The peptide array attached to cellulose was prewashed in binding buffer (50 mM Hepes, pH 7.2/0.15 M NaCl/5 mM DTT/5% sucrose/0.05% Tween 20). The p53 constructs were incubated with the array in binding buffer for 30 min at 10°C with gentle shaking. The cellulose sheet was washed rapidly in ice-cold binding buffer four times and transferred to polyvinylene difluoride membrane (Bio-Rad) with the peptide side against the membrane. Bound p53 was transferred to membrane (anode side) by semidry blotting at a current of 0.9 mA/cm2 for 20 min. The filters in the blotting sandwich were soaked with different buffers at the cathode (75 mM Tris base/120 mM 6-amino-hexanoic acid/0.01% SDS), on the anode side of the membrane (90 mM Tris base), and on the far anode side (300 mM Tris base). Immunodetection with antibody Pab 240 (Santa Cruz Biotechnology) was used to identify transferred p53. The procedure was adopted according to the manufacturer of enhanced chemiluminescence or enhanced chemifluorescence (Amersham Pharmacia).
Synthesis and Purification of Peptides.
The peptides were synthesized by using a Pioneer peptide synthesizer (Perseptive, Foster City, CA). Standard 9-fluorenylmethoxycarbonyl (Fmoc) chemistry was used, with double couplings when needed. The labeled amino acid derivative Fmoc-Lys (Mca)-OH was purchased from Nova Biochem (18). The peptides were cleaved from the resin by using a mixture of trifluoroacetic acid/H2O/triisopropylsilane 95:2.5:2.5, precipitated from cold ethyl ether, washed 2–3 times with cold ethyl ether, dissolved in H2O/acetonitrile 50:50, and lyophilized. The peptides were purified by using reverse-phase HPLC (Waters 600) on a preparative C8 column (Vydac, Hesperia, CA). The peptides were analyzed by using analytical HPLC and matrix-assisted laser desorption ionization/time-of-flight mass spectrometry (Voyager instrument, Perseptive).
Fluorescence Anisotropy.
p53c (10–160 μM) was titrated into 0.45 μM labeled HIF-1α-derived peptides in 50 mM Hepes, pH 7.2/1 mM DTT at 10°C. For each data point, the mixture was incubated for 1 min with 20 sec of stirring before measurement of fluorescence. Dissociation constants were obtained by fitting both anisotropy and total fluorescence data to equations of one-site binding and additional linear drift. In competition experiments, a stock solution of unlabeled peptide (50–700 μM) was injected stepwise into 800 μl of preincubated mixture of 10 μM p53c and 0.45 μM-labeled peptide. For every step, the new equilibrium was reached before measuring the total fluorescence and anisotropy. The affinity for the unlabeled peptide to p53c was estimated by using the treatment of data described elsewhere (15).
NMR Spectroscopy.
Samples for NMR experiments contained 200–230 μM 15N-labeled p53c and peptides at a concentration of 0.8–1.6 μM. Different peptides were compared at identical concentrations. The samples were dialyzed into 25 mM sodium phosphate, pH 7.2/0.15 M KCl/3.5 mM DTT/2% 2H2O). The 1H-15N heteronuclear single quantum coherence spectra were acquired at 20°C (19). Hp51/52XHyp was analyzed instead of Hp51/52X because of higher solubility of the first peptide.
Results
p53c Binds to Sequence Motifs of HIF-1α.
To see whether and where particular sequences of HIF-1α bind to p53c, we assayed the binding of p53c to immobilized peptides derived from HIF-1α. A peptide array was designed such that 75 immobilized 18-mers covered the complete HIF-1α sequence (Fig. 1a). Neighboring peptides, which overlapped in sequence by seven residues, were synthesized and attached to the membrane through their C termini. p53c was incubated with the peptide array, and any bound p53 was immunodetected after electrotransfer to a membrane (Fig. 1b). Peptides known to bind the native state of p53c (CDB2 and CDB3; ref. 15) were applied in the corner positions of the array as controls.
p53c bound to a number of HIF-1α contiguous overlapping peptides (Fig. 1b), which consist of extended binding motifs (i.e., contiguous overlapping peptides in the array) rather than random nonspecific binding sites. A larger construct of p53c with its tetramerization domain produced a very similar binding pattern. The four major sequence motifs were HIF-1α-derived peptide (Hp)10–13, Hp23–26, Hp37–39, and Hp47–52 (Fig. 1) spanning residues 100–150, 243–293, 397–436, and 507–579, respectively. The two first sequence motifs (Hp10–13 and Hp23–26) mapped to the two PAS domains, which are folded dimerization domains in HIF-1α (Fig. 1a). The third motif (Hp37–39) had the highest affinity, and the fourth motif (Hp47–52) was the longest. These two latter motifs both are situated in the ODDD of HIF-1α (partly described as the N-terminal transactivation domain; ref. 20). One motif covers P402 (in Hp36/37), and the other contains P564 (in Hp51/52), which both act as switches for oxygen-dependent degradation of HIF-1α (21). The two sequences are similar; both contain a patch of negatively charged residues followed by proline (ETDDQQLEEVP in Hp39 and DTDLDLEMLAP in Hp51/52; Table 1).
Table 1.
HIF-1α-derived peptides and their binding affinity to p53 core domain
| Name | Sequence | Dissociation constant, Kd, μM |
|---|---|---|
| Hp38X | Ac–XI408ISLDFGSNDTETDDQQL425-NH2 | 4 ± 1 |
| Hp39X | Ac–XE419TDDQQLEEVPLYNDVML436-NH2 | 3.8 ± 0.5 |
| Hp39 | Ac–E419TDDQQLEEVPLYNDVML436-NH2 | 7 |
| Hp39-1 | Ac–E419TDDQQLEEVPL430-NH2 | ND |
| Hp39-2 | Ac–L425EEVPLYNDVML436-NH2 | ND |
| Hp52X | Ac–XL562APYIPMDDDFQLRSFDQ579-NH2 | >40 |
| Hp51/52X | Ac–XS551TQDTDLDLEMLAPYIPMDDDFQLRSFDQ579-NH2 | 1.7 ± 0.2 |
| Hp51/52XHyp | Ac–XS551TQDTDLDLEMLA-Hyp-YIPMDDDFQLRSFDQ579-NH2 | 2.7 ± 0.3 |
Ac, acetylated N terminus; X, Nɛ-(7-methoxy-coumarin)-lysine residue; ND, not determined.
The binding affinity of unlabeled Hp39 was estimated by binding competition with Hp39X under the assumption that peptides bind to the same site.
p53c Binds to HIF-1α-Derived Peptides in Solution.
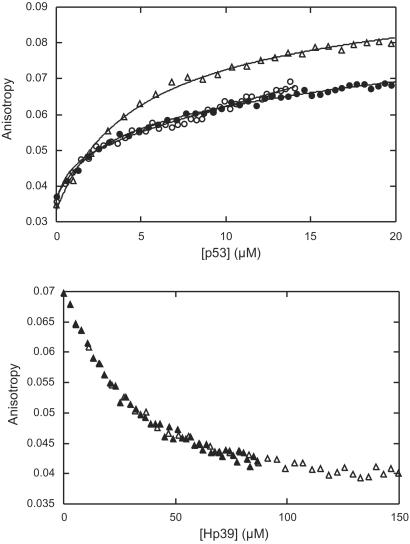
We measured the affinity of the peptides for p53c by fluorescence anisotropy. The peptides were labeled by 7-methoxycoumarin-lysine as the N-terminal residue (denoted X; ref. 18). In addition, the N termini of all peptides were acetylated as in the peptide array. p53c was titrated into a submicromolar concentration of labeled peptide, and both total fluorescence and anisotropy were analyzed (Fig. 2 and Table 1). The soluble peptides had affinities in the low micromolar range. Under screening conditions but with no salt added, the peptides of the first and strongest ODDD sequence motif (Hp38X and Hp39X) had dissociation constants (Kd) of 3–5 μM (Table 1). The combined Hp51 and Hp52 (Hp51/52X), derived from the second ODDD motif, had a Kd of ≈2 μM, whereas Hp52X bound much more weakly (Kd > 40 μM). Hp51/52X contains the full consensus sequence for hydroxylation of P546 (21). We synthesized the hydroxyproline derivative (Hp51/52XHyp) to investigate whether p53c distinguishes between normal proline and hydroxyproline. There was no significant difference between the affinities of Hp51/52X and Hp51/52XHyp. The p53 interaction thus was not dependent on the hydroxylation state of HIF-1α.
Fig 2.
Fluorescence anisotropy studies of binding of HIF-1α-derived peptides to p53c. (Upper) Binding titration (anisotropy) of p53c into Hp39X (triangles), Hp51/52X (open circles), and Hp51/52XHyp (closed circles). (Lower) Binding competition titration (anisotropy), where unlabeled Hp39 was titrated into a mixture of 10 μM p53c and 0.45 μM labeled peptide Hp39X. Two stock concentrations of Hp39 are displayed (open triangles for 633 μM and closed triangles for 317 μM).
The Peptides Bind to the DNA-Binding Site of p53c.
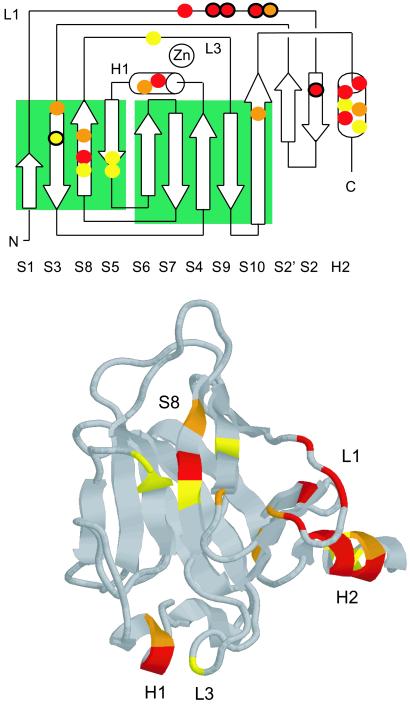
Binding competition experiments indicated that Hp39X (18-mer) and Hp51/52X (29-mer) had overlapping binding sites on p53c. Unlabeled Hp39 displaced a labeled peptide (Hp39X or Hp51/52X) bound to p53c. The relative affinity of Hp39 was estimated from the reduction in anisotropy, assuming the same binding site (Fig. 2 Lower). The affinity of unlabeled Hp39 was ≈7 μM in competition with both Hp39X and Hp51/52X, which showed that the longer Hp51/52X covered the p53c-binding site of the shorter Hp39X. NMR analysis confirmed a common binding site for the two HIF-1α sequence motifs and localized it to the DNA-binding site of p53c (Fig. 3). After binding, changes in the chemical shifts of the p53c backbone amides (1H-15N) were measured by heteronuclear single quantum coherence NMR spectroscopy. Unlabeled Hp39 was measured at two concentrations (0.8 and 1.5 mM) and showed a gradual increase in shift for a set of residues (Fig. 3 Upper). This set describes a continuous site that covers the DNA-binding site of p53c (loop 1 and helix 2; ref. 22) in combination with residues in only one of the β-sheets, forming a line perpendicular to the direction of the β-strands (strands 2, 3, 8, and 5). Residues in helix 1 together with E244 in loop 3 constituted a second site for Hp39. These structural elements harbor residues for coordination of Zn2+ and DNA binding in the minor groove.
Fig 3.
Binding of HIF-1α-derived peptides to p53c analyzed by NMR. (Upper) Topology diagram (22) displaying residues in p53c with difference in shift after binding Hp39. Red, strong shifts (δ-1H > 0.034 ppm or δ-15N > 0.15 ppm) by H115, G117, T118, V122, Y126, H178, H233, G279, R280, and T284; orange, medium shifts (δ-1H > 0.022 ppm or δ-15N > 0.135 ppm) by T123, T140, H179, T231, V272, and R283; yellow, weak shifts (δ-1H > 0.014 ppm or δ-15N > 0.075 ppm) by V143, E198, G199, Y234, G244, R282, and E285; black circles, Hp39 and Hp39-1 showed identical binding, whereas Hp39-2 was significantly weaker, G117, T118, V122, T123, Y126, and V143. (Lower) Picture of p53c with residues colored as described for Upper.
Narrowing Down the Binding Sites.
Two 12-mer fragments of the 18-mer Hp39 were synthesized (Hp39-1 and Hp39-2; Table 1) to narrow down the binding sites. Hp39-1 caused stronger changes in chemical shift than did the Hp39-2, but the full Hp39 gave the most significant changes, indicating that its second part did contribute to the affinity. For a set of six residues mainly found in loop 1 (Fig. 3 Upper), Hp39-1 and Hp39 showed identical binding results, whereas Hp39-2 was weaker. These data defined a region of p53c at which the major interaction with Hp39 took place and also showed that this binding motif of HIF-1α can be pinpointed to the overlap between Hp38 and Hp39, which Hp39-1 described. Hp51/52XHyp derived from the second ODDD site affected the same set of structural elements of p53c as did Hp39 but with larger changes in shifts, which reflected its higher affinity (Table 1). In summary, the NMR experiments defined loop 1 and helix 2 as being the major binding site for the two ODDD motifs in HIF-1α.
Discussion
Here we presented putative binding interfaces between the two transcription factors p53 and HIF-1α. Two homologous peptides derived from the regulatory domain of HIF-1α bound p53c by micromolar affinity.
Peptides as Models.
The interaction between the two proteins has been analyzed previously by cell biology methods, and no study has provided data on the molecular level. In the present work, we used peptides to explore this protein interaction. Peptides were useful in mapping the binding sites as well as in narrowing the interface by reduced peptide length.
There is always a question of whether peptide fragments that are largely unstructured are representative of the equivalent sequences in the native, folded protein. Because of this question mark, we concentrated on just two of four sequence motifs in the screening, Hp51/52 and Hp38/39. Hp51/52 resides in the transactivation domain of HIF-1α (Fig. 1). Because transactivation domains are known to be largely unstructured (23–25), peptide fragments are likely to be particularly good models for such regions. Further, a peptide fragment of Hp51/52 is a substrate for the hydroxylase (9), and also the second region, Hp38/39, is in a region accessible to the hydroxylase (9).
Comparison with Other Binding Proteins.
The identified binding regions appear to be shared with other binding proteins. p53c bound two sequence motifs corresponding to residues 397–436 and 507–576 of HIF-1α. The two proline residues (P402 and P564) crucial to the regulation of HIF-1α are situated in these two motifs. Prolyl hydroxylases as well as VHL–E3 ubiquitin ligase bind specifically to these sites, which in the presence of oxygen results in degradation of HIF-1α (10, 11, 21). The complex with VHL depends on Hyp564. On the contrary, p53c bound a 29-mer peptide containing P564 (Hp51/52X) as tightly as the peptide with Hyp564 (Hp51/52XHyp) (Table 1), which suggests that p53 binds HIF-1α under conditions of hypoxia.
Two proteins have been solved in complex with p53c, and both bind in the DNA-binding region of p53c. The p53-binding protein 1 (53BP1) forms a network of bonds to residues in loop 3 and helix 1 (26), and two loops of the p53-binding protein 2 (53BP2) interact with loops 2 and 3 of p53c (27). Because our results suggest that HIF-1α binds to the same region (Fig. 3), this may be a general binding site of the p53c. It should be noted that a large fraction of tumor-derived mutations map to this region of p53c (13).
Biological Implications.
The biological significance of the studied interaction between p53 and HIF-1α is intriguing and has caused a controversy of whether they act in cooperation or antagonize each other. p53 accumulates during hypoxia (1, 28), and it has been proposed that the phosphorylation status of HIF-1α is important for forming the complex of HIF-1α and p53 (3). After prolonged and severe hypoxia, a dephosphorylated form of HIF-1α is formed that binds p53 and thereby promotes apoptosis (3). It has been proposed also that apoptosis caused by hypoxia is caused by p53-dependent transcriptional repression where p53 interacts with corepressors, and the p53-dependent transcriptional activation is lost (29). Our data suggest that HIF-1α would bind to the DNA-binding site of p53 and thereby neutralize the specific DNA binding of p53, which fits with a cooperative transcriptional repression model.
HIF-1α is induced in many cancer tumors because of hypoxia (see review in ref. 7). HIF-1α is activated in hypoxic cells, because the prolines are not hydroxylated, and the VHL-mediated degradation pathway is then avoided. It has been proposed that p53 controls the level of HIF-1α via an alternative degradation mechanism of HIF-1α (4); the p53 in the p53–HIF-1α complex may recruit Mdm2 for ubiquitination and thus degrade the HIF-1α. Our results are consistent with this proposal, because the nonhydroxylated and hydroxylated peptides bind equally well to p53, whereas hydroxylation is essential for the binding to VHL. It seems that p53 has a tumor-suppressor activity under hypoxia by binding to the transactivation domain of HIF-1α, thereby inhibiting HIF-1-dependent angiogenesis.
The binding sites identified in this study are in agreement with different proposed biological mechanisms. Taken together, it is likely that HIF-1α binds the DNA-binding site of p53 via the two p53-binding motifs in the ODDD. As the two motifs are separated by ≈120 residues, we speculate that two subunits of one p53 tetramer can bind the two motifs in one HIF-1α molecule simultaneously, resulting in a tight cooperative binding. A large fraction of the tumor-derived mutations in p53 map to the DNA-binding site (13) and thus may have impact on the p53–HIF-1α interaction.
Acknowledgments
We thank Karoly von Glos for help in peptide synthesis, and Dr. Dmitry Veprintsev for assistance in fluorescence anisotropy measurements. L.O.H. was supported by a fellowship from the Wenner–Gren Foundations (Stockholm), A.F. was supported by Long-Term Fellowship LT00056/2000-M from the Human Frontier Science Program, and S.R. was supported by an EMBO Long-Term Fellowship.
Abbreviations
HIF, hypoxia-inducible factor
VHL, von Hippel Lindau (protein)
PAS, Per-ARNT-Sim
ODDD, oxygen-dependent degradation domain
p53c, p53 core domain
References
- 1.An W. G., Kanekal, M., Simon, M. C., Maltepe, E., Blagosklonny, M. V. & Neckers, L. M. (1998) Nature (London) 392, 405-408. [DOI] [PubMed] [Google Scholar]
- 2.Blagosklonny M. V., An, W. G., Romanova, L. Y., Trepel, J., Fojo, T. & Neckers, L. (1998) J. Biol. Chem. 273, 11995-11998. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki H., Tomida, A. & Tsuruo, T. (2001) Oncogene 20, 5779-5788. [DOI] [PubMed] [Google Scholar]
- 4.Ravi R., Mookerjee, B., Bhujwalla, Z. M., Sutter, C. H., Artemov, D., Zeng, Q., Dillehay, L. E., Madan, A., Semenza, G. L. & Bedi, A. (2000) Genes Dev. 14, 34-44. [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond E. M., Denko, N. C., Dorie, M. J., Abraham, R. T. & Giaccia, A. J. (2002) Mol. Cell. Biol. 22, 1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G. L., Jiang, B. H., Rue, E. A. & Semenza, G. L. (1995) Proc. Natl. Acad. Sci. USA 92, 5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza G. L. (2000) Genes Dev. 14, 1983-1991. [PubMed] [Google Scholar]
- 8.Bruick R. K. & McKnight, S. L. (2001) Science 294, 1337-1340. [DOI] [PubMed] [Google Scholar]
- 9.Epstein A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., et al. (2001) Cell 107, 43-54. [DOI] [PubMed] [Google Scholar]
- 10.Ivan M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S. & Kaelin, W. G., Jr. (2001) Science 292, 464-468. [DOI] [PubMed] [Google Scholar]
- 11.Jaakkola P., Mole, D. R., Tian, Y. M., Wilson, M. I., Gielbert, J., Gaskell, S. J., Kriegsheim Av, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., et al. (2001) Science 292, 468-472. [DOI] [PubMed] [Google Scholar]
- 12.Huang L. E., Gu, J., Schau, M. & Bunn, H. F. (1998) Proc. Natl. Acad. Sci. USA 95, 7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hainaut P. & Hollstein, M. (2000) Adv. Cancer Res. 77, 81-137. [DOI] [PubMed] [Google Scholar]
- 14.Woods D. B. & Vousden, K. H. (2001) Exp. Cell Res. 264, 56-66. [DOI] [PubMed] [Google Scholar]
- 15.Friedler A., Hansson, L. O., Veprintsev, D. B., Freund, S. M., Rippin, T. M., Nikolova, P. V., Proctor, M. R., Rüdiger, S. & Fersht, A. R. (2002) Proc. Natl. Acad. Sci. USA 99, 937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullock A. N., Henckel, J., DeDecker, B. S., Johnson, C. M., Nikolova, P. V., Proctor, M. R., Lane, D. P. & Fersht, A. R. (1997) Proc. Natl. Acad. Sci. USA 94, 14338-14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rüdiger S., Germeroth, L., Schneider-Mergener, J. & Bukau, B. (1997) EMBO J. 16, 1501-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight C. G., Willenbrock, F. & Murphy, G. (1992) FEBS Lett. 296, 263-266. [DOI] [PubMed] [Google Scholar]
- 19.Wong K. B., DeDecker, B. S., Freund, S. M., Proctor, M. R., Bycroft, M. & Fersht, A. R. (1999) Proc. Natl. Acad. Sci. USA 96, 8438-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugh C. W., O'Rourke, J. F., Nagao, M., Gleadle, J. M. & Ratcliffe, P. J. (1997) J. Biol. Chem. 272, 11205-11214. [DOI] [PubMed] [Google Scholar]
- 21.Masson N., Willam, C., Maxwell, P. H., Pugh, C. W. & Ratcliffe, P. J. (2001) EMBO J. 20, 5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346-355. [DOI] [PubMed] [Google Scholar]
- 23.Gauthier J. M., Dillner, J. & Yaniv, M. (1991) Nucleic Acids Res. 19, 7073-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz M. L., Silva, M. A. D., Altmann, H., Czisch, M., Holak, T. A. & Baeuerle, P. A. (1994) J. Biol. Chem. 269, 25613-25620. [PubMed] [Google Scholar]
- 25.Dyson H. J. & Wright, P. E. (2002) Curr. Opin. Struct. Biol. 12, 54-60. [DOI] [PubMed] [Google Scholar]
- 26.Joo W. S., Jeffrey, P. D., Cantor, S. B., Finnin, M. S., Livingston, D. M. & Pavletich, N. P. (2002) Genes Dev. 16, 583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorina S. & Pavletich, N. P. (1996) Science 274, 1001-1005. [DOI] [PubMed] [Google Scholar]
- 28.Graeber T. G., Peterson, J. F., Tsai, M., Monica, K., Fornace, A. J., Jr. & Giaccia, A. J. (1994) Mol. Cell. Biol. 14, 6264-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koumenis C., Alarcon, R., Hammond, E., Sutphin, P., Hoffman, W., Murphy, M., Derr, J., Taya, Y., Lowe, S. W., Kastan, M. & Giaccia, A. (2001) Mol. Cell. Biol. 21, 1297-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]