Abstract
A competitive growth assay has been used to identify yeast genes involved in the repair of UV- or MMS-induced DNA damage. A collection of 2,827 yeast strains was analyzed in which each strain has a single ORF replaced with a cassette containing two unique sequence tags, allowing for its detection by hybridization to a high-density oligonucleotide array. The hybridization data identify a high percentage of the deletion strains present in the collection that were previously characterized as being sensitive to the DNA-damaging agents. The assay, and subsequent analysis, has been used to identify six genes not formerly known to be involved in the damage response, whose deletion renders the yeast sensitive to UV or MMS treatment. The recently identified genes include three uncharacterized ORFs, as well as genes that encode protein products implicated in ubiquitination, gene silencing, and transport across the mitochondrial membrane. Epistatsis analysis of four of the genes was performed to determine the DNA damage repair pathways in which the protein products function.
DNA is a labile molecule that can undergo spontaneous hydrolysis or modification by physical and chemical agents within the cellular environment (1). Modified DNA must be rapidly recognized and efficiently repaired, thus both prokaryotes and eukaryotes have evolved complex surveillance and repair mechanisms. The response to DNA damage has been well characterized in Saccharomyces cerevisiae through the isolation of mutants that are hypersensitive to specific DNA damaging agents (1). These mutation studies led to the definition of three groups of proteins involved in different types of DNA repair, termed the RAD3, RAD52, and RAD6 epistasis groups, based on phenotypic sensitivity to UV, ionizing radiation, or both.
The cellular DNA damage response depends on the type of damage incurred. The UV response is largely mediated by two epistasis groups, the RAD3 group, which includes genes of the nucleotide excision and repair pathway, and the RAD6 group, encoding proteins involved in postreplication repair and damage-induced mutagenesis. Treating cells with the methylating agent methyl methanesulfonate (MMS) results in alkylated DNA, which is poorly replicated by DNA polymerases in vitro and in vivo (1, 2), and must be efficiently repaired. Base excision repair proteins (3), as well as proteins encoded by the MEC1 and RAD53 genes (2), and the RAD6 and the RAD52 epistasis group genes (4), have all been implicated in the response to MMS damage. Although excision repair and recombination repair pathways are relatively well understood, much less is known about the RAD6 mediated pathway. The response to both UV irradiation and MMS methylation likely involve additional genes, especially of the RAD6 pathway, which remain to be identified.
Toward this end, we have used a collection of S. cerevisiae deletion strains, wherein each strain has had an individual gene replaced by a unique DNA sequence, to phenotypically screen the yeast genome for proteins involved in the DNA damage response. As reported (5), the collection may be used to identify, in a single competitive fitness assay, the genes whose deletion render the yeast inviable or less competitive under a specific set of experimental conditions. Specifically, we identified yeast genes whose protein products are important for the repair and/or tolerance of UV- and MMS-induced DNA damage: three genes crucial in the UV response and four in the MMS response. None of these genes were formerly known to be involved in DNA damage repair or tolerance, and three correspond to annotated ORFs whose protein products have not previously been characterized in any context. Epistasis analysis of four of the identified genes connects three to the RAD6 repair pathway and one to the RAD52 pathway.
Materials and Methods
Materials.
Yeast strains used in the competitive growth assays and data confirmation include BY4741 (haploid), BY4743 (diploid), and individual deletion strains in these genetic backgrounds (American Type Culture Collection). These deletion strains contain a pair of unique 20-mer sequences, an UPTAG and DOWNTAG “barcode,” which serve as identifiers for individual gene deletions. (See http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html.) Deletion strains were pooled as described (5) to afford a homozygous diploid deletion pool of 2,827 individual strains.
UV irradiations were performed by using a G8T5 germicidal tube (Ushio America, Cypress, CA), and intensities were measured by using a UVX radiometer with a UVX-25 sensor (Ultraviolet Products). UV intensities reported herein are direct readings from the radiometer. Microarrays were 266 × 266 sense and antisense oligonucleotide probes, purchased from Affymetrix (TAG-ARRAY-3). The 6× SSPE-T contained 1 M NaCl, 66 mM NaH2PO4, 6.6 mM EDTA (pH 7.4), and 0.005% Triton X-100. Streptavidin phycoerythrin stain was 1 M NaCl, 100 mM Mes, 0.01% Triton X-100, 0.5 mg/ml BSA, and 10 μg/ml phycoerythrin-streptavidin (Molecular Probes). URA−-selective media consisted of 0.67% yeast nitrogen base without amino acids (Difco), 0.1% casamino acids (Difco), and 2% dextrose. Yeast extract/peptone/dextrose (YPD) media and synthetic dropout media, supplemented with various nutrients as required, were prepared as described (6).
Competitive Growth Assays.
The homozygous diploid deletion pool was UV treated in PBS with seven acute irradiation intensities ranging from 110 to 270 J/m2. After irradiation, the various samples were resuspended in YPD and allowed to recover at 30°C in the dark. MMS treatment of the pool was conducted by adding 0.001% or 0.01% MMS to YPD media and growing at 30°C. Samples of 1 × 108 cells were removed from the logarithmically growing pools at various times for up to 20 generations of growth. Control cultures were mock treated and sampled along with the experimental cultures. Genomic DNA was extracted from the samples, and UPTAGS and DOWNTAGS corresponding to the surviving individual strains were amplified in separate PCRs. PCR conditions and primers were identical to those used by Winzeler et al. (5), except that all primers were biotinylated. The resulting PCR products were hybridized to microarrays as described (5), with the exception that arrays were hybridized for 16 h at 42°C, washed five times with 6× SSPE-T, and then stained at 37°C for 15 minutes in streptavidin phycoerythrin stain. All strains in the pool were represented on the array with sense and anti-sense probes for each UPTAG and DOWNTAG. The hybridization intensities from multiple samples were normalized, and the probes for each strain were quantitated over time. The relative growth rate of each strain was determined as described (5), except that the hybridization signal was fit to a linear dependence on time, with the slope used to determine the growth rate. The relative fitness (RF) of each strain is the ratio of the growth rates for treated and untreated samples. (See Acute UV Sensitivity Assay and MMS Sensitivity Assay, which are published as supporting information on the PNAS web site, www.pnas.org.)
Verification of Strain Sensitivity.
Individual strains were grown to stationary phase in YPD, diluted to 5 × 106 cells/ml, aliquoted, and immediately treated with the respective DNA-damaging agent. UV strains were plated in triplicate on YPD–agar, irradiated at UV intensities of 0–80 J/m2, and incubated at 30°C in the dark for 38–44 h. Digital pictures of the plates were taken, and colonies were counted with BioRad quantity one software. MMS strains were plated in triplicate on YPD–agar containing 0–0.025% MMS and were incubated at 30°C for 64–70 h before colonies were counted. Colony counts were used to construct survival curves for each strain.
Complementation Assays.
ORFs were PCR amplified from S288C genomic DNA, isogenic to BY4743, by using 32- to 35-nt long primers. Primers were designed to amplify a product with a SalI site located 500–800 bp upstream from the start codon of the gene of interest and a NotI site located after the stop codon (see Table 3, which is published as supporting information on the PNAS web site, for primer sequences). PCR amplified products were digested and ligated into the low copy, centromeric plasmid pRS416 (7). Each deletion strain was independently transformed by using published protocols (8), with both pRS416 and pRS416 harboring the corresponding gene. Transformants from each reaction were grown to logarithmic phase and UV irradiated or treated with MMS as described above for the individual strains, with the exception that URA− media was used in all steps.
Genetic Analysis.
rad9Δ, rad14Δ, rad18Δ, and rad24Δ strains were constructed by replacing the kanMX4 module of BY4742 (MATα) strains (ATCC) with the heterologous HIS3MX6 module. Plasmid pFA6a-HIS3MX6 (9) was digested with BglII/BsmI and the resulting linear cassette was transformed into each recipient strain by using the lithium acetate method (8). Proper integration of each RAD:HIS3MX6 allele was verified. Strains carrying double mutations of the RAD genes (except rad52Δ, see below) with either DOA1, ESC4, YPL055c, or YLR376c were constructed by crossing the various ORFΔ:kanMX4 MATa haploids with the radΔ:HIS3MX6 MATα haploids and selecting for diploid cells by nutrient complementation on synthetic dropout media. The resulting diploids were sporulated, and spore clones were randomly analyzed for histidine prototrophy and G418 resistance (10). Ploidy was verified by standard procedures (11), and disruption of chromosomal genes was further confirmed by PCR analysis (9).
Double mutant strains of DOA1, ESC4, YPL055c, and YLR376c with RAD52 were constructed by single step gene disruption. A rad52Δ:LEU2 cassette was released by digesting pBR322-rad52Δ:LEU2 (the generous gift of D. Livingston, University of Minnesota) and transforming the linear DNA into the various ORFΔ:kanMX4 MATa strains (12). Disruption of the chromosomal gene was confirmed by PCR analysis. (Table 4, which is published as supporting information on the PNAS web site, contains further description of strains.)
Epistasis analyses of the double mutants were performed by comparing the phenotypic sensitivity of the various double mutants with those of the corresponding single mutants. Several independent spores or transformants were analyzed for each double mutant to rule out the possibility that observed sensitivities were caused by clonal artifacts.
Results
Identification of UV- or MMS-Sensitive Deletion Strains.
In competitive fitness assays, a pool of homozygous diploid deletion strains was subjected to multiple irradiation conditions or MMS concentrations and then allowed to recover for up to 20 generations of growth. During this recovery time, pools were sampled at various generations, genomic DNA was extracted from the cells, tags corresponding to individual deletion strains were PCR amplified, and hybridization intensities from oligonucleotide arrays were collected. Strains that did not exhibit at least one hybridized tag signal that was greater than 3 times the background signal were not considered for further analysis. The pool contained 20 viable deletion strains known to be UV sensitive and 13 deletion strains known to be MMS sensitive. All 20 UV-sensitive and 12 of 13 MMS-sensitive strains were correctly identified by the assay (see Tables 5 and 6, which are published as supporting information on the PNAS web site). The hybridization data for the remaining strains was used to calculate the RF for each deletion mutant in the pool. Strains were divided into three categories based on their RF values: highly, moderately, or weakly sensitive. The ORFs from 164 sensitive deletion strains identified in the UV assay and 147 strains from the MMS assay are tabulated in Tables 5 and 6. The strains listed in Tables 1 and 2 correspond to genes not previously characterized as UV or MMS sensitive, which displayed the most robust RF trends with relative fitness ratios decreasing as irradiation intensities or MMS concentrations increased.
Table 1.
Growth response of UV-treated strains determined by competition assays
| Sensitivity of strains with deleted ORFs | |||
|---|---|---|---|
| High | Moderate | Weak | |
| CKB2 | BEM3 | YML003W | ERG5 |
| CCR4 | ELM1 | YML005W | ERV25 |
| DOA1 | FMS1 | YML029W | HXT2 |
| ELC1 | HAP2 | YML030W | SOK2 |
| FPS1 | MCK1 | YMR003W | SSN3 |
| HAP4 | MRP49 | YMR018W | STB4 |
| NUP60 | MSS18 | YMR030W | TSA1 |
| PHO4 | OST6 | YNL201C | VPS30 |
| RTF1 | PEP12 | YPL191C | YPT7 |
| SRO9 | ROM1 | YER030W | |
| UBP15 | SIN3 | YLR257W | |
| UME1 | SWD3 | YML013W | |
| YBR174C | VPS8 | YMR009W | |
| YDR409W | YAL027W | YMR010W | |
| YER092W | YAR003W | YOR083W | |
| YKL002W | YBL089W | ||
| YML011C | YGR122W | ||
| YMR263W | YHL005C | ||
| YPL055C | YML002W | ||
High sensitivity, three or more relative fitness ratios below 0.92; moderate sensitivity, two ratios below 0.92; weak sensitivity, one ratio below 0.92.
Strains chosen for individual analysis.
During the preparation of this manuscript, these strains were identified as γ irradiation sensitive (33).
Strains displaying increased sensitivity during individual treatment.
Successfully complemented strains.
Table 2.
Growth response of MMS-treated strains determined by competition assays
| Sensitivity of strains with deleted ORFs | |||
|---|---|---|---|
| High | Moderate | Weak | |
| COQ4* | CKB2 | YGR182C | FPS1 |
| COX7 | CTR1 | YHR045W | PCD1 |
| COX18 | DOA1 | YKL098W* | PET111 |
| ESC4* | DRS2 | YLR135W | RIM101 |
| HOM3* | ILV1 | YNL201C | RTG2 |
| IKI3 | ISC1 | YOR284W | SCS7 |
| IRA2* | LST4 | YPL170W | SNF7 |
| ISM1 | MAC1* | YPR123C | SPO7 |
| PET309 | PHB1 | YPR153W | STP22 |
| RSM22 | POS5 | TOM6 | |
| TIM13* | PPH3* | UBI4 | |
| YCL061C* | SHU1 | VPS36 | |
| YER049W* | SLT2 | YDR540C | |
| YGR228W | TOF1 | YGR122W | |
| YHR207C | TOS10 | YHL005C | |
| YLR368W | TRP3 | YHR189W | |
| YLR376C* | YAL011W | YLR239C | |
| YOR338W | YBR223C* | YLR412W | |
| YDR078C | YOR192C | ||
| YER083C | YPL047W | ||
| YGR136W* | YPL184C | ||
High sensitivity, both relative fitness ratios below 0.88; moderate sensitivity, one ratio below 0.88; weak sensitivity, one ratio below 0.92.
* Strains chosen for individual analysis.
Strains displaying increased sensitivity during individual treatments.
Successfully complemented strains. None of these mutant strains were reported as γ irradiation sensitive (33).
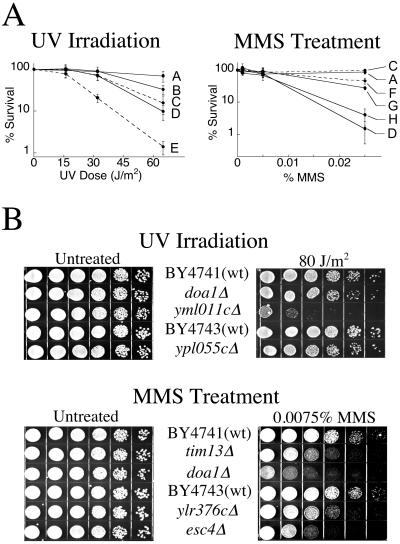
Twenty-five putative UV-sensitive strains and 13 putative MMS-sensitive strains were tested individually to substantiate the hybridization data. Growth rates in rich media were monitored and compared with that of BY4743/41 strains. One strain, yal021cΔ was omitted from further analysis because of slow growth rates and general poor health. UV irradiations were carried out on YPD–agar to ensure that results were not complicated by a lack of nutrients. Each UV strain was subjected to irradiations corresponding to 95%, 90%, and 70% survival of plated BY4743 cells, and control plates were left untreated. The MMS strains were plated onto YPD–agar containing 0, 0.001%, 0.005%, or 0.025% MMS. Three strains, doa1Δ, ypl055cΔ, and yml011cΔ, were verified as phenotypically more sensitive to UV irradiation than wild-type BY4743/41 cells (Fig. 1). The three deletion strains, esc4Δ, ylr376cΔ, and tim13Δ, were verified as exhibiting increased sensitivity to MMS compared with wild-type BY4743/41 cells (Fig. 1). Each strain identified as sensitive to either UV or MMS was analyzed for sensitivity to the other mutagen, doa1Δ showed sensitivity to both UV- and MMS-induced damage.
Fig 1.
Phenotypes of deletion strains as determined from individual treatment. (A) Survival curves for strains subjected to UV or MMS treatment. A, BY4743 (wild type); B, ypl055cΔ; C, BY4741 (wild type); D, doa1Δ; E, yml011cΔ; F, tim13Δ; G, ylr376cΔ; H, esc4Δ. Strains denoted by solid lines are diploid, whereas dotted lines are haploid. Error bars indicate standard deviations from a minimum of three treatments. (B) Dilution plates of deletion strains. Diploid strains are located below the parental BY4743, all other strains are haploid. Five-fold serial dilutions of ≈6 × 104 logarithmically growing cells were plated in duplicate. For UV treatment, one plate was irradiated at 80 J/m2 and incubated in the dark for 42 h at 30°C. MMS-sensitive strains were plated onto YPD–agar with or without 0.0075% MMS and incubated for 64 h at 30°C. Note that the dilution assay was conducted with doa1Δ haploid cells, whereas survival curves were constructed with the doa1Δ homozygous diploid strain, which became available during the course of this study.
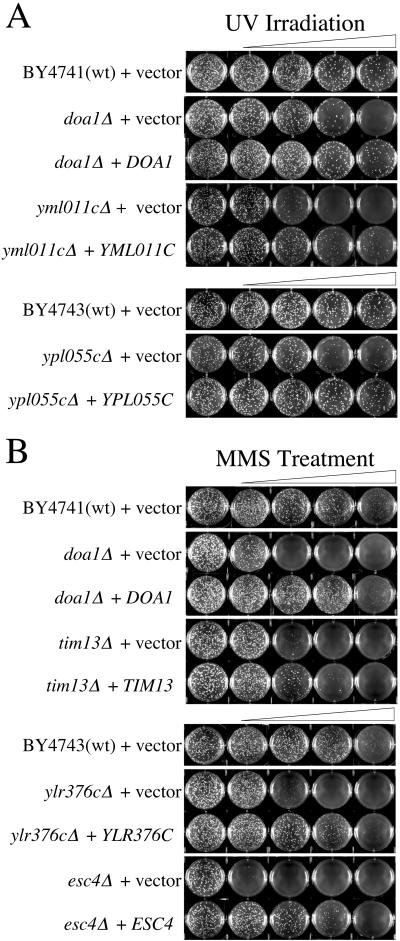
Complementation assays were conducted to verify that increased sensitivity to UV or MMS was caused by the ORF deletion and was not an artifact of strain construction. The region of the chromosome containing the gene and an upstream region containing the endogenous promoter were cloned into the pRS416 vector and transformed into the corresponding deletion strain. doa1Δ, ypl055cΔ, yml011cΔ, esc4Δ, ylr376cΔ, and tim13Δ, strains transformed with pRS416 containing the corresponding amplified PCR product were less sensitive to the damaging agent than the strains transformed with the empty pRS416 vector (Fig. 2).
Fig 2.
Complementation of deletion strains sensitive to DNA damaging agents. Strains were transformed with empty pRS416 (+vector) or with pRS416 containing the gene that the deletion strain is lacking (+gene). The BY4743 strain, ypl055cΔ, ylr376cΔ, and esc4Δ are diploid, whereas and all other strains are haploid. Pictured are representative plates from treatments repeated a minimum of three times. (A) UV-sensitive strains. Cells were irradiated at 0, 16, 32, 65, and 80 J/m2. (B) MMS-sensitive strains. Cells were treated with 0, 0.005%, 0.0075%, 0.010%, and 0.015% MMS.
Epistasis Analysis of DOA1, ESC4, YPL055c, and YLR376c.
Further genetic analyses of the identified deletion strains were performed to gain insight into the role of the respective protein products in the DNA damage response. Double mutant strains were constructed with genes from the major DNA repair and checkpoint response pathways. The completely nucleotide excision repair-deficient strain, rad14Δ, was used for the RAD3 epistasis group, the rad18Δ strain was chosen for the RAD6 epistasis group because it is fully deficient in postreplication repair and mutagenesis, and rad52Δ was chosen for the recombination repair epistasis group. Double mutants were constructed with RAD9 and RAD24 to address the potential role of the identified genes in DNA damage-mediated cell cycle control.
The sensitivities of double mutants of DOA1 with RAD14, RAD52, RAD9, and RAD24 to MMS are each additive relative to the corresponding single mutants (Fig. 3A). However, doa1Δ is epistatic to rad18Δ. This finding suggests that DOA1 is involved in the RAD6 pathway. Qualitatively identical results were obtained when performing the epistasis analysis of DOA1 with UV-induced DNA damage (data not shown). The MMS sensitivity of ESC4 double mutants with RAD14, RAD18, RAD52, RAD9, or RAD24 were additive relative to each single mutant. However, the esc4Δrad18Δ double mutant showed a strong synergistic sensitivity (Fig. 3B). This genetic interaction suggests that Esc4p acts on a substrate that is also processed in the RAD6 pathway. The corresponding epistasis analysis of ypl055cΔ strains treated with UV irradiation also identified YPL055c as uniquely epistatic to RAD18 (Fig. 3C). Double mutants of YLR376c with the same set of genes identified an epistatic relationship between YLR376c and RAD52, whereas all other double mutants showed additive sensitivity (Fig. 3D). This finding implies that the YLR376c protein product mediates a component of the recombination repair pathway.
Fig 3.
Genetic epistasis analyses of DOA1, ESC4, YLR376c, and YPL055c. Five-fold serial dilutions of ≈1 × 106 logarithmically growing cells were plated on YPD–agar containing 0, 0.0025%, 0.005%, 0.0075%, or 0.010% MMS, or UV irradiated at 0, 5, or 65 J/m2. Pictured are representative plates that best visually display the sensitivity of the double deletion strains in comparison to the single mutants. These sensitivities were consistent over the range of MMS concentrations and irradiations tested. (A) doa1Δ strains. (B) esc4Δ strains. (C) ypl055cΔ strains. (D) ylr376cΔ strains.
Discussion
Twenty of 20 and 12 of 13 genes known to be UV- and MMS-sensitive, respectively, were correctly identified by the competitive growth assays. The gene not correctly identified was YKU70, which was previously reported as hypersensitive to MMS (13). Analysis of the yku70Δ strain in the BY4743 genetic background showed that the deletion strain was more sensitive than wild-type, but not as sensitive as the strains identified herein (Fig. 4, which is published as supporting information on the PNAS web site). From the microarray analysis, 62 UV- and 69 MMS-sensitive strains were identified as most likely to possess the phenotype, because their RFs decreased with increasing UV or MMS exposure (Tables 1 and 2). From these candidate strains, individual killing curves were constructed for 25 UV and 13 MMS strains. These studies verified the UV sensitivity of five strains, doa1Δ, pho4Δ, ume1Δ, yml011cΔ, and ypl055cΔ, each originally categorized as highly sensitive; and the MMS-sensitivity of three strains, esc4Δ, tim13Δ, and ylr376cΔ, also originally categorized as highly sensitive. To confirm that the identified deletions represent genes involved in the damage response, the deleted ORFs were cloned into a low-copy vector, which was shown to complement six of the eight original deletion strains, verifying that the phenotype of these six strains resulted from loss of the specific protein product (Fig. 2). Each gene is discussed in detail, including a survey of the pertinent literature to place the gene in the larger context of the DNA damage response.
DOA1.
DOA1 deletion resulted in sensitivity to both UV and MMS. DOA1 is thought to encode a regulatory component of the proteasome pathway, which involves ubiquitin (Ub)-dependent protein degradation (14, 15). Doa1p is a 715-aa protein containing five tandem WD repeats, which are thought to mediate protein–protein interactions. Cells with mutated copies of DOA1 have abnormally low levels of free Ub, are resistant to several anesthetics (16), and are deficient in the degradation of a variety of proteins, including the MATα2 repressor and a variety of Ub–protein fusions (14, 17, 18). The Doa1p Ub pathway is regulated by de novo synthesis of sphingolipids (19) and requires Cdc48p, an essential cell cycle checkpoint protein in S. cerevisiae (20–22). Doa1p has been shown to physically associate with Cdc48p in vitro and in vivo (15). It is therefore tempting to speculate that Doa1p may function by coupling DNA damage to a cell cycle checkpoint through ubiquitination, possibly via Cdc48p. However, preliminary results indicate that doa1Δ mutant cells possess an intact S-phase checkpoint, based on treatment with MMS and subsequent analysis of DNA content by flow cytometry, and that the deletion strain is also proficient for Rad53 phosphorylation in response to MMS treatment in asychronous culture (data not shown).
Genetic analysis of DOA1 double mutants assigns the gene to the RAD6 epistasis group. Two separate modes of repair are mediated by RAD18 within this pathway and both involve ubiquitination; error-prone repair, involving translesion synthesis across a damaged template base, and error-free repair, which may use information from an undamaged sister chromatid for correct nucleotide insertion (23). The specific role of Doa1p in these pathways is not yet clear. Doa1p may play an important role in cellular Ub concentration and/or substrate specificity, and thus facilitate degradation or modification of proteins involved in error-free gap filling or mutagenesis.
ESC4.
The esc4Δ strain was sensitive to MMS. This gene encodes a protein of 1,070 aa containing six copies of the BRCT motif found in proteins associated with DNA checkpoint pathways (24). It is 21% identical over 973 aa to Schizosaccharomyces pombe Brc1p, a protein that is required for chromosome stability (25). ESC4 is known to interact genetically with SGS1, a recQ-like helicase, as deletion of ESC4 is lethal in an sgs1Δ background (26). This synthetic lethality implies that the functions of the two protein products play redundant or competitive roles. Sgs1p is thought to promote maturation of recombination intermediates formed during replication of damaged DNA (27, 28). ESC4 is also capable of silent information regulator (SIR)-dependent silencing of the HMR locus when appropriately targeted to the DNA (Rolf Sternglanz, personal communication).
Double mutant strains of esc4Δ with rad14Δ, rad52Δ, rad9Δ, or rad24Δ were each more sensitive, with the magnitude being additive, than the corresponding single mutants. However, even at low MMS concentrations, the esc4Δrad18Δ strain was approximately 104 more sensitive than either single mutant. This synergistic sensitivity implies that Rad18p and Esc4p have overlapping functions, possibly by each acting on a common substrate.
This connection between ESC4 and the RAD6 epistasis group is particularly interesting considering that both are involved in DNA repair and silencing. In addition to the repair process discussed above, RAD6 plays a role in the silencing of mating type loci, telomers, and ribosomal DNA (29). These functions are at least partially dependent on SIR2 and RAD52. RAD18 also plays a role in silencing by recruiting RAD6 to appropriate chromosomal locations, but this function is only critical in the absence of functional chromatin assembly factor I protein (30). These genetic and biochemical data imply that, after appropriate localization to a stalled replication fork, Esc4p may function by processing abortive recombination intermediates that are toxic in the absence of Sgs1p, possibly through silencing of damaged regions of DNA, which may act to repress deleterious recombination.
YPL055c.
The ypl055cΔ strain was UV sensitive. This protein product is predicted to be 332 aa in length and 30% identical over 130 aa to the Lin-1p of Caenorhabditis elegans, which is thought to regulate cell cycle progression (31). When overexpressed in Escherichia coli, Ypl055p was found to induce SOS in a RecA-dependent fashion (32). Recently, Resnick and colleagues (33) identified ypl055cΔ as sensitive to γ irradiation. Further testing for the ability of this strain to undergo recombination showed an increased rate of targeted recombination at the HIS3 locus as compared with wild type. However, ypl055cΔ did not show a defect in repair of a homothallic switching endonuclease-induced double strand break, which is characteristic of the RAD52 group genes (33). Consistent with this data, our epistasis analysis associated this protein with the RAD6 postreplication repair and mutagenesis pathway, not the RAD52 group.
YLR376c.
Deletion of YLR376c conferred sensitivity to MMS. The gene encodes a protein of 242 aa that is known to interact with Csm2p (involved in segregation of chromosomes during meiosis) and Dal80p (involved in RNA polymerase II transcription) (34). The protein product of YLR376c also interacts with Shu1p, whose mutation renders cells sensitive to MMS and also suppresses Hydroxy Urea sensitivity of sgs1Δ mutants (see YPD, http://www.incyte.com/proteome/mainmenu.jsp). Epistasis analysis determined that YLR376c is a member of the RAD52 epistasis group and must play an important role in the recombination repair of damaged DNA.
TIM13.
Tim13p is a 105-residue protein that is localized to the mitochondrial intermembrane space, where it forms a soluble complex with Tim8p and Tim9p (35). Tim13p is required for the most efficient import of a variety of proteins, including Tim23p, under normal cellular conditions (36, 37). The sensitivity of tim13Δ strains to MMS suggests that this protein may be required for a rapid assembly of Tim23p inner membrane translocase pores, which could import proteins to repair damaged DNA. For example, Tim23p is known to interact with Hmi1p, a DNA helicase with homology to E. coli Rep and UvrD proteins, as well as the damage-specific Srs2p of Sch. pombe (1).
YML011c.
The yml011cΔ strain was UV sensitive. This gene encodes a protein of 177 aa, which is known to interact with several other proteins that are likely to be involved in the damage response, including Ade2p, Std1p, Prr2p, Ydr314p, and Yll059p (see YPD). Although further investigation is necessary, these interactions suggest that the YML011c protein product has an important role in the DNA damage response.
Conclusion
We have used a nearly genome-wide phenotypic screen to identify previously uncharacterized genes whose protein products are required for the response to UV- and MMS-induced DNA damage. After verification of the sensitivity, and complementation with a plasmid expressing the protein of interest, six genes were identified, DOA1, ESC4, YPL055c, YLR376c, TIM13, and YML011c.
The identified genes code for proteins involved in postreplication and recombination repair. The fact that three of the genes are linked to the RAD6-mediated postreplication repair and mutagenesis pathway highlights that this pathway remains poorly characterized. These proteins also reveal the growing importance of ubiquitination and DNA silencing as components of the damage response, and further emphasize the mechanistic diversity of the RAD6 damage repair pathway.
Supplementary Material
Acknowledgments
We thank Prof. Steve Reed (The Scripps Research Institute) for helpful discussion. This work was supported by The Donald E. and Delia B. Baxter Foundation.
Abbreviations
MMS, methyl methanesulfonate
RF, relative fitness
YPD, yeast extract/peptone/dextrose
Ub, ubiquitin
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nickoloff J. A. & Hoekstra, M. F. (1998) in Contemporary Cancer Research, ed. Nickoloff, J. A. (Humana, Totowa, NJ), Vol. 2.
- 2.Tercero J. A. & Diffley, J. F. X. (2001) Nature (London) 412, 553-557. [DOI] [PubMed] [Google Scholar]
- 3.Chen J., Derfler, B. & Samson, L. (1990) EMBO J. 9, 4569-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao W., Chow, B. L. & Rathgeber, L. (1996) Curr. Genet. 30, 461-468. [DOI] [PubMed] [Google Scholar]
- 5.Winzeler E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 6.Sherman F., Fink, G. R. & Hicks, J., (1983) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 7.Sikorski R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agatep R., Kirkpatrick, R. D., Parchaliuk, D. L., Woods, R. A. & Gietz, R. D. (1998) Technical Tips Online 1, 51. [Google Scholar]
- 9.Wach A., Brachat, A., Alberti-Segui, C., Rebischung, C. & Philippsen, P. (1997) Yeast 13, 1065-1075. [DOI] [PubMed] [Google Scholar]
- 10.Sherman F. & Hicks, I. (1991) Methods Enzymol. 194, 21-37. [DOI] [PubMed] [Google Scholar]
- 11.Sprague G. (1991) Methods Enzymol. 194, 77-93. [DOI] [PubMed] [Google Scholar]
- 12.Dornfield K. J. & Livingston, D. M. (1991) Mol. Cell Biol. 11, 2013-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin S. G., Laroche, T., Suka, N., Grunstein, M. & Gasser, S. M. (1999) Cell 28, 621-633. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. & Varchavsky, A. (1990) Cell 61, 697-708. [DOI] [PubMed] [Google Scholar]
- 15.Ghislain M. R., Dohmen, R. J., Levy, F. & Varchavsky, A. (1996) EMBO J. 15, 4884-4899. [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe D., Reiner, T., Keeley, J. L., Pizzini, M. & Keil, R. L. (1999) Mol. Cell. Biol. 19, 8254-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P., Johnson, P., Sommer, T., Jentsch, S. & Hochstrasser, M. (1993) Cell 74, 357-369. [DOI] [PubMed] [Google Scholar]
- 18.Johnson E. S., Ma, P. C. M., Ota, I. M. & Varshavsky, A. (1995) J. Biol. Chem. 270, 17442-17456. [DOI] [PubMed] [Google Scholar]
- 19.Chung N., Jenkins, G., Hannun, Y. A., Heitman, J. & Obeid, L. M. (2000) J. Biol. Chem. 275, 17229-17232. [DOI] [PubMed] [Google Scholar]
- 20.Schnall R., Mannhaupt, G., Stucka, R., Tauer, R., Ehnle, S., Schwarzlose, C., Vetter, I. & Feldman, H. (1994) Yeast 10, 1141-1155. [DOI] [PubMed] [Google Scholar]
- 21.Feiler H. S., Desprez, T., Santoni, V., Kronenberger, J., Caboche, M. & Traas, J. (1995) EMBO J. 14, 5626-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frohlich K. U., Fries, H. W., Rudiger, M., Erdmann, R., Botstein, D. & Mecke, D. (1991) J. Cell. Biol. 114, 443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence C. (1994) BioEssays 16, 253-258. [DOI] [PubMed] [Google Scholar]
- 24.Bork P., Hofmann, K., Bucher, P., Neuwald, A. F., Altschul, S. F. & Koonin, E. V. (1996) FASEB J. 11, 68-76. [PubMed] [Google Scholar]
- 25.Verkade H. M., Bugg, S. J., Lindsay, H. D., Carr, A. M. & O'Connell, M. J. (1999) Mol. Biol. Cell 10, 2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- 27.Saffi J., Pereira, V. R. & Henriques, J. A. P. (2000) Curr. Genet. 37, 75-78. [DOI] [PubMed] [Google Scholar]
- 28.Gangloff S., Soustell, C. & Fabre, F. (2000) Nat. Genet. 25, 192-194. [DOI] [PubMed] [Google Scholar]
- 29.Huang H., Kahana, A., Gottschling, D. E., Prakash, L. & Liebman, S. W. (1997) Mol. Cell Biol. 17, 6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Game J. C. & Kaufman, P. D. (1999) Genetics 151, 485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White-Cooper H., Leroy, D., MacQueen, A. & Fuller, M. T. (2000) Development (Cambridge, U.K.) 127,Suppl., 5463-5473. [DOI] [PubMed] [Google Scholar]
- 32.Perkins E. L., Sterling, J. F., Hashem, V. I. & Resnick, M. A. (1999) Proc. Natl. Acad. Sci. USA 96, 2204-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett C. B., Lewis, L. K., Karthikeyan, G., Lobachev, K. S., Jin, Y. H., Sterling, J. F., Snipe, J. R. & Resnick, M. A. (2001) Nat. Genet. 29, 426-434. [DOI] [PubMed] [Google Scholar]
- 34.Ito T., Tashiro, K., Muta, S., Ozawa, R., Chiba, T. N., M., Yamamoto, K., Kuhara, S. & Sakaki, Y. (2000) Proc. Natl. Acad. Sci. USA 97, 1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace D. C. & Murdock, D. G. (1999) Proc. Natl. Acad. Sci. USA 96, 1817-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leuenberger D., Bally, N. A., Schatz, G. & Koehler, C. M. (1999) EMBO J. 18, 4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paschen S. A., Rothbauer, U., Káldi, K., Bauer, M. F., Neupert, W. & Brunner, M. (2000) EMBO J. 19, 6392-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.