Abstract
CCR5 encodes a cell surface chemokine receptor molecule that serves as the principal coreceptor, with CD4, for HIV-type 1 (HIV-1). Varied HIV-1 susceptibility and time to progression to AIDS have been associated with polymorphisms in CCR5. Many of these polymorphisms are located in the 5′ cis-regulatory region of CCR5, suggesting that it may have been a target of natural selection. We characterized CCR5 sequence variation in this region in 400 chromosomes from worldwide populations and compared it to a genome-wide analysis of 100 Alu polymorphisms typed in the same populations. Variation was substantially higher than expected and characterized by an excess of intermediate-frequency alleles. A genealogy of CCR5 haplotypes had deep branch lengths despite markedly little differentiation among populations. This finding suggested a deviation from neutrality not accounted for by population structure, which was confirmed by tests for natural selection. These results are strong evidence that balancing selection has shaped the pattern of variation in CCR5 and suggest that HIV-1 resistance afforded by CCR5 5′ cis-regulatory region haplotypes may be the consequence of adaptive changes to older pathogens.
More than 30 million adults are infected with HIV type 1 (HIV-1), and AIDS is the leading cause of death in some nations (1). However, not everyone exposed to HIV-1 becomes infected, and of those infected, the time of progression to AIDS and death varies substantially. This variation is caused, in part, by a combination of environmental, viral, and host factors that influence exposure and mediate the host response to HIV-1. Understanding the genetic determinants of the host response has been one of the major areas of investigation of AIDS researchers over the last decade.
The initial entry of HIV-1 into a cell requires the expression of two receptors on the cell surface, CD4 and CC chemokine receptor 5 (CCR5), a 7-transmembrane G-protein-coupled chemokine receptor (2). The role of CCR5 in the pathogenesis of AIDS was highlighted by the observation that individuals who were homozygous for a 32-bp deletion in the ORF of CCR5 (i.e., CCR5-Δ32) were relatively resistant to infection with HIV-1 (3). Subsequent studies confirmed that polymorphisms in the coding region of CCR5 that cause reduced or absent cell-surface expression of CCR5 lead to diminished susceptibility to HIV-1 infection (4). Yet, the few polymorphisms that affect cell-surface expression of CCR5 exist at very low frequencies and are virtually population-specific (5, 6). Thus, they account for little of the variation in disease risk in most individuals and populations.
This observation, in part, motivated further characterization of polymorphisms in the 5′ cis-regulatory region of CCR5 (7). Several of these were associated with varying susceptibility to HIV-1 infection and disease progression (8). However, some of them were in strong linkage disequilibrium (LD) with one another. As a consequence, assigning function to a particular nucleotide site was challenging, and single-site association studies sometimes came to contradictory conclusions (9). To overcome this problem, we and others defined CCR5 5′ cis-regulatory region haplotypes and used them to demonstrate that common CCR5 haplotypes were associated with varying HIV-1 disease progression (10–12). Some of these haplotypes were associated with similar effects across populations, whereas others were associated with delayed or accelerated disease progression depending on the population in which they were tested (11). Collectively, these findings suggested that the 5′ cis-regulatory region of CCR5 might have been a target of natural selection.
To test for a signature of selection, we sequenced ≈1.1 kb of the CCR5 5′ cis-regulatory region (Fig. 1) from each of 180 chromosomes from the National Institutes of Health (NIH) DNA Polymorphism Discovery Resource, hereafter called the “NIH” panel and from 224 chromosomes sampled from Africans, Asians, and Europeans, hereafter called the “Old World” panel. To assess interspecies phylogenetic relationships, we compared these data to CCR5 sequences obtained from chimpanzees (56 chromosomes) and a gorilla.
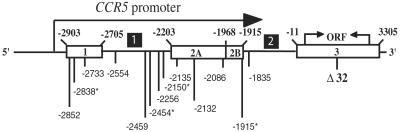
Fig 1.
Schematic drawing of CCR5 loci on chromosome 3 (not to scale). The CCR5 exons (open boxes) and introns are numbered. Polymorphisms found in the 5′ cis-regulatory region of CCR5 spanning from −2867 to −1745 are indicated and numbered. Sequencing revealed 13 SNPs in the Old World panel for an average SNP density of 1 SNP per 86 bp, and 9 SNPs in the NIH panel for an average SNP density of 1 per 125 bp. Four SNPs, all of which had frequencies of <1%, were found only in the Old World panel. No SNP was found only in the NIH panel. Thus, all of the SNPs with a frequency of >1% were found in both panels. Four singletons were found in the Old World panel (asterisks). See Appendix for further information.
Methods
Populations Samples.
For the Old Word panel, DNA sequence was obtained from 112 individuals including 31 Africans, 27 non-Indian Asians, 24 Europeans, and 30 South Indians. Further details on the identities of each individual sampled are in the Appendix, which is published as supporting information on the PNAS web site, www.pnas.org. Informed consent was obtained from all subjects whose blood was drawn at the University of Utah or in India. Sequencing was also carried out on the 90-member DNA Polymorphism Discovery Resource available from the NIH. This collection of individuals consists of roughly equal numbers of European Americans, African Americans, Hispanic Americans, Native Americans, and Asian Americans. In addition to the human subjects, DNA sequencing was carried out for 1 gorilla (Gorilla gorilla), 3 bonobos (Pan paniscus), and 25 chimpanzees (Pan troglodytes). The chimpanzee samples included all 3 recognized subspecies.
CCR5 Sequencing.
The CCR5 numbering system used identifies the first nucleotide of the CCR5 translational start site as +1, and the nucleotide immediately upstream as −1 (13). The region corresponding to human CCR5 −2867 to −1745 was PCR amplified and sequenced on both strands. This region includes the 3′ end of exon 1, intron 1, exons 2A and 2B, and the 5′ end of intron 2 (Fig. 1). All of these exons are noncoding. PCR primer sequences were derived from human genomic sequence (GenBank accession nos. AF031236 and AF031237). The primer sequences and detailed information on the reaction conditions are available in the Appendix. Sequence trace files were evaluated by using the phred, phrap, and consed programs (13). Potential heterozygotes were identified by using the polyphred version 3.5 program (14). Polymorphisms were verified by manual evaluation of the individual sequence traces. For most polymorphisms, it was possible to evaluate both the forward and the reverse sequences.
Statistical Analysis.
Allele frequencies for each single-nucleotide polymorphism (SNP) were determined by gene counting, and the significance of deviations from Hardy–Weinberg equilibrium (HWE) was tested by using a random permutation procedure implemented in the Arlequin package (15), which is available at http://anthropologie.unige.ch/arlequin. None of the SNPs showed a significant departure from HWE in any of the continental populations. Genetic differentiation among population samples was estimated with the FST statistic, which measures the fraction of total genetic variation that is distributed among rather than within populations.
Two measures of diversity were estimated for each panel and the continental populations (Africans, Asians, and Europeans). Because of their distinct geographic location and history, the samples from the Indian subcontinent were treated as a fourth population. Genetic diversity under mutation-drift equilibrium model was assessed by using θ, an estimate of the expected per-site heterozygosity (16), and π, the direct estimate of per-site heterozygosity derived from the average pairwise sequence difference (17). A difference in diversity estimates between Africans and non-Africans is notable in θW because rare variants, of which there were 3 in Africans, contribute more to θW than to π. Genetic diversity measured by the number of segregating sites and θW is higher in the Old World panel compared with the NIH panel (Table 1). However, the average nucleotide diversity (π) is higher in the NIH panel than the Old World panel (Table 1). This finding probably reflects the greater sampling of Africans, in whom singletons are more common, in the Old World panel (62 chromosomes) versus the NIH panel (≈36 chromosomes). Additionally, African and Asian samples in the NIH panel were ascertained from self-identified African Americans and Asian Americans. Thus, the level of admixture between individuals from different continental populations is higher in the NIH panel. This could further reduce the pool of African chromosomes sampled.
Table 1.
Summary of sequence variation in the cis-regulatory region of CCR5
| Population | n | S | θW | π ± SD, % | Tajima's D | Fs |
|---|---|---|---|---|---|---|
| NIH panel | 176 | 9 | 1.57 | 0.29 ± 0.17 | — | — |
| Old World panel | 224 | 13 | 2.18 | 0.21 ± 0.13 | 0.667 (0.37) | 0.02 (0.38) |
| Africans | 62 | 12 | 2.56 | 0.22 ± 0.13 | 0.292 (0.38) | −0.81 (0.57) |
| Non-Africans | 162 | 8 | 1.42 | 0.21 ± 0.12 | 2.08 (0.03) | 2.57 (0.10) |
| Asians | 54 | 6 | 1.32 | 0.20 ± 0.12 | 2.52 (0.01) | 3.45 (0.06) |
| Europeans | 48 | 7 | 1.58 | 0.22 ± 0.13 | 2.20 (0.02) | 1.61 (0.17) |
| Indians | 60 | 7 | 1.54 | 0.20 ± 0.12 | 1.85 (0.04) | 2.34 (0.12) |
Number of chromosomes.
P value is given in parentheses.
Ethnic identity unlinked to samples, therefore haplotypes could not be estimated reliably.
The test statistic D was used to compare these summary statistics. Under the standard neutral model, the expectations of θW and of π are equal. The difference between these values is reflected by Tajima's D, the expected value of which is 0 under neutrality. Significance values for these test statistics were computed by comparison to a distribution of estimates computed for 1,000 random samples of the same sample size and level of polymorphism as the observed data. It should be noted that all of the above computations require segregating site data but do not require haplotype data. Another test statistic, Fs (18), was used to compare the observed number of haplotypes in each sample to the number expected under the assumption of an infinite sites model of mutation without recombination. Statistical significance was based on 5,000 simulated samples.
Autosomal Alu polymorphisms used for estimating a genome-wide FST value were amplified by using conditions specifically optimized for each system. Further information on these conditions is available at www.genetics.utah.edu/∼swatkins/pub/Alu_primers.htm. The complete data set of genotypes from all 100 autosomal loci is available in the Appendix. It would also be insightful to compare the FST from the CCR5 5′ cis-regulatory region to the distribution of single-locus FST estimates from regions resequenced in continental populations. However, data are available from fewer than a handful of resequencing studies.
Pairwise LD between each pair of SNPs was measured with the parameter, D, calculated as D = Pij – pipj, where Pij is the frequency of the most common gametic type for a pair of sites. and pi and pj are the frequencies of the nucleotides in that haplotype and D′, an estimate of D standardize by Dmax, D′ = D/Dmax using arlequin (15). All pairs of SNPs exhibited significant LD. Haplotype frequencies were estimated by using a version of the expectation maximization (EM) algorithm provided in the Arlequin package (15). This procedure has been shown to yield reliable estimates of haplotype frequencies (19). In particular, the EM approach should work well in small regions with a high degree of LD, a high proportion of homozygotes, and a sample size of >100 chromosomes (20). All of these conditions are present in the data analyzed here. Additionally, all haplotypes were confirmed by identification of individuals who were homozygous for each SNP or sequencing of clones containing CCR5 haplotypes from individuals heterozygous for a singleton.
The mutational relationships among the CCR5 haplotypes were depicted by constructing rooted networks using the chimp as an outgroup species. Networks constructed by using the neighbor-joining, parsimony, and maximum likelihood methods had a similar topology to the minimum-spanning (MS) network (15). The MS algorithm unambiguously links haplotypes that differ by one or more mutation steps and employs a frequency-based compatibility criterion to choose among equally likely mutational paths linking haplotypes. When parallelisms cannot be resolved, the ambiguity is depicted as a reticulation.
Results and Discussion
The density of SNPs in the CCR5 5′ cis-regulatory region was ≈1.5–2 times higher than in most previously sequenced noncoding regions and regulatory regions (ref. 21 and references therein). Based on inference of the ancestral state from chimpanzee sequence, 5 derived SNPs were found at frequencies of 0.25 or greater in both the Old World and NIH panels. This is substantially higher than the expected density of 1 SNP every 1,124 bp for minor alleles with a frequency of 0.25 to 0.5 (22) assuming a standard neutral model (i.e., panmictic population of constant size in which genetic variation is neutral and follows an infinite sites model). Estimates of nucleotide diversity (π) for both the NIH panel (0.0029) and the Old World panel (0.0021) were about twice as high as the average π (0.001) for 4-fold degenerate sites in humans (23); twice as high as the π (0.0011) of the CCR5 ORF (24); and higher than has been reported in most studies of noncoding autosomal regions (22).
There are several possible explanations for these observations. There is evidence that π and SNP density are positively correlated with region-specific recombination rates (25). Yet, the recombination rate at 3p21.31, the region in which CCR5 is located, is estimated to be 0.66 centimorgan (cM)/Mb, well below the genomic average of 1.0–1.5 cM/Mb (26). The higher π observed for the 5′ cis-regulatory region of CCR5 could be caused by a higher mutation rate or a lower level of selective constraint. This is unlikely, however, because the divergence between the human and chimpanzee CCR5 sequence, 7 fixed differences, is roughly half the average 1.4% divergence estimated from other loci (27). Thus, none of these explanations is adequate.
Higher than expected levels of π can also be found in regions influenced by balancing selection. In this circumstance, genetic variation accumulates in alleles that are the target(s) of selection as a consequence of genetic hitchhiking. Because balancing selection can maintain an excess of alleles at intermediate frequencies, the variation accrued on these alleles also accumulates in a population (28). For example, the elevated estimates of π within the antigen-presenting HLA class I and II genes and flanking noncoding regions have been attributed to balancing selection (29).
If the elevated estimates of π for CCR5 were the result of balancing selection favoring intermediate-frequency alleles, the pattern of diversity among populations should be atypical of human loci as well. In most studies of neutral markers, including mitochondrial DNA (30), Y chromosome polymorphisms (31), autosomal short tandem repeats (32), and Alu polymorphisms (33), genetic diversity is substantially greater in Africans. However, π in the CCR5 5′ cis-regulatory region is virtually the same in each population (Table 1), though the number of segregating sites and θW (expected per-site heterozygosity) was highest in Africans. Nearly equal π values in Africans and Asians have been found in a few regions of the genome. This includes a 3-kb interval encompassing the β-globin gene, for which there is compelling evidence that balancing selection has influenced the surrounding genetic variation (34).
Multiallelic loci under balancing selection favoring intermediate-frequency alleles are expected to show a very different pattern of sequence diversity compared with neutral loci (35). Balancing selection increases within-deme diversity relative to total diversity because alleles are kept in more equal frequencies compared with a neutral locus, and an allele newly introduced into a population by migration will be positively selected. This increases its chances of survival compared with a neutral allele. Both of these processes are expected to lower population differentiation, as measured by FST, compared with neutral loci.
The FST for the 5′ cis-regulatory region of CCR5 among Africans, Asians, and Europeans was 0.016, not significantly different from zero (P > 0.09). This was nearly 10-fold lower than typical estimates of FST (0.15) among continental populations in large surveys of neutral loci (22, 33) and lower than all reported resequenced regions (see selected references in ref. 13). Among groups within each continent, FST was highest for the pooled African populations (0.032). Even in Africans, however, FST was considerably below the within-continent FST values typical of neutral loci (22) and was not significantly different from zero (P > 0.14). Thus, there is no evidence of significant differentiation within or among the continental populations.
The magnitude of the reduction in FST caused by balancing selection depends on the selection intensity and can be estimated by comparing the locus of interest to neutral loci. When the average FST for neutral loci among populations is <0.1, only strong selection on any particular locus is likely to be detectable (35). We compared the FST in the 5′ cis-regulatory region of CCR5 to the FST estimated from 100 selectively neutral Alu polymorphisms assayed in the same individuals. The FST for the Alu polymorphisms among populations was 0.073, significantly different from zero (P < 0.001). This is similar to a recently reported FST of 0.071 based on resequencing of 40 loci on 80 chromosomes from different continental populations (D. J. Cutler, personal communication). These FST estimates are probably an accurate reflection of the evolutionary and demographic forces averaged across the entire genome, and both are roughly 5-fold higher than the FST for the CCR5 sequences. Thus, it is difficult to attribute the very low FST estimates for the CCR5 5′ cis-regulatory region to population structure alone, suggesting that another force, such as balancing selection, has affected the distribution of diversity in the CCR5 5′ cis-regulatory region.
Population genetics theory predicts that natural selection should lead to a molecular signature in the region surrounding the target of selection. One such signature is a shift in the allele frequency spectrum. If the action of selection is directional, mutations that do occur are expected to exist at low frequency, skewing the spectrum of allele frequencies to the left. Because rare alleles are more likely to be retained in an expanding population, such a shift can also be caused by population growth. The allele frequency spectrum of most genetic systems is skewed to the left, a trend that has been interpreted as evidence for a population expansion (21). In contrast, balancing selection can result in an excess of intermediate-frequency alleles and a shift in the spectrum of allele frequencies to the right.
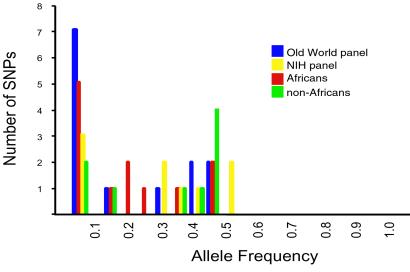
The allele frequency distributions for the total Old World panel, NIH panel, and subdivided Old World panel (i.e., Africans and non-Africans) are shown in Fig. 2. For each of the SNPs, the major allele is fixed in chimpanzees and gorilla, so the minor allele was inferred to be the derived state. For both total panels and the Africans, the site frequency spectrum has a bimodal distribution consistent with the presence of predominantly two types of polymorphisms, rare variants and intermediate-frequency variants. However, most of the rare variants are found only in Africans. For non-African populations, the distribution is skewed to the right by an apparent excess of intermediate-frequency polymorphisms.
Fig 2.
Allele frequency spectrum for 13 SNPs found in the Old World and NIH panels, and for Africans and non-Africans. The frequency of the derived allele of each SNP is shown.
The allele frequency distribution can be used to test hypotheses about the action of natural selection by applying two test statistics, Tajima's D and Fu's Fs. A negative value of D or Fs reflects a relative excess of rare variants, whereas a positive value of D or Fs reveals a relative excess of intermediate-frequency alleles as expected under a model of population subdivision or balancing selection. A recent survey of more than 300 human genes found that 90% of them had a negative Tajima's D value (36). In contrast, estimates of D for the CCR5 5′ cis-regulatory region were positive for the Old World panel, non-Africans, and each individual population. These values were significantly greater than zero in Asians, Europeans, Indians, and pooled non-Africans (Table 1), and higher than 99% of the more than 320 D values reported in humans (26, 32). A similar deviation from neutrality was observed by using the Fs statistic (Table 1).
Tajima's D and Fu's Fs are known to lack power to detect selection (37). For example, they failed to detect a departure from neutrality for at least 2 loci for which there is compelling evidence of selective effects, β-globin (34) and the Duffy blood group locus (38). The observation that these test statistics did detect a departure from neutrality in the CCR5 5′ cis-regulatory region underscores the strength of the signal of selection.
A more powerful test of selection compares the frequencies of polymorphisms within a species to the level of divergence between species. If the level of interspecific divergence between two species is similar for two genes (suggesting the mutation rates are the same) and each is undergoing neutral evolution so that the fate of new mutations is determined by only genetic drift and effective population size, then their levels of intraspecific polymorphism are also expected to be similar. This comparison has been developed formally as the Hudson/Kreitman/Aguade test (39).
We performed the Hudson/Kreitman/Aguade test (using a program kindly provided by Jody Hey, Rutgers Univ.) by comparing polymorphism and divergence levels in the 5′ cis-regulatory region of CCR5 against those found in the 3.7-kb noncoding sequence 5′ of CYP1A2 assayed in the same individuals. The null hypothesis was that the same neutral parameters fit both loci. The test yielded a χ2 = 4.8 (P < 0.03), reflecting a significant departure from neutrality across both loci. This result indicates that CYP1A2 and the 5′ cis-regulatory region of CCR5 have different patterns of polymorphism and divergence, so variation in either gene could have been affected by natural selection while the other remained neutral. However, the pattern of genetic variation in the CYP1A2 promoter is typical of most human loci (i.e., π = 0.00047 and highest in Africans; D = −1.15), compared with an average π = 0.0006 and D = −0.97 estimated from 313 genes (36). Hudson/Kreitman/Aguade tests comparing the CCR5 5′ cis-regulatory region to recently published data from 10 kb of noncoding Xq13.3 (χ2 = 4.131; P < 0.0421) and ≈65 kb from 3 loci on the Y chromosome (χ2 = 10.758; P < 0.001) also indicated a significant departure from neutrality (40, 41).
One effect of balancing selection is to preserve two or more lineages over an extended period, resulting in a genealogy with two or more clusters separated by long branch lengths (42). In contrast, population growth leads to the retention of new lineages and a star-like genealogy. If population growth has occurred in addition to balancing selection, then evidence of both may be present. To examine the genealogy of CCR5, we identified 13 CCR5 5′ cis-regulatory region haplotypes (Table 2) and built a MS network. The topology of this network (Fig. 3) was relatively unambiguous (i.e., there are only two reticulations, both of which involve low-frequency haplotypes), a pattern consistent with the high levels of LD between sites (data not shown). Each haplotype with a frequency >1% in the Old World panel (Table 2) was recognized previously in our classification of CCR5 haplotypes (11, 12).
Table 2.
Haplotype frequencies in each population
| Haplotype | Africans | Asians | Europeans | Indians |
|---|---|---|---|---|
| 1 (HHA) | 0.258 | 0.074 | 0.104 | 0.150 |
| 2 | 0.016 | |||
| 3 (HHF) | 0.177 | 0.130 | 0.125 | 0.133 |
| 4 (HHF) | 0.016 | |||
| 5 (HHF) | 0.016 | |||
| 6 (HHB) | 0.032 | 0.019 | 0.042 | 0.017 |
| 7 (HHC) | 0.161 | 0.500 | 0.313 | 0.367 |
| 8 (HHD) | 0.113 | |||
| 9 (HHE) | 0.161 | 0.280 | 0.292 | 0.217 |
| 10 | 0.016 | |||
| 11 (HHG) | 0.032 | 0.104 | ||
| 12 | 0.021 | |||
| 13 | 0.017 |
Haplotype nomenclature of Mummidi et al. (12).
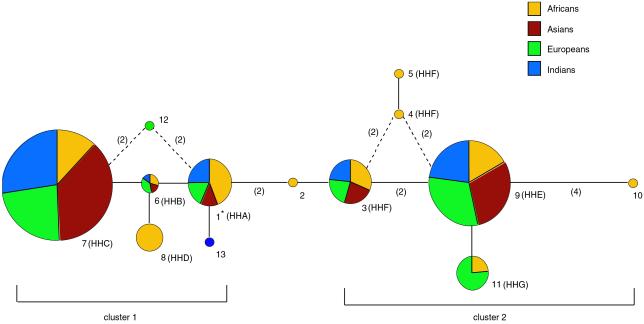
Fig 3.
A reduced median network of the 13 unique CCR5 5′ cis-regulatory region haplotypes found in the Old World panel. Haplotypes are also labeled according to the nomenclature we reported previously (refs. 11 and 12, and see Table 2). The size of each node is proportional to the haplotype frequency in the Old World panel, and the frequency of each haplotype within each continental population is indicated by the varied colors within each node. Branch lengths represent one nucleotide substitution, except where noted in parentheses. Dashed lines indicate alternate topologies of equal length. Haplotype 1, indicated with an asterisk, had the highest number of ancestral character states, differing from the chimpanzee consensus sequence by 7 nucleotide sites. This finding suggests that it may be the oldest lineage in the human sample and the root of the genealogy. It is also more common in Africans than any other continental population (Table 2), it is the most frequent haplotype in Africans, and it reaches a peak frequency of more than 70% (n = 68) in Mbuti Pygmies (11). Fifty percent of the CCR5 haplotypes in Asians are within 2 mutation steps of haplotype 1. Thus, although the CCR5 haplotypes in Africans are the most diverse and closest to the root, there is a substantial amount of haplotype diversity in Asians that is close to the root as well.
The MS network (Fig. 3) illustrates that 5 SNPs separate the CCR5 haplotypes into 2 major clusters: SNPs −2554, −2132, and −2086 define cluster 1, and SNPs −2459 and −2135 define cluster 2. One of each of the two most common haplotypes (i.e., 7 and 9) is found in each cluster, and these haplotypes are separated by a relatively long branch of 7 mutation steps. The mean pairwise sequence divergence between these clusters is 4.8. Thus, the estimated divergence age between clusters is ≈2.1 × 106 years. This deep genealogical structure is extraordinary in light of evidence from analyses of many other genetic systems that reveal a star-like genealogy consistent with the hypothesis of a human population expansion at the end of the Pleistocene (43). One explanation for this finding is that the effects of balancing selection at some loci have been obscured by population growth (43), a view supported by an analysis of synonymous versus nonsynonymous SNPs in more than 50 genes (50). Nevertheless, at CCR5, the signature of balancing selection remains apparent.
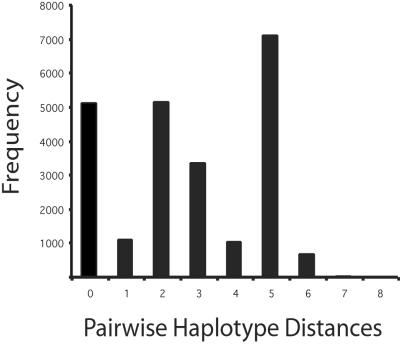
Finally, we used Rogers' mismatch test (44) to analyze the pairwise differences among the CCR5 5′ cis-regulatory region haplotypes. Under population growth or directional selection, the distribution of pairwise differences, or mismatch distribution, is smooth and unimodal (44). In contrast, the mismatch distribution is multimodal or ragged under balancing selection or a stationary population size. For most loci studied to date in continental populations, the frequencies of pairwise haplotype distances exhibit this unimodal distribution (21). The distribution of pairwise haplotype distances between CCR5 haplotypes, however, is multimodal (Fig. 4). The level of raggedness of this distribution was highly significant (P < 0.001). This finding is consistent with two or more divergent haplotypes being maintained within a population by balancing selection.
Fig 4.
Pairwise mismatch distribution CCR5 haplotypes, constructed by counting the number of differences between all pairs of the 224 haplotypes in the Old World panel. Mismatch distributions from populations that have expanded in the absence of natural selection and/or population subdivision are expected to be unimodal. In contrast, the pairwise mismatch distribution of CCR5 haplotypes is multimodal, consistent with the two major clusters containing haplotypes existing at appreciable frequencies in all continental populations. The mode at 0 nucleotide differences is caused by comparisons within haplotypes 7 and 9, the two most common haplotypes. The mode at haplotype 2 is caused by comparisons among haplotypes within clusters 1 and 2 in the MS network (Fig. 3), and the mode at haplotype 5 resulted from comparisons of haplotypes between clusters 1 and 2. Counts of haplotypes in each cluster for each of the continental populations were nearly identical (χ2 = 2.85; P > 0.42). Thus, geographic isolation does not explain the depth of the split between clusters 1 and 2.
Balancing selection necessarily involves some type of rare-allele advantage. Two major types of selection that feature rare-allele advantage are generalized over-dominance and negative frequency-dependent selection. In generalized over-dominance, heterozygotes maintain a selective advantage over homozygotes, and thus the rare alleles benefit from their representation in the heterozygotes. This type of selection is thought to maintain the high levels of allelic variation observed at the MHC locus (29), an insight derived largely from functional data demonstrating that MHC heterozygotes can present an expanded spectrum of antigens to T cells compared with MHC homozygotes. This is germane in light of the observation that heterozygosity of CCR5 5′ cis-regulatory region haplotypes from different clusters is strongly associated with a delay in HIV-1 disease progression, whereas homozygosity is associated with accelerated disease progression (10, 11).
The introduction of HIV-1 into human populations occurred too recently to explain the observed pattern of variation in CCR5. It is plausible, however, that heterozygosity for the CCR5 5′ cis-regulatory region conferred selective advantages to hosts exposed to other pathogens over the gene's long history. This observation is consistent with the finding that chemokine receptors such as CCR5 and the Duffy antigen on the surface of blood cells are targets that appear to have been exploited by many different pathogens (45). Thus, HIV-1 resistance provided by CCR5 5′ cis-regulatory region haplotypes might be the result of adaptive changes to older pathogens (e.g., smallpox) (46).
This analysis of the CCR5 5′ cis-regulatory region underscores the complexity of testing for the effects of selection at multiple levels (i.e., on a single polymorphism, between species, within species). Previous studies have demonstrated that directional selection has rapidly increased the frequency of at least one polymorphism in the coding region of CCR5 (i.e., CCR5-Δ32) in Northeastern Europeans (47), and we reported evidence of directional selection acting on the NH2 terminus of CCR5 among different species of primates (12). The overall impact of selection on the CCR5 5′ cis-regulatory region among human populations appears to have been different. This is reminiscent of the pattern of selective forces acing on the MHC locus where directional selection has influenced the frequency of individual alleles, particularly new alleles, while overall allelic diversity is maintained by balancing selection (48). This finding also has different implications for designing and interpreting association studies using CCR5 5′ cis-regulatory haplotypes. For example, it may be important to compare haplotype type pairs from cluster 1 vs. cluster 2 (Fig. 3) or weight haplotypes by their evolutionary distance from one another.
The impact of selection may have also varied between coding and noncoding regions of CCR5. Such disparate effects of selection on coding versus noncoding regions have also been reported for the MHC, although in contrast to the pattern at CCR5, it is the coding regions that have been influenced by balancing selection (48). It is possible, however, that despite the differences in transcriptional activity conferred by CCR5 5′ cis-regulatory region haplotypes (12), they are in LD with other polymorphisms that may also be targets of selection.
Noncoding regions such as cis-regulatory sequences perform important functions, and there is evidence that they evolve rapidly by point mutations (49). These point mutations can be associated with important phenotypic effects that help to drive evolutionary change. Yet, little is known about the relative importance of variation in the regulatory sequences of human genes, how they differ between populations, and what insights they can reveal about human evolutionary history. Deeper investigation of these regions should provide new insights about the design of association studies, the relationship between genetic variation and phenotypes (normal and disease-related), and the evolutionary history of humans.
Supplementary Material
Acknowledgments
We thank M. Batzer and C. E. Ricker for assistance with the analysis of Alu elements. We thank H. Harpending and A. R. Rogers for comments and suggestions on the manuscript. We thank K. Bulayeva, T. Jenkins, H. Soodyall, and E. Zietkewicz for some of the DNA samples. This research was supported by National Institutes of Health Grants GM-59290, RR-00064, ES-10058, AI43279, and AI46326, and by National Science Foundation Grants SBR-9514733, BC5-0073871, and SBR-9818215. S.K.A., S.S.A., S.M., and E.G. are supported by the Veterans Administration Center for AIDS and HIV Infection in San Antonio. S.K.A. is a recipient of the Elizabeth Glaser Scientist Award (Pediatric AIDS Foundation) and the Burroughs Wellcome Clinical Scientist Award in Translational Research.
Abbreviations
HIV-1, HIV type 1
CCR5, CC chemokine receptor 5
LD, linkage disequilibrium
SNP, single nucleotide polymorphism
MS, minimum spanning
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Morgan D. & Whitworth, J. (2001) Nat. Med. 7, 143-145. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G., Combadiere, C., Broder, C. C., Feng, Y., Kennedy, P. E., Murphy, P. M. & Berger, E. A. (1996) Science 272, 1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Dean M., Carrington, M., Winkler, C., Huttley, G. A., Smith, M. W., Allikmets, R., Goedert, J. J., Buchbinder, S. P., Vittinghoff, E., Gomperts, E., et al. (1996) Science 273, 1856-1862. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien S. J. & Moore, J. P. (2000) Immunol. Rev. 177, 99-111. [DOI] [PubMed] [Google Scholar]
- 5.Blanpain C., Lee, B., Tackoen, M., Puffer, B., Boom, A., Libert, F., Sharron, M., Wittamer, V., Vassart, G., Doms, R. W. & Parmentier, M. (2000) Blood 96, 1638-1645. [PubMed] [Google Scholar]
- 6.Carrington M., Dean, M., Martin, M. P. & O'Brien, S. J. (1999) Hum. Mol. Genet. 8, 1939-1945. [DOI] [PubMed] [Google Scholar]
- 7.Mummidi S., Ahuja, S. S., McDaniel, B. L. & Ahuja, S. K. (1997) J. Biol. Chem. 272, 30662-30671. [DOI] [PubMed] [Google Scholar]
- 8.Martin M. P., Dean, M., Smith, M. W., Winkler, C., Gerrard, B., Michael, N. L., Lee, B., Doms, R. W., Margolick, J., Buchbinder, S., et al. (1998) Science 282, 1907-1911. [DOI] [PubMed] [Google Scholar]
- 9.Garred P. (1998) Lancet 351, 2-3. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez E., Dhanda, R., Bamshad, R., Mummidi, S., Geevarghese, R., Catano, G., Anderson, S. A., Walker, E. A., Stephan, K. T., Hammer, M. F., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez E., Bamshad, M., Sato, N., Mummidi, S., Dhanda, R., Catano, G., Caberea, S., McBride, M., Cao, X.-H., Merrill, G., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 12004-12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mummidi S., Bamshad, M., Ahuja, S. S., Gonzalez, E., Feuillet, P. M., Begum, K., Galvis, M. C., Kostecki, V., Valente, A. J., Murthy, K. K., et al. (2000) J. Biol. Chem. 275, 18946-18961. [DOI] [PubMed] [Google Scholar]
- 13.Ewing B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 14.Nickerson D. A., Tobe, V. O. & Taylor, S. L. (1997) Nucleic Acids. Res. 25, 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider S., Roessli, D. & Excoffier, L., (2000) arlequin: A Software For Population Genetics Data Analysis (Univ. of Geneva), Version 2.000.
- 16.Watterson G. (1975) Theor. Popul. Biol. 7, 256-276. [DOI] [PubMed] [Google Scholar]
- 17.Nei M., (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York), pp. 261–266.
- 18.Fu Y.-X. (1997) Genetics 147, 915-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tishkoff S. A., Pakstis, A. J., Ruano, G. & Kidd, K. K. (2000) Am. J. Hum. Genet. 67, 518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fallin D. & Schork, N. J. (2000) Am. J. Hum. Genet. 67, 947-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorde L. B., Watkins, W. S. & Bamshad, M. J. (2001) Hum. Mol. Genet. 10, 2199-2207. [DOI] [PubMed] [Google Scholar]
- 22.Przeworski M., Hudson, R. R. & Di Rienzo, A. (2000) Trends Genet. 16, 296-302. [DOI] [PubMed] [Google Scholar]
- 23.Li W. H. & Sadler, L. A. (1991) Genetics 129, 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakravarti A. (1999) Nat. Genet. 21, Suppl. 1, 56-60. [DOI] [PubMed] [Google Scholar]
- 25.Nachman M. W. (2001) Trends Genet. 17, 481-485. [DOI] [PubMed] [Google Scholar]
- 26.Yu N., Zhao, Z., Fu, Y.-X., Sambuughin, N., Ramsay, M., Jenkins, T., Leskinen, E., Patthy, L., Jorde, L. B., Kuromori, T. & Li, W. H. (2001) Mol. Biol. Evol. 18, 214-222. [DOI] [PubMed] [Google Scholar]
- 27.Takahata N. & Satta, Y. (1997) Proc. Natl. Acad. Sci. USA 94, 4811-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richman A. (2000) Mol. Ecol. 9, 1953-1963. [DOI] [PubMed] [Google Scholar]
- 29.Grimsley C., Mather, K. A. & Ober, C. (1998) Mol. Biol. Evol. 15, 1581-1588. [DOI] [PubMed] [Google Scholar]
- 30.Ingman M., Kaessmann, H., Paabo, S. & Gyllensten, U. (2000) Nature (London) 408, 708-713. [DOI] [PubMed] [Google Scholar]
- 31.Ke Y., Su, B., Song, X., Lu, D., Chen, L., Li, H., Qi, C., Marzuki, S., Deka, R., Underhill, P., et al. (2001) Science 292, 1151-1153. [DOI] [PubMed] [Google Scholar]
- 32.Jorde L. B., Rogers, A. R., Bamshad, M., Watkins, W. S., Krakowiak, P., Sung, S., Kere, J. & Harpending, H. C. (1997) Proc. Natl. Acad. Sci. USA 94, 3100-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watkins W. S., Ricker, C. E., Bamshad, M. J., Carroll, M. L., Nguyen, S. V., Batzer, M. A., Harpending, H. C., Rogers, A. R. & Jorde, L. B. (2001) Am. J. Hum. Genet. 68, 738-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harding R. M., Fullerton, S. M., Griffiths, R. C., Bond, J., Cox, M. J., Schneider, J. A., Moulin, D. S. & Clegg, J. B. (1997) Am. J. Hum. Genet. 60, 772-789. [PMC free article] [PubMed] [Google Scholar]
- 35.Schierup M. H., Vekemans, X. & Charlesworth, D. (2000) Genet. Res. 76, 51-62. [DOI] [PubMed] [Google Scholar]
- 36.Stephens J. C., Schneider, J. A., Tanguay, D. A., Choi, J., Acharya, T., Stanley, S. E., Jiang, R., Messer, C. J., Chew, A., Han, J. H., et al. (2001) Science 293, 489-493. [DOI] [PubMed] [Google Scholar]
- 37.Simonsen K. L., Churchill, G. A. & Aquadro, C. F. (1995) Genetics 141, 413-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamblin M. T. & Di Rienzo, A. (2000) Am. J. Hum. Genet. 66, 1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson R. R., Kreitman, M. & Aguade, M. (1987) Genetics 116, 153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaessmann H., Heissig, F., von Haeseler, A. & Paabo, S. (1999) Nat. Genet. 22, 78-81. [DOI] [PubMed] [Google Scholar]
- 41.Thomson R., Pritchard, J. K., Shen, P., Oefner, P. J. & Feldman, M. W. (2000) Proc. Natl. Acad. Sci. USA 97, 7360-7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marjoram P. & Donnelly, P. (1994) Genetics 136, 673-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harpending H. & Rogers, A. (2000) Ann. Rev. Genomics Hum. Gen. 1, 361-385. [DOI] [PubMed] [Google Scholar]
- 44.Rogers A. R. & Harpending, H. (1992) Mol. Biol. Evol. 9, 552-569. [DOI] [PubMed] [Google Scholar]
- 45.Pease J. E. & Murphy, P. M. (1998) Semin. Immunol. 10, 169-178. [DOI] [PubMed] [Google Scholar]
- 46.Lalani A. S., Masters, J., Zeng, W., Barrett, J., Pannu, R., Everett, H., Arendt, C. W. & McFadden, G. (1999) Science 286, 1968-1971. [DOI] [PubMed] [Google Scholar]
- 47.Stephens J. C. (1998) Am. J. Hum. Genet. 62, 1507-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeager M. & Hughes, A. L. (1998) Immunol. Rev. 167, 45-58. [DOI] [PubMed] [Google Scholar]
- 49.Stone J. R. & Wray, G. A. (2001) Mol. Biol. Evol. 18, 1764-1770. [DOI] [PubMed] [Google Scholar]
- 50.Wooding, S. & Rogers, A. R. (2002) Genetics, in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.