Abstract
Molecular dynamics simulations have been used to derive the structures of ground (orotidine-5′-monophosphate decarboxylase⋅orotidine 5′-monophosphate; ODC⋅OMP) and intermediate (ODC⋅intermediate; ODC⋅I−) states in the ODC-catalyzed decarboxylation of OMP. For comparison, a molecular dynamics simulation of the conformers of OMP dissolved in water was also studied. This structural information is unavailable from present crystal structures. The electrostatic network in the active site around the carboxylate moiety of OMP exhibits remarkable stability. The conformation of enzyme-bound OMP is very similar to the conformation of OMP in water. Thus, the proposed Circe effect mechanism for ODC catalysis is unlikely. Comparison of ground state and intermediate state structures shows that on decarboxylation C6 takes the position of the carboxylate O8. This significant movement of the ligand is accompanied by a placement of the C6 carbanion in the vicinity of the protonated Lys-93 and is enforced by a change of the 203–218 loop from an unstructured form to an ordered β-hairpin. Previously proposed mechanisms involving protonation at O2, O4, or C5 have in common internal stabilization of the anionic intermediate by conjugation with positive charge on the pyrimidine ring. These mechanisms are not supported because there are no proton sources near O2, O4, and C5. We propose that the stabilization of intermediate ODC⋅I− is achieved by movement of the carbanion toward the external cation Lys-93 on decarboxylation and organization of the 203–218 loop. Because the intermediate and transition state are energetically similar, stabilization of the former decreases the free energy content of the latter.
Orotidine-5′-monophosphate decarboxylase (ODC; EC 4.1.1.23) catalyzes the decarboxylation of orotidine 5′-monophosphate (OMP), the final step in the de novo biosynthesis of uridine 5′-monophosphate (UMP). The chemistry of OMP decarboxylation is unusual in that the electron pair developing on formation of the carbanion intermediate (I−) cannot be stabilized by delocalization through π bonds. This inability to stabilize I− results in a very slow reaction in water (t1/2 = 8 × 107 years) (1). ODC accelerates this reaction by 17 orders of magnitude without using any metal ions or cofactors, or without forming a covalent intermediate. These features have attracted much attention to the mechanism of this enzyme, which is still not known.
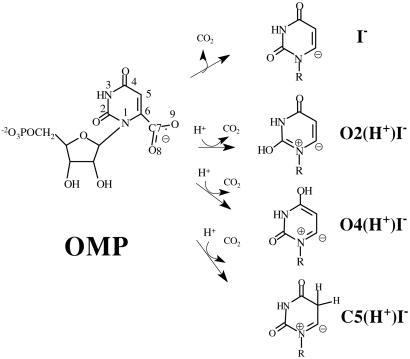
Mechanisms proposed to date can be categorized according to the protonation state of the intermediate (Scheme S1): the intermediate that does not follow any protonation step before decarboxylation (I−), the intermediate protonated at O2 (O2(H+)I−), of the intermediate protonated at O4 (O4(H+)I−) or at C5 (C5(H+)I−). Although there have been abundant experimental and theoretical studies, each study has appeared to support a different mechanism.
Scheme 1.
Decarboxylation of OMP without prior protonation has been suggested on the basis of the examination of crystal structures of ODC⋅inhibitor complexes (2, 3). In the following quantum mechanics/molecular mechanics study, electrostatic repulsion between the substrate carboxylate and Asp carboxylate has been proposed to be the driving force for the decarboxylation (2). Such ground state destabilization was rationalized in terms of the Circe effect, where the binding energy of the nonreactive part of the substrate is used to gear the reactive part of the substrate toward the transition state. This mechanism, however, could not explain the profile of inhibitor binding affinities where the negative charge at the C6 position is preferred to a neutral one (4). Ab initio calculations have shown that O2- or O4-protonation of OMP lowers the activation barrier for decarboxylation by 22–24 kcal/mol in the gas phase (5). Experimentally determined normal 15N kinetic isotope effects (KIEs) at N1 suggested no change in the bond order of N1 before decarboxylation (6), but in the following computational study, calculated 15N KIEs at N1 were rather insensitive to the bond order change (7). Recently, a mechanism involving an enamine protonation at C5 has been proposed as the first step in the decarboxylation reaction (8).
Choices between the proposed mechanisms are made difficult mainly by the lack of reliable information regarding the ground state geometry of enzyme⋅substrate complexes. Crystal structures with inhibitors or products have played an important role, but the exact location of the catalytic sites relative to the substrate is elusive from the available crystal information. Recently, the crystal structure with OMP bound to the active site was solved for an Asp-70/Lys-73 double mutant of Methanobacterium thermoautotrophicum enzyme (corresponding to Asp-91/Lys-93 in yeast enzyme), but the position of the Asp-70/Lys-73 relative to the OMP's carboxylate remains to be solved. Long-term molecular dynamics (MD) simulation is appropriate to obtain such information. In this paper, we describe results based upon a 2.5-ns simulation of ODC⋅OMP, 1.5.ns simulations of intermediate ODC⋅I−, and 700-ps simulation of OMP free in water.
Methods
Yeast enzyme [PDB ID code 1DQX (9)] was chosen for all MD simulations to take advantage of the enzyme structure complexed with 1-(5′-phospho-β-d-ribofuranosyl)barbituric acid (BMP), which most resembles the geometry of ODC⋅OMP among crystallized ODC⋅inhibitor complexes. All four residues of the catalytic tetrad (Lys-59–Asp-91–Lys-93–Asp-96*) were modeled assuming both Lys entities to be protonated and Asp carboxylates to be ionized. The force fields and atomic charges for the ribosyl and phosphate moieties of OMP and I− were used directly from the standard CHARMM27 parameters. The charges and parameters for the pyrimidine base were obtained by MP2/6–31+G*//RHF/6–31+G* calculations. Final charges of the pyrimidine portions are summarized in Table 5, which is published as supporting information on the PNAS web site, www.pnas.org.
OMP was docked into the active site by replacing the C6-hydroxide group of the BMP inhibitor in the crystal structure by a C6-carboxylate group. To model the decarboxylated intermediates the C6–C7 bond force was eliminated. After completion of docking of each ligand, the ODC⋅ligand complex with crystallographic water was energy-minimized and solvated into a sphere of TIP3P water molecules (18) (sphere radius = 45 Å). This water sphere solvates the whole dimeric enzyme, and the sphere boundary is more than 25 Å apart from the closest active site. For the simulation of OMP free in water, a TIP3P water sphere of radius 25 Å was used and the position of O1′ of OMP was fixed at the center of the sphere during the simulation to prevent the molecule from drifting toward the boundary. The stochastic boundary condition was used for all simulations and the program CHARMM25 was used for implementation of the simulations. Details of the methods are also included in the supporting information on the PNAS web site.
Results
ODC is catalytically active as a dimer. Each active site is located at the opening end of the triose-phosphate isomerase (TIM)-barrel and is surrounded by several loops from the same unit and two loops from the other subunit. Comparison of single-turnover and steady-state turnover kinetics has shown that the two active sites are catalytically independent (10). In this paper, only active site A will be discussed for all three simulations, and residues from B subunit will be designated with * (i.e., Asp-96*).
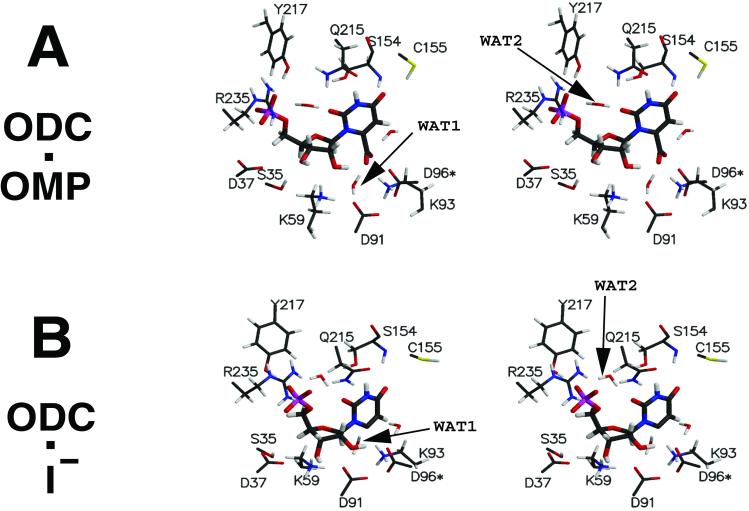
The dynamics structure of ODC⋅OMP exhibits several features quite different from the crystal structure of the enzyme⋅inhibitor complex (ODC⋅BMP). The crystal structure of ODC⋅BMP was found, however, to be quite similar to the MD simulation structures of ODC⋅I−. A large structural change in ODC occurs when OMP undergoes decarboxylation. This is mainly due to the positional movement of I− by ≈2.2 Å and the accompanying change in the protein loop structure (residues 203–218). These changes alter the hydrophobic contact regions of the pyrimidine ring and the solvent-accessible part of the ligand. The interaction mode of ODC⋅OMP and ODC⋅I− are presented in stereoview in Fig. 1 A and B, respectively. The average interaction distances are provided in Table 1, in comparison with those found in the crystal structure of ODC⋅BMP. To understand the following material the reader should refer to Fig. 1.
Figure 1.
Stereoviews of the active site. The structures are averaged structures of each simulation. The interaction distances are provided in Table 1. WAT, water molecule.
Table 1.
Electrostatic interaction distances between ligand and ODC
| Interaction | Ligand ⋯ Enzyme | Distance, Å
|
||
|---|---|---|---|---|
| ODC⋅OMP | ODC⋅I− | ODC⋅BMP† | ||
| Carboxylate binding | O8 ⋯ Asp-91(Oδ2) | 3.49 ± 0.23 | 4.43 ± 0.83 | |
| O8 ⋯ Asp-96(Oδ1) | 4.08 ± 0.30 | 7.86 ± 0.98 | ||
| O8 ⋯ Lys-93(Nζ) | 2.70 ± 0.10 | 5.38 ± 1.33 | ||
| C6 ⋯ Asp-91(Oδ2) | 5.79 ± 0.22 | 4.02 ± 0.18 | 2.82 | |
| C6 ⋯ Asp-96(Oδ1) | 5.34 ± 0.27 | 3.99 ± 0.20 | 4.24 | |
| C6 ⋯ Lys-93(Nζ) | 4.89 ± 0.15 | 2.82 ± 0.10 | 2.70 | |
| Ribose binding | O2′ ⋯ Lys-59(Nζ) | 2.99 ± 0.44 | 4.48 ± 0.23 | 4.84 |
| O3′ ⋯ Lys-59(Nζ) | 3.83 ± 0.47 | 2.87 ± 0.14 | 3.16 | |
| O3′ ⋯ Ser-35(Oγ) | 2.89 ± 0.20 | 3.45 ± 0.26 | 3.58 | |
| O3′ ⋯ Asp-37(Oδ2) | 6.92 ± 1.48 | 2.84 ± 0.34 | 2.86 | |
| Phosphate binding | Phosphate O ⋯ Arg-235(Nη2) | 2.85 ± 0.35 | 2.74 ± 0.12 | 2.84 |
| Phosphate O ⋯ Arg-235(Nη1) | 2.94 ± 0.34 | 2.90 ± 0.25 | 2.82 | |
| Phosphate O ⋯ Tyr-217(OH) | 2.60 ± 0.09 | 2.62 ± 0.09 | 2.68 | |
| Phosphate O ⋯ Gln-215(Nɛ2) | 4.30 ± 0.36 | 3.28 ± 0.60 | 5.19 | |
| Capping loop | O2 ⋯ Gln-215(Nɛ2) | 3.23 ± 0.45 | 3.43 ± 0.39 | 4.01 |
| O4 ⋯ Ser-154(N) | 2.96 ± 0.20 | 3.31 ± 0.33 | 2.78 | |
| O4 ⋯ Ser-154(Oγ) | 3.41 ± 0.22 | 3.97 ± 0.24 | 3.76 | |
| O4 ⋯ Cys-155(Sγ) | 3.56 ± 0.47 | 6.05 ± 0.49 | 5.88 | |
ODC⋅BMP data are taken from the crystal structure of ref. 9.
Electrostatic Interaction Between ODC and OMP.
Except for two hydrophobic residues (Pro-202 and Ile-97*), the substrate is bound mostly by electrostatic interactions. These electrostatic interactions are divided into four categories: (i) the carboxylate-binding pocket; (ii) the ribosyl ring-binding pocket; (iii) the 5′-phosphate-binding pocket; and (iv) the pyrimidine-capping loop.
The carboxylate-binding pocket consists of Lys-93, Asp-91, and Asp-96*, where the carboxylates of the two Asp residues rigidly position the protonated amino group of Lys-93. These Asp carboxylates do not face directly with the carboxylate of OMP. Lys-93 intervenes between the Asp-96* and OMP carboxylates at an O8⋅⋅⋅Asp-96(Oδ1) distance of ≈4.0 Å. A water molecule (WAT1) is bridged between the Asp-91 and OMP carboxylates and, along with Lys-93, alleviates the electrostatic repulsion.
The ribosyl-binding pocket is formed by Lys-59 and Ser-35. The protonated amino group of Lys-59 is positioned between the ribose C2′- and C3′-hydroxyl groups, and the Ser-35's hydroxyl group is hydrogen bonded to the C3′-hydroxyl O. Although Asp-37 is initially hydrogen bonded to the C3′-hydroxyl group, the backbone of Asp-37 moves away from the active site at ≈500 ps after the initiation of the dynamics. This O3′⋅⋅⋅Asp-37(Oδ2) interaction remains in the MD simulations of ODC⋅I−. In kinetic experiments the Asp-37 → Ala mutant showed a 6-fold increase in Km and a 50-fold decrease in kcat compared with the wild type (11). The importance of this interaction for the catalysis appears to be in intermediate binding, rather than in substrate binding.
The doubly negatively charged phosphate group of OMP forms double salt bridges with the guanidino group of Arg-235 as well as a hydrogen bond with the hydroxyl group of Tyr-217. Although one would suppose the ionic interaction of the phosphate⋅⋅⋅Arg-235 to be strong, the interaction suffers rather large fluctuations. The guanidino group of Arg-235 appears to be in an unfavorable conformation when OMP is bound, having an ≈30° distorted improper angle around Nη1 (see Fig. 1A). The guanidino group of Arg-235 is also hydrogen bonding to a water molecule (WAT2) that is bridging between the phosphate and O2 of the substrate. Although this region is highly exposed to solvent, the bridging water does not exchange with other waters. Also, the positions of all three atoms of WAT2 are well defined by maintaining three distinct hydrogen bonds: WAT2(H1)⋅⋅⋅O2, WAT2(H2)⋅⋅⋅phosphate group, and WAT2(O)⋅⋅⋅Arg-235 (Fig. 1A).
Loops formed by residues 203–218 and 153–160 envelop the pyrimidine ring. The amide group of the Gln-215 in the loop 203–218 is hydrogen bonded with N3. From the loop 153–160, the backbone amide N and the hydroxyl of Ser-154 interact with O4, and the thiol group of Cys-155 weakly interacts with O4.
The four protein regions described exhibit different flexibilities during the dynamics. Table 2 shows the positional fluctuation of each residue when Cα atoms of the active sites are matched. Asp-91–Lys-93–Asp-96* from the carboxylate-binding pocket and Lys-59 from the ribosyl-binding region have the smallest fluctuation values (≈0.3 Å) and are almost perfectly superimposable when matching each of the ten 100-ps-averaged structures. This tetrad (Lys-59–Asp-91–Lys-93–Asp-96*) has been shown to be perfectly conserved during evolution (3). Going farther from the carboxylate-binding pocket, the degree of mismatch increases. Arg-235 frequently changes the interacting mode with the substrate phosphate group and has a large thermal fluctuation value of 0.71 Å in its side chain. Tyr-217 in the phosphate binding domain and residues from the ligand-capping regions show significant motions in the backbones. This backbone fluctuation is seen in the whole loops of 203–218 and of 153–160.
Table 2.
Positional fluctuation of protein residues involved in electrostatic interactions with OMP
| Interaction | Residue | Positional fluctuation, Å
|
|
|---|---|---|---|
| Cα | Side chain† | ||
| Carboxylate-binding domain | Asp-91 | 0.28 | 0.37 |
| Lys-93 | 0.31 | 0.36 | |
| Asp-96* | 0.38 | 0.43 | |
| Ribosyl-binding domain | Lys-59 | 0.28 | 0.37 |
| Ser-35 | 0.37 | 0.53 | |
| Asp-37 | 0.81 | 0.37 | |
| Phosphate-binding domain | Tyr-217 | 0.46 | 0.55 |
| Arg-235 | 0.38 | 0.71 | |
| Capping loop | Ser-154 | 0.37 | 0.45 |
| Cys-155 | 0.44 | 0.57 | |
| Gln-215 | 0.49 | 0.52 | |
Average values over all heavy atoms of each amino acid except for the backbone atoms Cα, N, C, and O.
Electrostatic Interaction Between ODC and I−.
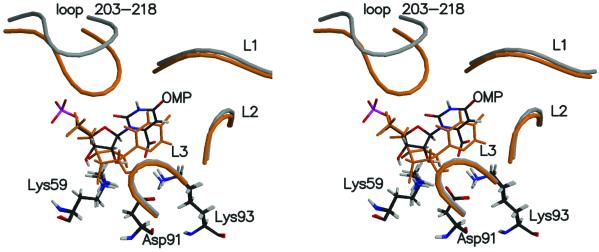
When superimposing the active site structure of ODC⋅I− (Fig. 1B) to the active site of ODC⋅OMP, one observes the following. Residues that are rigid in ODC⋅OMP, such as Lys-59, Asp-91, Lys-93, and Asp-96*, preserve positions on going from ODC⋅OMP to ODC⋅I− while the ribosyl- and phosphate-binding domains rearrange and the structure of the loop 203–218 changes (Fig. 2).
Figure 2.
Stereoview of two structures [ODC⋅OMP (gray) and ODC⋅I− (orange)] superimposed by matching the backbones of the enzyme. Lys-59–Asp-91–Lys-93 are perfectly well matched in these two structures and colored by standard atomic color definition along with OMP. L1 is formed by residues 154–160, L2 is by 125*–130*, and L3 is by 95*–103*.
When CO2 is released from OMP, the CO2 molecule drifts away from the Asp-91–Lys-93–Asp-96* region and is captured by the Phe-89 aromatic hydrogen and the Ile-183 methyl hydrogen. Concurrently with removal of the CO2 from the carboxylate-binding pocket, the O8⋅⋅⋅Lys-93 interaction in ground state is replaced by the C6⋅⋅⋅Lys-93 interaction in intermediate states (Fig. 2). C6 is 4.89 Å apart from Lys-93(Nζ) in ODC⋅OMP, whereas it is 2.82 Å in ODC⋅I−. Because Lys-93 is rigidly held by Asp-91 and Asp-96*, the above change occurs by repositioning of the pyrimidine ring closer to Lys-93. This, in turn, necessarily involves the movement of the whole ligand, including the ribosyl and phosphate moieties, in the same direction as the pyrimidine movement (Fig. 2). This shifting of the position results in the ribosyl 3′-hydroxyl interacting with Asp-37 instead of Ser-35 and rearrangement of ligand capping loop 203–215 in the intermediate state.
Loop Conformation Change.
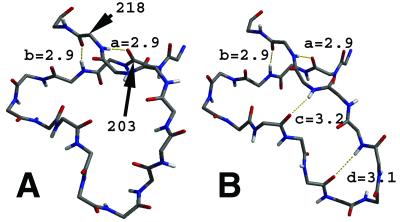
The loop 203–218 is formed by crossing the backbone over itself (Fig. 3). At the intersection, the hydrogen bonds of 203(O)⋅⋅⋅218(HN) and of 205(HN)⋅⋅⋅218(O) fix the edges of the loop in place, whereas the remainder is free to move. When ODC⋅OMP is converted into ODC⋅I−, this loop becomes a β-hairpin (Fig. 3). This conformational change appears to relate to the size of the ligand. When the carboxylate group is eliminated from OMP the space between the capping loop and the carboxylate-binding pocket reduces by ≈2.2 Å. As the capping loop of 203–218 becomes buried into the protein in the intermediate state, it forms new contacts with other protein domains (Table 3). The increases in contact result in a more closed packing of the 203–218 loop and its conversion to a β-hairpin structure. It has been previously shown by crystallography that this loop is in a completely open form in a ligand-free enzyme without having any contact with L1, L2, and L3, but becomes closed upon inhibitor binding (9). In our simulation, the loop structure not only depends on the absence or presence of a ligand but also is sensitive to the size of the ligand. This finding suggests significant motion during the catalysis.
Figure 3.
Structure of the loop 203–218 in ODC⋅OMP (A) and ODC⋅I− (B). Alphabet symbols identify interactions as follows: a, 203(O)⋅⋅⋅218(HN); b, 205(HN)⋅⋅⋅218(O); c, 209(O)⋅⋅⋅217(HN); d, 211(O)⋅⋅⋅215(HN). Hydrogen bonds a and b are very strong for both structures. In A no other hydrogen bonds between the loop backbones are observed. This is different from B, where β-hairpin structure is formed by additional c and d interactions. The distances shown (Å) are between heavy atoms.
Table 3.
Contact areas between the loop 203–218 and surrounding loops (L1, L2, and L3 are defined in Fig. 2)
| Contact | Area, Å2
|
|
|---|---|---|
| OMP | I− | |
| Loop 203–218/L1 | 68 | 64 |
| Loop 203–218/L2 | 1 | 10 |
| Loop 203–218/L3 | 17 | 35 |
Solvent-Accessible Surface Area (SASA) and Hydrophobic Contact.
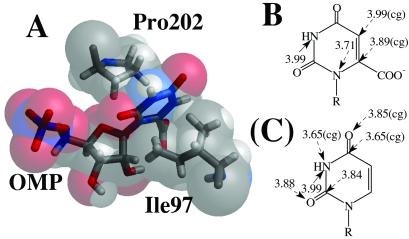
SASA study shows that the enzyme-bound OMP is extensively exposed to solvent water, but the decarboxylated intermediate is not (Table 4). Significant decrease of SASA in the ribosyl moiety can be attributed to the change in the capping loop, which envelops the ring more extensively in ODC⋅I− than in ODC⋅OMP. Unlike the ribose ring, the pyrimidine ring and the carboxylate group are fairly protected from solvent even in the ground state. This protection is achieved by hydrophobic contacts with Pro-202 and Ile-97. These residues flank the two faces of the pyrimidine ring (Fig. 4A). Only certain parts of the pyrimidine ring are subject to these hydrophobic interactions. Most interesting is the observation that the regions of contact become quite different on going from ODC⋅OMP to ODC⋅I− (Fig. 4). N1 and C6 suffer strong pressure from Pro-202 and Ile-97 in ODC⋅OMP, whereas C2 and C4 form strong contacts in ODC⋅I−. This shifting is due to the movement of the ligand as depicted in Fig. 2, while Pro-202 and Ile-97 remain stationary in the protein structure.
Table 4.
Solvent-accessible surface area (SASA) of each moiety from different ligands
| Moiety | SASA, Å2
|
|
|---|---|---|
| OMP | I− | |
| CO2 | 1.9 | 0.1 |
| C6 | † | 0.1 |
| Ribose | 15.7 | 3.4 |
| Phosphate | 5.1 | 6.3 |
| Pyrimidine | 2.4 | 0.6 |
In OMP, SASA of C6 is included in the SASA of the pyrimidine.
Figure 4.
(A) Corey–Pauling–Koltun (CPK) model of OMP with Pro-202/Ile-97. Heavy atom distances of hydrophobic contacts (less than 4 Å) are shown for OMP (B) and I− (C). Contacts with Pro-202 are designated by dashed arrows and contacts with Ile-97 are by solid arrows. The interaction distances marked by “(cg)” are interactions with Pro-202(Cγ) and the rest are with Pro-202(Cβ).
Conformation of OMP in Water and Enzyme.
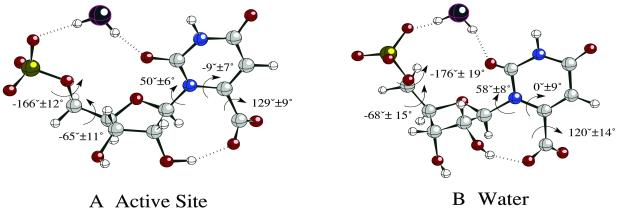
The conformation of OMP bound to the active site was compared with the conformer free in water (Fig. 5). All of the dihedral angles in the active site were reproduced in one of the water-phase conformers. This water conformer occupies ≈10% of the total ground state population in water and geometrical resemblance of this conformer to the one in the active site suggests no external strain by enzyme residues. In particular, the dihedral angles around the carboxylate group are similar in the two environments because the intramolecular hydrogen bond between the 2′-hydroxyl group and the carboxylate favors such a geometry. Lys-93 in the active site merely stabilizes this angle so that no exchange of positions between O8 and O9 occurs. The water-bridged hydrogen bond between the phosphate and O2 are observed in both environments.
Figure 5.
MD simulation snapshot of OMP geometry in the active site (A) and in a water sphere (B). The dihedral angles shown are average values and standard deviations.
Discussion
Two MD simulations of ODC in aqueous solvent have been performed. In one simulation ODC is complexed with the substrate OMP and in the other simulation ODC is complexed to the carbanion intermediate I− formed by OMP decarboxylation (Scheme S1). The comparison of ground state and intermediate states shows that decarboxylation of OMP in the ODC active site occurs with significant changes in the position of ligand and the conformation of the protein loop involving residues 203–218. Also, the structure of ODC⋅OMP in the MD simulation is noticeably different from the crystal structure of ODC with the inhibitor BMP. On the basis of the observation of our simulations, we have reexamined the previously proposed mechanisms in Scheme S1.
Reexamination of Circe Effect.
One of the explanations for OMP decarboxylation involves the binding energy of the nonreactive part of the OMP being used to force the OMP carboxylate toward the carboxylate of Asp-91 (Circe effect) (2). This juxtaposition of the carboxylates is proposed to induce shifting of the electron density away from the OMP carboxylate to catalyze decarboxylation. This mechanism requires the following conditions. First, the direction that the protein attracts the nonreactive part of OMP (i.e., ribose-phosphate moiety) should be opposite to the direction that Asp-91 repels the OMP's carboxylate. Also, the binding domain for the nonreactive part of OMP should be stiff enough to exert a strain. Second, the Circe effect mechanism presumes that the ODC-bound structure of OMP is unstable.
In our simulation of ODC⋅OMP, the active site surrounding the ribose-phosphate regions is very flexible, as shown in the positional fluctuations in Table 2. Arg-235 has been proposed to be essential in providing the binding energy for distortion, but the guanidino group enjoys significant motions. In addition, Arg-235 does not attract the phosphate moiety, as proposed, in such a direction that Arg-235 can bring the substrate's carboxylate into the proximity of Asp-91 (Fig. 1A). By mutant experiments Tyr-217 has also been shown to contribute to catalysis as much as does Arg-235 (12). However, orientation of Tyr-217 relative to the OMP's phosphate makes it unlikely that Tyr-217 pushes the substrate's carboxylate toward Asp-91.
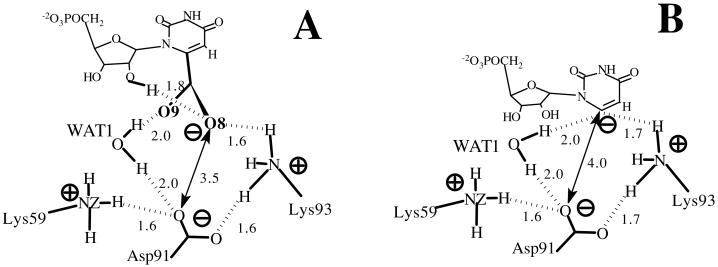
Indeed, the OMP's carboxylate does not directly confront the carboxylate of Asp-91 in its equilibrium state of ODC⋅OMP complex (Fig. 6 and Fig. 1A). Lys-93 and WAT1 shield the negative charges. In experiments, mutation of Asp-91 into Ala did not enhance the binding affinity of the substrate (13) and inhibitors with negative charges at C6 position show greater binding affinities compared with neutral ones (4). Most importantly, the enzyme-bound OMP is not in an unstable form. This is shown by the fact that all of the dihedral angles of OMP when in the active site are exactly reproduced when OMP is in aqueous solution (Fig. 5).
Figure 6.
Electrostatic interaction network near the carboxylate moieties. The distances (Å) are averaged values from ODC⋅OMP (A) and ODC⋅I− (B) simulation, respectively.
The importance of the phosphate–enzyme interaction (12) can be attributed to correct positioning of the OMP's carboxylate. Without the phosphate bound to the Arg-235/Tyr-217, the substrate's carboxylate might prefer more readily accessible Lys-59 to deeply buried Lys-93.
Reexamination of O4 or C5 Protonation Mechanism.
Ab initio calculations have shown that O4 and C5 have high proton affinities and that protonation at either of these moieties reduces the activation energy in the gas phase for decarboxylation by a significant amount because of delocalizing the electron pair through π bonds (Scheme S1). However, the absence of required acidic moieties in the vicinity of O4 and C5 makes these proposals unlikely. Our simulation of ODC⋅OMP shows that O4 constantly forms several hydrogen bonds with the protein residues, none of which are acidic (Fig. 1A). Although there is a water molecule within ≈3.7 Å of C5, which is hydrogen bonded to the carboxylate of OMP (Fig. 1A), the same water is present when OMP is in aqueous solution. There is an absence of any acidic group that could pass a proton through this water. Also, C5 is mainly protected by hydrophobic contact with Pro-202 and Ile-97 (Fig. 4).
Reexamination of O2 Protonation.
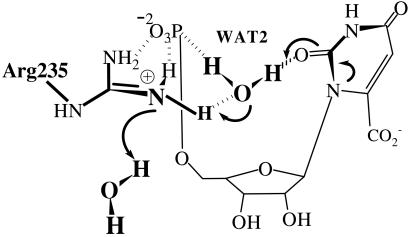
In gas phase the proton affinity of O4 is ≈15 kcal/mol higher than that of O2, but the activation energies for decarboxylation are similar when starting from either protonated OMP (7). One might conclude that the reaction path via O2(H+)I− might be favored in certain environments where O2 becomes basic and the proton source is activated. In the ODC⋅OMP structure specific hydrophobic interactions with the pyrimidine ring increase the basicity of O2 (Fig. 4). A water molecule (WAT2) exists as a bridge between O2 and the terminal phosphate, and the WAT2 oxygen is hydrogen bonded to the protonated guanidino group of Arg-235 in the active site (Fig. 1A). WAT2 is the only source from which O2 can be protonated. Transfer of H+ from WAT2 to O2 must be concerted with transfer of H+ from Arg-235 to the incipient HO−. This must be in turn followed by H+ uptake by the neutral Arg-235 from bulk water solvent. It should be noted that Arg-235 is exposed to solvent. This hypothetical proton relay from bulk water solvent → Arg-235 → WAT2 → O2 of OMP (Fig. 7) could be proposed to be involved in decarboxylation reaction by means of O2 protonation. However, the Arg235Ala mutant shows only 100-fold drop in kcat, resulting in kcat/knon = 1015 (12). Short simulation of ODC⋅O2-protonated OMP (data not presented) showed that carboxylate moiety becomes quite remote from Lys-93 and Asp-91 (>5 Å away), which are experimentally characterized to be important for catalysis. These results do not support an O2 protonation mechanism. Deuterium solvent isotope effects (14) and substrate substituent (O2 → S2) experiment (15) have been presented in support of the mechanism involving O2 protonation. However, such D2O effects, as pointed out by Rishavy and Cleland (6), do not prove the existence of a protonation step before the decarboxylation. Loss of activity on replacement of O2 by S can be a result of poor binding of the substrate, as bulkier S2 may cause a steric conflict with the phosphoester O when 2-thio-OMP binds to the active site (O2⋅⋅⋅phosphoester O = 3.5 ± 0.2 Å in ODC⋅OMP).
Figure 7.
Electrostatic network near O2 of OMP, showing the unlikely proton transfer relay from solvent water → Arg-235 → WAT2 → O2.
Intermediate Stabilization.
Theoretical calculations show that the decarboxylation of OMP is endothermic and the activation barrier (ΔG‡) is very similar to the endothermicity (ΔG°) of the gas-phase reaction (16). In such situations stability of product (decarboxylated intermediate) can determine the stability of the transition state according to the Leffler–Hammond postulate. In the ODC active site, the C6 carbanion of I− moves into the position formerly occupied by O8 of OMP, bringing the whole ligand toward Lys-93 by ≈2.2 Å (Fig. 6). The position of the 203–218 loop follows this movement toward the interior of the TIM-barrel (Fig. 2) and the conformation of the 203–218 loop changes from an ill-defined structure to a well-ordered β-hairpin (Fig. 3) along with increasing contacts with other protein residues (Table 3). Positional fluctuation of the backbone of this loop decreases from ≈0.8 Å (ODC⋅OMP) to ≈0.4 Å (ODC⋅I−).
Conclusion
Proposed reaction steps for decarboxylation of OMP by ODC are shown in Scheme S1. On the basis of our 2.5-ns MD simulation of ground state ODC⋅OMP and the intermediate state ODC⋅I−, we have reexamined previously proposed mechanisms and have suggested consideration of a catalytic role of enzyme dynamics by means of the loop of residues 203–218. Flexibility of the phosphoribosyl-binding domain and stability of the enzyme-bound substrate conformation do not support a concept that the enzyme uses the binding energy to distort the carboxylate moiety. Although the catalytic antibody, which has hydrophobic surroundings near the OMP carboxylate, shows remarkable rate enhancement of 108, ODC does not appear to select such a ground state destabilization mechanism (17). Lack of available acidic residues near O2, O4, and C5 also makes it hard to conceive of those moieties undergoing protonation before decarboxylation.
The comparison of ground state and intermediate state shows that OMP moves toward Lys-93 upon decarboxylation. This positional shift is accompanied by the movement of the 203–218 loop position and a conformational change from this unstructured loop to an ordered β-hairpin. These changes are suggested to enhance intermediate stabilization and lowering of the endothermicity of the reaction. Because I− and transition state are energetically quite similar, enzyme stabilization of the former would provide lower free energy content for the latter.
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation Grant MCB-9727937. We gratefully acknowledge computer time at the National Partnership for Advanced Computational Infrastructure (NPACI) (San Diego Supercomputer Center).
Abbreviations
- ODC
orotidine-5′-monophosphate decarboxylase
- OMP
orotidine 5′-monophosphate
- I−
carbanion intermediate
- MD
molecular dynamics
- BMP
1-(5′-phospho-β-d-ribofuranosyl)barbituric acid
References
- 1.Radzicka A, Wolfenden R. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 2.Wu N, Mo Y R, Gao J L, Pai E F. Proc Natl Acad Sci USA. 2000;97:2017–2022. doi: 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris P, Poulsen J C N, Jensen K F, Larsen S. Biochemistry. 2000;39:4217–4224. doi: 10.1021/bi992952r. [DOI] [PubMed] [Google Scholar]
- 4.Levine H L, Brody R S, Westheimer F H. Biochemistry. 1980;19:4993–4999. doi: 10.1021/bi00563a010. [DOI] [PubMed] [Google Scholar]
- 5.Lee J K, Houk K N. Science. 1997;276:942–945. doi: 10.1126/science.276.5314.942. [DOI] [PubMed] [Google Scholar]
- 6.Rishavy M A, Cleland W W. Biochemistry. 2000;39:4569–4574. doi: 10.1021/bi000376p. [DOI] [PubMed] [Google Scholar]
- 7.Phillips L M, Lee J K. J Am Chem Soc. 2001;123:12067–12073. doi: 10.1021/ja0117332. [DOI] [PubMed] [Google Scholar]
- 8.Lee T S, Chong L T, Chodera J D, Kollman P A. J Am Chem Soc. 2001;123:12837–12848. doi: 10.1021/ja011096f. [DOI] [PubMed] [Google Scholar]
- 9.Miller B G, Hassell A M, Wolfenden R, Milburn M V, Short S A. Proc Natl Acad Sci USA. 2000;97:2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter D J T, Short S A. Biochemistry. 2000;39:11788–11800. doi: 10.1021/bi001199v. [DOI] [PubMed] [Google Scholar]
- 11.Miller B G, Butterfoss G L, Short S A, Wolfenden R. Biochemistry. 2001;40:6227–6232. doi: 10.1021/bi0028993. [DOI] [PubMed] [Google Scholar]
- 12.Miller B G, Snider M J, Short S A, Wolfenden R. Biochemistry. 2000;39:8113–8118. doi: 10.1021/bi000818x. [DOI] [PubMed] [Google Scholar]
- 13.Miller B G, Snider M J, Wolfenden R, Short S A. J Biol Chem. 2001;276:15174–15176. doi: 10.1074/jbc.M011429200. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich J I, Hwang C-C, Cook P F, Blanchard J S. J Am Chem Soc. 1999;121:6966–6967. [Google Scholar]
- 15.Smiley J A, Hay K M, Levison B S. Bioorg Chem. 2001;29:96–106. doi: 10.1006/bioo.2001.1201. [DOI] [PubMed] [Google Scholar]
- 16.Feng W Y, Austin T J, Chew F, Gronert S, Wu W M. Biochemistry. 2000;39:1778–1783. doi: 10.1021/bi992553w. [DOI] [PubMed] [Google Scholar]
- 17.Smiley J A, Benkovic S J. Proc Natl Acad Sci USA. 1994;91:8319–8323. doi: 10.1073/pnas.91.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen W L, Chandrasekhar J, Madura J D, Impey R W, Klein M L. J Chem Phys. 1983;79:926–935. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.