Abstract
The structure of the adeno-associated virus (AAV-2) has been determined to 3-Å resolution by x-ray crystallography. AAV is being developed as a vector for gene therapy to treat diseases including hemophilia, cancer, and cystic fibrosis. As in the distantly related autonomous parvoviruses, the capsid protein has a β-barrel fold, but long loops between the β-strands share little structural homology with other parvoviruses, leading to unique surface features. Most prominent are groups of threefold-related peaks, each an intimate association of loops from two neighboring subunits. Mutations affecting cell entry and receptor binding are clustered near the positively charged side of each peak, implicating the region in attachment to the cellular receptor, heparan sulfate proteoglycan. Amino acids involved in antibody binding are in the same general vicinity. The structure will guide rational engineering of vector capsids to tailor cellular targeting and to avoid immediate neutralization by an immune system sensitized by prior exposure to AAV.
Gene therapy is being developed as a treatment for a wide range of diseases for which a genetic cause or an inherited predisposition exists. Gene therapy involves the introduction of new genetic material to correct an abnormality, and one of the biggest challenges remains the development of suitable vectors for the delivery of DNA into targeted cells (1). Along with retroviruses and adenoviruses, recombinantly engineered adeno-associated viruses (rAAV) have emerged as leading candidates (2), in which the therapeutic DNA replaces much of the viral genome and is contained within the protein capsid shell. Each viral vector has its own utility. The advantages of AAV include transduction of nondividing cells and a mild immune reaction (3). Clinical trials with rAAV vectors are underway for treatment of several diseases including cystic fibrosis (4) and hemophilia (5), whereas success in animal models is encouraging development of treatments for high blood pressure (6), Parkinson's disease (7, 8), muscular dystrophy (9, 10), and many other afflictions.
The natural hosts of the wild-type virus are human. Indeed ≈80% of the population is seropositive because of natural exposure to AAV (11, 12). However, AAV has not been associated with disease (13). The replication of AAV depends on coinfection by a helper virus, and no pathogenic effects apparently exist beyond those caused by the helper virus. The helper is usually adenovirus (14). Hence the name “adeno-associated virus” and their classification as “dependoviruses,” although they share no other relation to adenoviruses. The dependence on a helper virus makes AAV a satellite virus, leading to technical challenges (see below) in this first structure determination of an animal satellite virus. Before replication, the natural life cycle involves latent integration of AAV's DNA into the host's. Wild-type AAV is the only known eukaryotic virus that integrates at a specific site in the human genome (15). Vector constructs have generally not taken advantage of this specificity, because the gene for the replication protein that encodes the site specificity is usually replaced.
Adeno-associated viruses are small single-stranded DNA viruses of the Dependovirus genus that constitute one of the four Parvoviridae family genuses (14). The other genuses have no requirement for a helper virus, replicating autonomously. Atomic structures have been obtained for several from the Parvovirus genus. Canine parvovirus (CPV), feline panleukopenia virus, and minute virus of mouse share 50–97% capsid sequence identity, and similar three-dimensional structures (16–20). The genomic DNA is surrounded by a near-spherical protein shell that comprises 60 capsid protein subunits arranged with T = 1 icosahedral symmetry. The subunit has a β-barrel core that is common in viral capsid proteins (21). The structure of an insect parvovirus (Densovirus) is also known: the subunit fold is the same, but the loops are a total of ≈130 residues shorter and most of the surface features of the capsid are truncated (22). Hitherto, high-resolution structure has not been available for Dependoviruses like AAV. The genetic organization is similar to other parvoviruses, but the capsid sequence identities are low (7–22%), less than between the rhino- and entero-subfamilies of picornaviruses (23), which indicates that homology between AAV and other parvoviruses might be limited. Indeed, recent electron microscopy images showed substantial differences in surface topology between AAV-2 and canine parvovirus (24). Although homology modeling has been attempted (see ref. 25, for example), an experimental atomic structure has been a high priority for development of viral gene therapy vectors.
AAV-2 is one of five distinct serotypes. Serotypes 1, 2, and 3 share ≈85% capsid sequence identity, with 2 and 3 likely using the same cellular receptor, whereas serotypes 4 and 5 are more distantly related (≈55% identity to each other and to AAV-2) and use different receptors (26, 27). The primary receptor for AAV-2 is heparan sulfate proteoglycan, but secondary receptors and alternative primary receptors may also be available (28–31). AAV-2 enters the cell through an endosomal route, is transported to the nucleus by motor proteins, and enters the nucleus, all within seconds (32, 33). Monoclonal antibodies have been used to study capsid assembly and to map sequence determinants of receptor attachment (see ref. 34, for example). Systematic mutagenesis has begun to yield the determinants of various phenotypes (see refs. 25, 35, and 36, for example), providing an emerging wealth of data to be integrated in mapping function to structure.
Materials and Methods
AAV-2 was propagated in cultured human cells that were infected with helper adenovirus. HeLa cells were transfected with the pAV2 clone then infected with adenovirus-2 (37, 38). Production was scaled-up by optimization of the timing of transfection/infection and by switching to suspension culture. AAV was purified by three CsCl-gradient sedimentations by ultracentrifugation for ≈24 h in an SW55 swinging-bucket rotor at 38,000 rpm. Solubility at high concentration/low ionic strength was maintained through the addition of >5% glycerol. Crystallization was by vapor diffusion with AAV at ≈8.5 mg/ml in 25% glycerol, equilibrated against a reservoir also containing 4–5% polyethylene glycol 6000.
Diffraction data were collected at the Cornell High Energy Synchrotron Source F1 beam line. Data from the best two cryo-frozen form II crystals were processed with HKL (39) initially with Laue group R3 with a unit cell of a = b = c = 648.1 Å and α = β = γ = 22.2° (hexagonal equivalent: a = b = 250.3 Å, c = 1,895.1 Å). The data set was expanded 3-fold and reindexed to P1 on determination that the exact threefold in the diffraction was the result of a noncrystallographic 31 axis between particles exactly along the pseudo-hexagonal axis.
Phases were determined at low resolution by molecular replacement, then extended by noncrystallographic symmetry averaging. Virus orientations were determined by the glrf self-rotation function, locked according to the icosahedral symmetry (40). At 3-Å resolution, it showed icosahedral threefolds tilted 6° from the noncrystallographic screw axis. Approximate particle positions were found from a native Patterson at 10-Å resolution that showed interparticle vectors caused by the near alignment of icosahedral threefolds with the interparticle noncrystallographic 31. Positions were improved by a home-programmed cross-translation function by using the structure of canine parvovirus (16), initially constraining screw symmetry between particle positions. Phases were extended from the average of superposed canine parvovirus and densovirus (22) models by using Rave (41), starting at 15-Å resolution and slowly extending to 3 Å in six-cycle steps of 1/250 Å. Amplitudes and phases for missing observations between 40 and 3.75 Å were filled—i.e., calculated by Fourier transformation of the averaged map. The resolution was then extended from 3.75 to 3.0 Å again, filling the unobserved reflections. The model was built by using o (42) and refined with simulated annealing in real space (43), then conjugate gradients in reciprocal space by using cns (44).
Results and Discussion
Structure Determination.
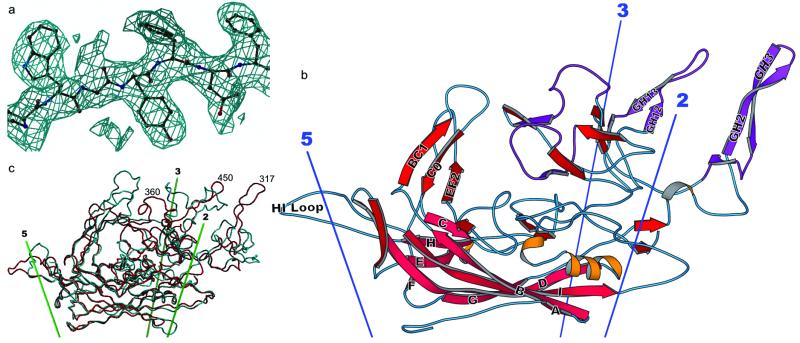
As a satellite virus, AAV-2 was challenging to produce in the required milligram quantities because of the cytopathic effects of the helper virus on the human cells in which AAV was propagated. The optimized protocol yielded 2–5 mg per preparation of 99% pure AAV-2, as judged by silver-stained SDS/PAGE, internally calibrated according to the known composition of VP1, 2, and 3. Crystallization was intractable until we found that a glycerol cosolvent could increase the solubility. The high-resolution crystal form eventually obtained presented challenges in data collection and analysis, because the asymmetric unit contained three complete virus particles or 180 proteins, totaling about 12 million daltons, which complicated the determination of viral positions and orientations, a prerequisite of phasing by symmetry averaging. The challenges of complete data collection and averaging led to statistics for the averaged map and final model (Table 1) that are similar to the poorer examples of prior successful viral structure determinations (e.g., refs. 45 and 46). Nevertheless, the dividend of the high-order noncrystallographic redundancy was an electron density map for the protein capsid shell that was of higher quality than usually obtained in protein crystallographic analyses at 3-Å resolution and that was interpretable without ambiguity (Fig. 1a).
Table 1.
Structure determination of AAV-2
| Space group | P1 | Unit cell | a = b = 249.7 Å, c = 644.8 Å; α = 90.0°, β = 101.2°, γ = 120.0° |
| Data processing (three-fold-expanded in pseudo-R3) | Noncrystallographic averaging | ||
| Observations | 747,988 | Redundancy | 180-fold |
| Unique reflections | 495,356 | Correlation coefficients | |
| Resolution | 100–3 Å (shell at 3.5 Å) | cc(2Fo − F〈map〉, φ〈map〉) | 0.78 |
| Completeness | 56% (43%) (80% to 3.5 Å) | cc(Fo, F〈map〉) | 0.65 |
| Rmerge | 0.156 (0.309) | R(Fo, F〈map〉) | 0.32 |
| Model | |||
| Amino acids | 180 × 519 | R/Rfree | 0.338/0.342 |
| Missing | 1–14 (VP3) | rms deviations from ideal bond lengths/angles | 0.02 Å/2.2° |
Rmerge = ∑h∑i[Ih,i − 〈Ih〉]/∑h〈Ih〉, where Ih,i is the ith observation of a symmetry equivalent of reflection h.
F〈map〉, φ〈map〉 are Fourier terms of the back-transformed final averaged map.
Correlation between the densities of all noncrystallographic symmetry equivalent points.
Rfree is not unbiased. Test-set reflections are correlated to working-set reflections mapped to the neighborhood with noncrystallographic symmetry. However, with the high ratio of data to parameters, over-fitting will be negligible.
Fig 1.
Structure of the AAV-2 subunit and comparison with related structures. (a) Experimental electron density for AAV-2. Phases for this 3-Å resolution electron density map are independent of the AAV-2 model, having been obtained by symmetry averaging and extension from a CPV model at 15-Å resolution. Density is clear and allows an unambiguous fitting of the chemical sequence throughout VP3. (b) Ribbon drawing of the AAV-2 subunit. The locations of the neighboring symmetry axes are shown. The β-barrel is on the inner surface of the capsid (pink) with strands of the two sheets labeled conventionally as A, B, I, D, and G, and C, H, E, and F. Loops are labeled according to the flanking strands—e.g., GH loop. Regions where the sequence differs greatest between the AAV serotypes are colored purple (61). (c) Comparison of the backbones of AAV-2 (red) and canine parvovirus (cyan). The loop structure, which is responsible for many of the viral-host interactions differs substantially between AAV-2 and canine parvovirus, is largely absent from insect densoviruses (not shown).
Each capsid comprises 60 subunits arranged with T = 1 icosahedral symmetry. Parvoviruses actually contain three capsid proteins (VP1–3) in proportions of about 1:1:10, that share overlapping sequence, differing only at their N termini. Electrophoresis showed that all were present in the crystals. VP2 is 137 residues shorter than VP1 and is the product of an alternative start codon, while VP3 is 65 residues shorter than VP2. Although we follow the convention of VP2 numbering for parvoviral structures, it is the 533-residue VP3 that is visualized, along with the corresponding regions of VP1 andVP2 occupying equivalent positions in the capsid, and not the N termini unique to the minority proteins. All except 14 N-terminal residues of VP3 were clearly traceable, and no ambiguity occurred in the alignment of the chemical sequence to the atomic structure.
Subunit Architecture.
Although they share only 7–23% sequence identity (20), the AAV structure shows that dependoviruses share the same subunit fold as the autonomously replicating parvoviruses, including insect densoviruses (22) and canine parvovirus (16). The motif, a jelly-roll β-barrel, comprising two antiparallel β-sheets, is common in virus capsids (21). However, in AAV and canine parvovirus long-loop insertions between the strands of the core β-barrel make the domain two to four times larger than in small RNA viruses. The densovirus subunit is 130 residues shorter, and has truncated loops relative to both AAV and canine parvovirus (22). In other ways the structure of AAV is closer to that of mammalian autonomous parvoviruses than insect parvoviruses. The β-barrels are oriented with respect to the icosahedral symmetry axes as they are in canine parvovirus, not rotated as in densovirus.
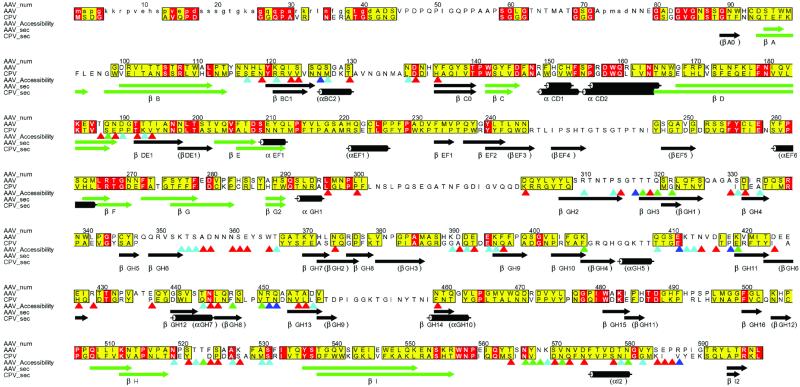
The long interstrand loops of AAV, which together comprise about 60% of the structure, contain β-ribbons and other additional elements of secondary structure, about half of which are found in the autonomous viruses like canine parvovirus (Fig. 2). Thus, the loops have quite different structures in the different parvovirus genuses, consistent with the lower sequence identity (<15%) outside the more conserved barrel β-strands. These loops constitute the surface features of the virus that govern interactions with antibodies and cellular receptors.
Fig 2.
Conservation of sequence and secondary structure. Sequences of AAV-2 and CPV were aligned according to structure, then improved in loop regions according to sequence [Programs STAMP, PILEUP, and ALSCRIPT (62–66)]. Residues that are identical in AAV-2 and CPV are red. Those with an ALSCRIPT similarity score >5 (measured from 0 to 10) are yellow. Secondary structures of AAV-2 and CPV are compared, the core β-barrel highlighted in green, and labeled according to AAV-2 (CPV in parentheses where different). Side chain (outer) surface accessibility was calculated with a 1.5 Å radius probe, and is shown with triangles shaded according to each residue's accessibility: 20 Å2 < red <50 Å2 < cyan <80 Å2 < green <100 Å2 < blue.
Surface Topology and Assembly.
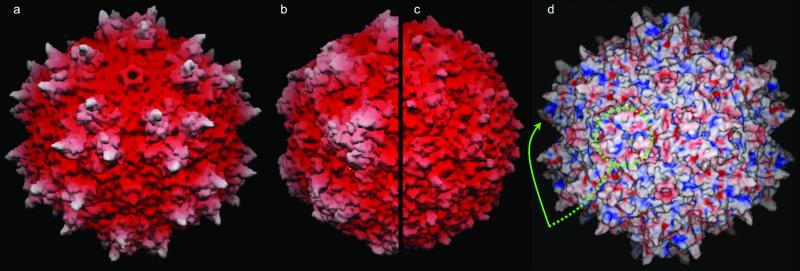
The surface of AAV has more distinctive topology than that of autonomous parvoviruses or densoviruses (Fig. 3). The most prominent features are “threefold-proximal” peaks clustering around each icosahedral threefold rotation axis. Here, loops from neighboring subunits interact intimately. At the center of each peak is a subloop (VP2 residues 348–379) that is part of the GH loop (between β-strands G and H) of one subunit. It is sandwiched by two other subloops (293–341 and 440–458) from the GH loop of a threefold symmetric subunit (Fig. 4a). The subloops of the second subunit are therefore folded away from their β-barrel to extend over a neighboring subunit. Somewhat similarly, a more modest interaction occurs between residues 517 and 531 of the HI loop extending over a fivefold-related neighbor to interact with residues 113–115 and 234–236 of the BC and EF loops, respectively. The loop structures that form these interactions between neighboring subunits are unique to AAV (Fig. 1c). Near the threefold axis, the long GH loops are missing in densoviruses and have very different structure in canine parvovirus. In canine parvovirus they layer over one another as subunits come together during assembly (17), achieving intimate contacts by very different means.
Fig 3.
Surface topology and electrostatics. (a–c) show grasp (67) surface renderings of AAV-2, canine parvovirus, and the insect densovirus, respectively, drawn to scale, and colored according to distance from the viral center. The view is down a twofold axis (center of the virus) with threefolds left and right of center, fivefolds above and below (see Fig. 4b). (d) shows the electrostatic surface potential of AAV-2 calculated with spock (68) running from −10 (red) to +10 (blue). The putative receptor-binding sites are positively charged patches on the side of each threefold-proximal peak, viewed edge-on (arrowed) or head-on where 3 equivalent patches join at each threefold (circled).
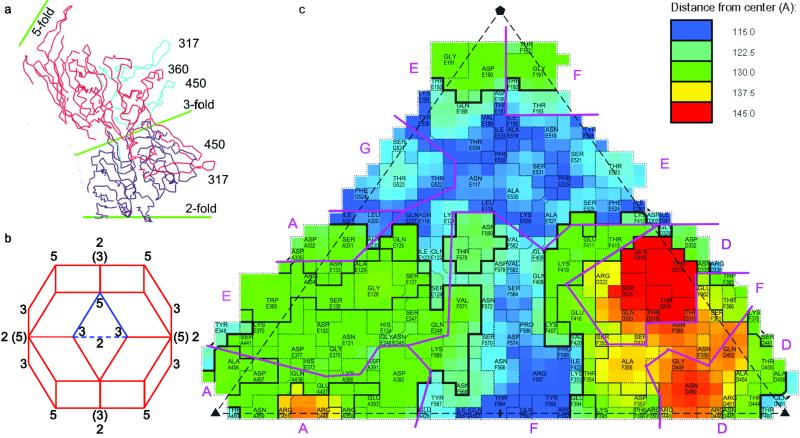
Fig 4.
Interdigitation of subunits: (a) shows a thin equatorial cross-section of AAV-2 (Cα traces) orthogonal to a twofold near the threefold axes. It emphasizes the elevation of the peaks, each of which comprises loops from two subunits that are colored differently. (b) a rhombic triacontahedron showing the viral orientation for Figs. 3 and 4. The three- and fivefold axes are at vertices joining three and five faces, respectively, and twofolds bisect neighboring threefolds, some of which are labeled. (Axes in parentheses superimpose, at an angle, upon twofolds behind.) The highlighted triangle (used in c) may be repeated sixtyfold with icosahedral symmetry operators to generate the entire capsid. (c) A schematic projection of one of the 60 triangular facets shows the surface amino acids with different subunits separated by purple lines. roadmap (69) was used to color as in a topographical map, with blue closest to the virus center, red farthest from the virus center. VP2 numbering is used with a letter prefix denoting the symmetry equivalent subunit.
The loop-to-loop contacts and maturation of the peak structure can only come about after the subunits associate. Assembly cannot be regarded simply as an association between rigid units. Although it is possible that these postencounter rearrangements do not have an impact on the kinetic pathways of assembly, once formed, they would provide a greater thermodynamic barrier to disassembly and therefore stabilize the threefold interactions.
Receptor Attachment and Antigenicity.
The primary receptor for AAV-2 on the cell surface is heparan sulfate proteoglycan, but other receptors may also be available as alternates or secondary receptors (28–31). Serotype 3 shares ≈85% capsid sequence identity with AAV-2 and likely uses the same cellular receptor, whereas serotypes 4 and 5 (≈55% identity) likely use different receptors (26, 27). With the structure, it is possible to map the sites of mutations that affect receptor binding that come from different parts of the primary sequence. Several of these sites come together in the three-dimensional structure, implicating the side of the threefold-proximal peaks in receptor binding (Figs. 3 and 4). Details follow. Of five hep− mutants that are deficient in heparan binding which were prepared by Wu et al. (25), two are located within one loop of the threefold-proximal peak: an alanine substitution at 448-RGNR-451, and a hemagluttinin epitope insertion at 454. They are on the side of the peak facing the valley separating this peak from its threefold-related neighbor (Fig. 3). The third mutation, an insertion at H372, is on the floor of the valley. The fourth mutation, an alanine substitution at 424–428 is at the base of the peak, facing the twofold axis. The fifth mutation, an insertion at 385, and the hep− mutation at 382 of Rabinowitz et al. (36) are buried underneath the peak and possibly affect binding indirectly through residues 390–395 of the valley floor. Further evidence comes from monoclonal antibody C37-B that inhibits cell attachment (34). Of scanned AAV peptides, it is 356–366 that best inhibits competitively the binding of antibody to virus, implicating residues at the crest of the peak adjacent to the hep− sites (448–451) of a symmetry-related subunit. Insertion mutations that block binding of antibody C37-B are also in the hep− loop (at 436 and 450) (34), suggesting that the epitope and receptor-binding regions overlap. Another mutant of similar phenotype at residue 397 (34) might act indirectly, because, like 436, it is buried at the base of the peak. In summary, regions previously implicated in receptor binding from different parts of the primary structure, come together in the three-dimensional structure, at or near the surface of the threefold-proximal peaks.
Positively charged patches are common in heparin-binding proteins (47) because of the predominance of ionic interactions with the sugar sulfates. Overall, the outside surface is positively charged with a prominent ring of symmetry-related patches in a depression surrounding the fivefold axes (Fig. 3d). Similarity to the rhinoviral canyon, the major group receptor-binding site (48), seems to be coincidental. The AAV depression is wider and more accessible to antibodies, and mutational data (previous paragraph) implicate not this region, but the side of the threefold-proximal spikes, which is the other prominently positively charged region, extending from the valley between peaks to merge at the threefold axis (Fig. 3d). It includes basic residues R448 and R451 (mentioned above), R347 and R350 which all point into the cleft, whereas K353 points to the valley floor. (K390 and K395 of the valley floor are counterbalanced by acidic residues 391–393.) The linear sequences resemble consensus motifs (49): 448–451 as a BTTB motif, and 347–353 as a BXXBXXB motif, where B and T signify basic and turn residues, respectively. The region 347–353 is conserved in the two serotypes that bind heparin sulfate, but has a R350G substitution in AAV-4 and -5 which do not. Residues 448-RGNR-451 on the valley rim are unique to AAV-2, offering a rationale for the 10-fold weaker inhibition of AAV-3 infectivity by heparin compared with AAV-2 (50). The electrostatic receptor interactions with these residues would be more subtle than in many heparin-binding proteins, but stronger than in foot-and-mouth-disease virus, where an additive effect of weak interactions at several symmetry-related sites was proposed (51), an argument that could apply equally to AAV. The separation between sites surrounding a threefold in AAV is 20 Å, consistent with binding neighboring disaccharides (52), whereas the separation between threefolds is ≈70 Å, commensurate with the spacing of highly sulfonated regions of the carbohydrate (51).
With regard to mapping the antigenic determinants of AAV, the binding of monoclonal antibody C37-B to the threefold-proximal peaks has already been discussed. Of other antibodies, monoclonal A20 is the best characterized, recognizing a conformational epitope specific to assembled particles (25). Three of four AAV regions implicated in its binding are colocalized on the surface midway between three- and fivefold axes. Residue A129 was implicated by scanning insertional mutagenesis (25) and is adjacent to two other regions implicated in A20 binding: 134–143 and 232–241 (34) (but 34 Å away from a 4th at 429–438). Other antigenic maps, based on polyclonal sera and peptide scanning (12), are less consistent with the structure: some implicated regions are on the outer surface, others not. The picture emerging from the structural interpretation of A20 and C37-B binding is that antigenic sites include, but are not restricted to the protruding threefold-proximal peaks. In fact, probing the structure with an antibody-sized sphere (10-Å radius) and analysis of B-factors, indicate that many parts of the surface are suitably accessible and flexible, two criteria that have previously been used to predict antigenic sites (53, 54).
Prospects for Gene Therapy.
The structure, combined with prior phenotypic mappings, provides initial pointers toward the engineering of improved gene therapy vectors (35, 55). It implicates the side of each threefold-proximal peak in cellular receptor binding, and shows that neighboring regions are also accessible to antibody binding. Comparing to picornaviruses, AAV-2 is more like foot-and-mouth-disease virus and minor receptor group rhinoviruses, with overlapping antigenic and receptor-binding sites (51, 56), than major-group rhinoviruses, poliovirus and some coxsackie A and B viruses, where surface topology restricts antibody access to the receptor-binding site (48, 57–60). The structure also provides a framework for rational site-directed mutagenesis, through which the molecular mechanisms of viral-host interactions can be probed. A rational, structure-assisted approach should accelerate the progress in mapping functional sites and modulating function, that has, to date, been attempted through scanning mutagenesis (25, 35, 55). This mapping, in turn, will provide a foundation for the engineering of recombinant AAV for the more efficient delivery of gene therapies to specifically targeted cells.
Acknowledgments
We thank the laboratory of Ken Berns for hospitality when we were learning to culture AAV. Data were collected at the Cornell High Energy Synchrotron Source. The work was funded by the American Cancer Society (RPG-99-356-01-GMC) and a predoctoral fellowship (to S.B.) from the Florida Division of the American Heart Association.
Abbreviations
AAV, adeno-associated virus
CPV, canine parvovirus
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Orkin S. H. & Motulsky, A. G., (1995) Report and Recommendations of the Panel to Access the NIH Investment in Research on Gene Therapy (National Institutes of Health, Bethesda).
- 2.Pfeifer A. & Verma, I. (2001) in Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 469–492.
- 3.Carter B. J. (2000) in Parvoviruses: From Molecular Biology to Pathology and Therapeutic Uses, eds. Faisst, S. & Rommelaere, J. (Karger, Basel), pp. 85–106.
- 4.Flotte T. R. & Carter, B. J. (1998) Methods Enzymol. 292, 717-732. [DOI] [PubMed] [Google Scholar]
- 5.Kay M. A., Manno, C. S., Ragni, M. V., Larson, P. J., Couto, L. B., McClelland, A., Glader, B., Chew, A. J., Tai, S. J., Herzog, R. W., et al. (2000) Nat. Genet. 24, 257-261. [DOI] [PubMed] [Google Scholar]
- 6.Phillips M. (1997) Hypertension 29, 177-187. [DOI] [PubMed] [Google Scholar]
- 7.Mandel R. J., Rendahl, K. G., Snyder, R. O. & Leff, S. E. (1999) Exp. Neurol. 159, 47-64. [DOI] [PubMed] [Google Scholar]
- 8.Mochizuki H., Hayakawa, H., Migita, M., Shibata, M., Tanaka, R., Suzuki, A., Shimo-Nakanishi, Y., Urabe, T., Yamada, M., Tamayose, K., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10918-10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greelish J. P., Su, L. T., Lankford, E. B., Burkman, J. M., Chen, H., Konig, S. K., Mercier, I. M., Desjardins, P. R., Mitchell, M. A., Zheng, X. G., et al. (1999) Nat. Med. 5, 439-443. [DOI] [PubMed] [Google Scholar]
- 10.Wang B., Li, J. & Xiao, X. (2000) Proc. Natl. Acad. Sci. USA 97, 13714-13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erles K., Sebokova, P. & Schlehofer, J. R. (1999) J. Med. Virol. 59, 406-411. [DOI] [PubMed] [Google Scholar]
- 12.Moskalenko M., Chen, L., van Roey, M., Donahue, B. A., Snyder, R. O., McArthur, J. G. & Patel, S. D. (2000) J. Virol. 74, 1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattison J. R. (1990) in Virology, eds. Fields, B. N. & Knipe, D. M. (Raven, New York), pp. 1765–1784.
- 14.Muzyczka N. & Berns, K. I. (2001) in Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 2327–2360.
- 15.Kotin R. M., Siniscalco, M., Samulski, R. J., Zhu, X. D., Hunter, L., Laughlin, C. A., McLaughlin, S., Muzyczka, M., Rocchi, M. & Berns, K. I. (1990) Proc. Natl. Acad. Sci. USA 87, 2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsao J., Chapman, M. S., Agbandje, M., Keller, W., Smith, K., Wu, H., Luo, M., Smith, T. J., Rossmann, M. G., Compans, R. W., et al. (1991) Science 251, 1456-1464. [DOI] [PubMed] [Google Scholar]
- 17.Xie Q. & Chapman, M. S. (1996) J. Mol. Biol. 264, 497-520. [DOI] [PubMed] [Google Scholar]
- 18.Agbandje-McKenna M., Llamas-Saiz, A. L., Wang, F., Tattersall, P. & Rossmann, M. G. (1998) Structure (London) 6, 1369-1381. [DOI] [PubMed] [Google Scholar]
- 19.Agbandje M., McKenna, R., Rossmann, M. G., Strassheim, M. L. & Parrish, C. R. (1993) Proteins 16, 155-171. [DOI] [PubMed] [Google Scholar]
- 20.Chapman M. S. & Rossmann, M. G. (1993) Virology 194, 491-508. [DOI] [PubMed] [Google Scholar]
- 21.Harrison S. C. (2001) in Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), pp. 53–86.
- 22.Simpson A. A., Chipman, P. R., Baker, T. S., Tijssen, P. & Rossmann, M. G. (1998) Structure (London) 6, 1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman M. S. & Rossmann, M. G. (1993) Virology 195, 745-765. [DOI] [PubMed] [Google Scholar]
- 24.Kronenberg S., Kleinschmidt, J. A. & Bottcher, B. (2001) EMBO Rep. 2, 997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu P., Xiao, W., Conlon, T., Hughes, J., Agbandje-McKenna, M., Ferkol, T., Flotte, T. & Muzyczka, N. (2000) J. Virol. 74, 8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davidson B. L., Stein, C. S., Heth, J. A., Martins, I., Kotin, R. M., Derksen, T. A., Zabner, J., Ghodsi, A. & Chiorini, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinowitz J. E., Rolling, F., Li, C., Conrath, H., Xiao, W., Xiao, X. & Samulski, R. J. (2002) J. Virol. 76, 791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summerford C. & Samulski, R. J. (1998) J. Virol. 72, 1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summerford C., Bartlett, J. S. & Samulski, R. J. (1999) Nat. Med. 5, 78-82. [DOI] [PubMed] [Google Scholar]
- 30.Qing K., Mah, C., Hansen, J., Zhou, S., Dwarki, V. & Srivastava, A. (1999) Nat. Med. 5, 71-77. [DOI] [PubMed] [Google Scholar]
- 31.Qiu J.-M., Mizukami, H. & Brown, N. S. (1999) Nat. Med. 5, 467. [DOI] [PubMed] [Google Scholar]
- 32.Bartlett J. S., Wilcher, R. & Samulski, R. J. (2000) J. Virol. 74, 2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seisenberger G., Ried, M. U., Endress, T., Buning, H., Hallek, M. & Brauchle, C. (2001) Science 294, 1929-1932. [DOI] [PubMed] [Google Scholar]
- 34.Wobus C. E., Hugle-Dorr, B., Girod, A., Petersen, G., Hallek, M. & Kleinschmidt, J. A. (2000) J. Virol. 74, 9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girod A., Ried, M., Wobus, C., Lahm, H., Leike, K., Kleinschmidt, J., Deleage, G. & Hallek, M. (1999) Nat. Med. 5, 1052-1056. [DOI] [PubMed] [Google Scholar]
- 36.Rabinowitz J. E., Xiao, W. & Samulski, R. J. (1999) Virology 265, 274-285. [DOI] [PubMed] [Google Scholar]
- 37.Yakobson B., Koch, T. & Winocour, E. (1987) J. Virol. 61, 972-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laughlin C. A., Tratschin, J. D., Coon, H. & Carter, B. J. (1983) Gene 23, 65-73. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z. & Minor, W. (2001) in International Tables for Crystallography: Crystallography of Biological Molecules, eds. Rossmann, M. G. & Arnold, E. (International Union of Crystallography, Dordrecht, The Netherlands), Vol. F.
- 40.Tong L. & Rossmann, M. G. (1997) Methods Enzymol. 276, 594-611. [PubMed] [Google Scholar]
- 41.Jones T. A. (1992) in Proceedings of the CCP4 Study Weekend, eds. Dodson, E., Gover, S. & Wolf, W. (Science and Engineering Research Council, Daresbury Laboratory, Warrington, U.K.), pp. 91–105.
- 42.Jones T. A. & Kjeldgaard, M. (1997) Methods Enzymol. 277, 173-208. [DOI] [PubMed] [Google Scholar]
- 43.Korostelev A., Bertram, R. & Chapman, M. S. (2002) Acta Crystallogr. D 58, 761-767. [DOI] [PubMed] [Google Scholar]
- 44.Brünger A. T., Adams, P. D., Clore, G. M., Gros, P., Gross-Kunstleve, R. W., Jiang, J.-S., Kurzewski, J., Nilges, M., Pannu, N. S., Read, R. J., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 45.Llamas-Saiz A. L., Agbandje-McKenna, M., Wikoff, W. R., Rossmann, M. G., Bratton, J. & Tattersall, P. (1997) Acta Crystallogr. D 53, 93-102. [DOI] [PubMed] [Google Scholar]
- 46.Filman D. J., Wien, M. W., Cunningham, J. A., Bergelson, J. M. & Hogle, J. M. (1998) Acta Crystallogr. D 54, 1261-1272. [DOI] [PubMed] [Google Scholar]
- 47.Mulloy B. & Linhardt, R. J. (2001) Curr. Opin. Struct. Biol. 11, 623-628. [DOI] [PubMed] [Google Scholar]
- 48.Olson N., Kolatkar, P., Oliveira, M. A., Cheng, R. H., Greve, J. M., McClelland, A., Baker, T. S. & Rossmann, M. G. (1993) Proc. Natl. Acad. Sci. USA 90, 507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hileman R. E., Fromm, J. R., Weiler, J. M. & Linhardt, R. J. (1998) BioEssays 20, 156-167. [DOI] [PubMed] [Google Scholar]
- 50.Handa A., Muramatsu, S., Qiu, J., Mizukami, H. & Brown, K. E. (2000) J. Gen. Virol. 81, 2077-2084. [DOI] [PubMed] [Google Scholar]
- 51.Fry E. E., Lea, S. M., Jackson, T., Newman, J. W., Ellard, F. M., Blakemore, W. E., Abu-Ghazaleh, R., Samuel, A., King, A. M. & Stuart, D. I. (1999) EMBO J. 18, 543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Margalit H., Fischer, N. & Ben-Sasson, S. A. (1993) J. Biol. Chem. 268, 19228-19231. [PubMed] [Google Scholar]
- 53.Novotny J., Bruccoleri, R. E., Carlson, W. D., Handschumacher, M. & Haber, E. (1987) Science 238, 1584-1586. [DOI] [PubMed] [Google Scholar]
- 54.Tainer J. A., Getzoff, E. D., Alexander, H., Houghten, R. A., Olson, A. J., Lerner, R. A. & Hendrickson, W. A. (1984) Nature (London) 312, 127-134. [DOI] [PubMed] [Google Scholar]
- 55.Rabinowitz J. E. & Samulski, R. J. (2000) Virology 278, 301-308. [DOI] [PubMed] [Google Scholar]
- 56.Hewat E. A., Neumann, E., Conway, J. F., Moser, R., Ronacher, B., Marlovits, T. C. & Blaas, D. (2000) EMBO J. 19, 6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He Y., Bowman, V. D., Mueller, S., Bator, C. M., Bella, J., Peng, X., Baker, T. S., Wimmer, E., Kuhn, R. J. & Rossmann, M. G. (2000) Proc. Natl. Acad. Sci. USA 97, 79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belnap D. M., McDermott, B. M., Jr., Filman, D. J., Cheng, N., Trus, B. L., Zuccola, H. J., Racaniello, V. R., Hogle, J. M. & Steven, A. C. (2000) Proc. Natl. Acad. Sci. USA 97, 73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rieder E., Gorbalenya, A. E., Xiao, C., He, Y., Baker, T. S., Kuhn, R. J., Rossmann, M. G. & Wimmer, E. (2001) Dev. Biol. 105, 111-122., 149–150. [PubMed] [Google Scholar]
- 60.He Y., Chipman, P. R., Howitt, J., Bator, C. M., Whitt, M. A., Baker, T. S., Kuhn, R. J., Anderson, C. W., Freimuth, P. & Rossmann, M. G. (2001) Nat. Struct. Biol. 8, 874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rutledge E., Halbert, C. & Russell, D. (1998) J. Virol. 72, 309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russell R. B. & Barton, G. J. (1992) Proteins 14, 309-323. [DOI] [PubMed] [Google Scholar]
- 63.GCG, (2001) gcg Wisconsin Package (Accelrys, San Diego).
- 64.Barton G. J. (1993) Protein Eng. 6, 37-40. [DOI] [PubMed] [Google Scholar]
- 65.Ruffing M., Heid, H. & Kleinschmidt, J. A. (1994) J. Gen. Virol. 75, 3385-3392. [DOI] [PubMed] [Google Scholar]
- 66.Parrish C. R., Aquadro, C. F., Strassheim, M. L., Evermann, J. F., Sgro, J.-Y. & Mohammed, H. O. (1991) J. Virol. 65, 6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicholls A., (1992) grasp: Graphical Representation and Analysis of Surface Properties (Columbia Univ. Press, New York).
- 68.Christopher J. A. & Baldwin, T. O., (1997) spock (Texas A&M Univ., College Station, TX).
- 69.Chapman M. S. (1993) Protein Sci. 2, 459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]