Abstract
Methylation, deletions, and amplifications of cancer genes constitute important mechanisms in carcinogenesis. For genome-wide analysis of these changes, we propose the use of NotI clone microarrays and genomic subtraction, because NotI recognition sites are closely associated with CpG islands and genes. We show here that the CODE (Cloning Of DEleted sequences) genomic subtraction procedure can be adapted to NotI flanking sequences and to CpG islands. Because the sequence complexity of this procedure is greatly reduced, only two cycles of subtraction are required. A NotI-CODE procedure can be used to prepare NotI representations (NRs) containing 0.1–0.5% of the total DNA. The NRs contain, on average, 10-fold less repetitive sequences than the whole human genome and can be used as probes for hybridization to NotI microarrays. These microarrays, when probed with NRs, can simultaneously detect copy number changes and methylation. NotI microarrays offer a powerful tool with which to study carcinogenesis.
Representational difference analysis (RDA) (1) and restriction fragment length polymorphism subtraction (2) were reproducibly successful in cloning deleted sequences. However, these methods are sensitive to minor impurities, are laborious, and suffer from a number of limitations (e.g., the inability to detect differences due to point mutations, small deletions, or insertions). Furthermore, the PCR amplification after the first hybridization step and before the nuclease treatment may give rise to artifacts. Excess driver DNA can result in reduced efficiency in amplification of the tester–tester duplexes because of the potential formation of residual driver–driver and driver–tester duplexes that act as competitors. As RDA is based mainly on the specific amplification of the desired products and requires 95–110 PCR cycles, it suffers from a “plateau effect” that is characterized by a decline in the exponential rate of accumulation of amplification products. However, the major problem results from the inefficiency of the multiple restriction digestion and ligation reactions that are used in this method, and which lead to the generation of false positives. Furthermore, these experiments result in the cloning of products that usually do not represent functional genes. Similarly, the methylation-sensitive representational difference analysis (3), aimed at CpG-rich sequences, suffers from the same limitations as the original RDA.
Recently, we developed a procedure for cloning deleted sequences (Cloning Of DEleted sequences, CODE) (4) that is free from some of the limitations inherent in the RDA and restriction fragment length polymorphism subtraction protocols. Our major objective was to improve the subtractive enrichment, thereby avoiding excessive PCR kinetic enrichment steps that often generate small DNA products.
It has been suggested and shown (5–8) that NotI sites are almost exclusively located in CpG islands and are closely associated with functional genes. Therefore, NotI sites can serve as very useful markers for both physical and genetic mapping. We have constructed high-density grids with 50,000 NotI clones and identified among them 22,551 unique sequences (each clone usually produced two sequences). The results of this work demonstrated again that NotI flanking sequences are a rich source from which to identify new genes. A rough estimation (based primarily on chromosomes 21 and 22) is that the human genome contains 15,000–20,000 NotI sites, of which 6,000–9,000 are unmethylated in any particular cell (9). Thus we suggested the development of a modification of CODE using only regions surrounding NotI sites.
Microarrays with immobilized P1- and BAC-cloned DNA were used for high-resolution analysis of DNA copy-number variation using comparative genome hybridization (referred to as “arrays-CGH”) (10). However, construction of these microarrays covering the whole human genome with mapped P1 and BAC clones is very expensive, laborious, and time-consuming. If small-insert NotI linking clones could fulfill the same function, this would allow the construction of microarrays for CGH analysis for use by a single research group or for the study of many organisms.
The overall goal of this work was to investigate whether it is possible to use, (i) the CODE procedure for restriction enzymes containing CG in their recognition sites; and (ii) NotI-flanking sequences for genome-wide screening for deleted, amplified, and methylated NotI sites.
Materials and Methods
Cell Lines and General Methods.
ACC-LC5, a small-cell lung carcinoma cell line, contains a homozygous 0.7-Mb deletion in 3p21.3–p22 (11). MCH903.1 (DNA A) is an MCH line that contains a single copy of human chromosome 3, derived from a normal human diploid cell line, as its only human component. This chromosome does not contain visible deletions. The MCH939.2 (DNA B) cell line originally contained a cytogenetically normal chromosome 3 derived from a normal human diploid cell line HHW1108 but now carries a deletion (3p14-p22) in the short arm of this chromosome (7). Paired normal (DNA A) and renal cell carcinoma (RCC; DNA B) tissue samples were collected immediately after resection and stored at −80°C before DNA extraction. Isolation of DNA, Southern transfer, hybridization, etc., was performed according to standard methods.
The construction of NotI linking libraries has been described previously (12). Plasmid DNA was purified by using the R.E.A.L. Prep kit (Qiagen, Chatsworth, CA). A standard protocol was used to prepare nylon filter replicas of the gridded NotI linking clones. Nylon filters contained 90 mapped chromosome-3-specific NotI linking clones (6) and five random unmapped human NotI linking clones (8). For hybridization to the nylon filters, the NotI representations (NRs; see below) probes were 32P-labeled by PCR.
Sequencing gels were run on ABI 310 Automated Sequencers (Applied Biosystems) according to the manufacturer's protocol.
The Modified NotI-CODE Procedure.
Two oligonucleotides, NotX: 5′-AAAAGAATGTCAGTGTGTCACGTATGGACGAATTCGC-3′ and NotY: 5′-AAACTTACAGTGTGTGTCACGTATGGCTGCTTAAGCGCCGG-3′, were used to create the NotI linker. Two micrograms of DNA A (tester) and DNA B (driver) at a concentration of 50 μg/ml were digested with 20 units of BamHI and 20 units of BglII (Roche Molecular Biochemicals) at 37°C for 5 h and then heat-inactivated for 20 min at 85°C.
Then 0.4 μg of the digested DNAs was circularized overnight with T4 DNA ligase (Roche Molecular Biochemicals) in the appropriate buffer in 1-ml reaction mixtures. The DNA was then concentrated with ethanol, partially filled in (12), and digested with 10 units of NotI at 37°C for 3 h.
After digestion, NotI was heat-inactivated, and the DNAs were ligated overnight in the presence of a 50 M excess of NotI linker at room temperature. All further steps were performed as described (4), but NotX primer was used for PCR amplification, and only two cycles were performed. We call these PCR-amplified tester and driver amplicons NRs.
Microarray Preparation, Hybridization, and Scanning.
Microarrays were constructed essentially as described (13). The microarrays described here contained 150 sequence-validated human chromosome-3-specific (6) sequence-tagged sites in six repetitions, representing 61 known genes and 49 unknown expressed sequence tags.
The NR probes were labeled in a PCR reaction with the NotX primer.
Arrays were scanned by using the GMS 418 Scanner (Genetic MicroSystems, Woburn, MA) and were analyzed and represented by imagene 3.02 software (Biodiscovery, Marina del Rey, CA).
Quantitative Real-Time PCR with TaqMan Probes.
Oligonucleotide primers and probes (sequences are available on request) were designed to amplify five NotI linking clones: NRL1–1 (3p21.2), NL3–001 (3p21.2–21.32), NL1–205 (3p21.2–21.32), NLJ-003 (3p21.33), and 924–021 (3p12.3). The human β-actin gene was used as a reference sequence. TaqMan probes and primers were obtained from Perkin–Elmer. Details concerning the theory and derivation of the comparative cycle threshold (CT) method (ΔΔCT method) for the quantitative assessment of target sequences has been published (ABI PRISM 7700 Sequence Detection System. User Bulletin no. 2. Relative quantitation of Gene Expression. PE Applied Biosystems, 1997).
Results and Discussion
NotI-CODE Subtraction.
The NotI-CODE technique reduces the complexity of the genome by using subtraction for only short regions surrounding NotI sites (NRs). To validate this approach, we have compared a lung carcinoma cell line, ACC-LC5, which contains a 0.7-Mb homozygously deleted region in 3p21–p22, with normal lymphocyte control DNA. It is not known whether this cell line contains homozygous deletions in other chromosomes. The “normal DNA” was not an entirely appropriate control, because it was isolated from another individual. Therefore, we may expect to clone polymorphic sequences as well as deleted regions. It is obvious from the NotI-CODE scheme that methylated NotI sites will behave as deleted NotI sites because they will not be digested. This means that this procedure will simultaneously detect genes that are either deleted or methylated.
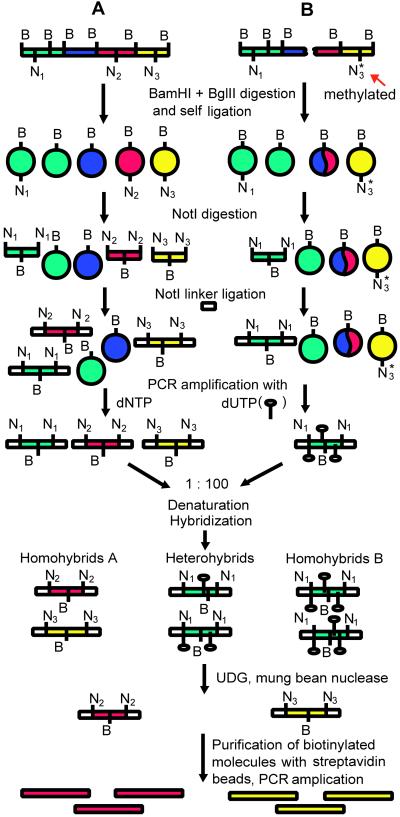
An overview of the subtractive procedure is shown in Fig. 1. Except the first steps, it is very similar to the original CODE procedure (4).
Fig 1.
A schematic outline of the NotI-CODE subtractive procedure. N and B represent NotI and BamHI/BglII sites, respectively. Methylated NotI sites are indicated by an asterisk. UDG, uracil–DNA glycosylase.
Tester and driver DNAs were digested with BamHI + BglII and self-ligated at very low concentrations of DNA to form circles. The remaining linear molecules were inactivated with Klenow fragment. Intermolecular ligation was not a problem here, because the vast majority of these ligated molecules are not PCR-amplified in the further steps. In the rare cases when these two ligated molecules contain closely located NotI sites and are PCR-amplified, they can be used to normalize the representativity of different surrounding NotI sequences. These circles were then digested with NotI. Nearly all of the circles will remain closed and will therefore be omitted from further reactions. The driver DNA, in contrast to the tester DNA, was amplified with dUTP. The products of DNA amplification (that is NRs) were denatured, hybridized, and then treated with uracil-DNA glycosylase (which destroyed all of the driver DNA) and mung bean nuclease (which digested single-stranded DNA and all of the nonperfect hybrids). The resulting tester homohybrids were concentrated with ethanol and subjected to one more round of subtraction. The final PCR product was amplified and cloned.
From previous experiments (V.K. et al., unpublished data), we know that the NLJ-003 and NL1-401 clones are deleted in this cell line. We have isolated DNA from 10 random clones and sequenced them (Southern analysis with these small inserts was impossible because of their high CG content). Two of these clones contained the NLJ-003 NotI site.
This experiment demonstrated that subtraction using NotI-surrounding sequences is quite efficient because, of the 15,000–20,000 NotI sites, only two were located in the homozygously deleted region, and one of these was found after the analysis of only 10 clones. Other clones can be either polymorphic or hemizygously deleted, because when the CODE procedure was applied to the same driver–tester pair, most informative clones (11 of 19) fell into this category (4).
Use of NR as a Probe for Hybridization.
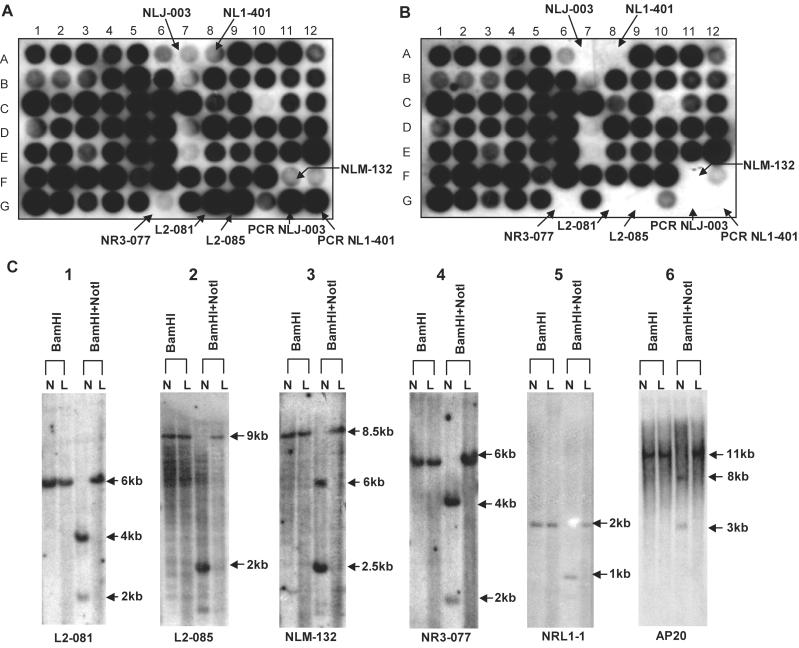
In the experiment described above, NRs were cloned after subtraction, but they may also be used as probes for hybridization to detect deleted, methylated, or polymorphic NotI sites. To this end, nylon filters with immobilized DNA from NotI linking clones were prepared. These filters were hybridized with NRs of normal lymphocyte DNA (NR-A) and ACC-LC5 (NR-B). These two NRs revealed different hybridization patterns (Fig. 2 A and B): several clones that hybridized to NR-A did not hybridize to NR-B. It is clear from this figure that homozygously deleted NLJ-003 and NL1–401 were detected easily. To understand why other clones failed to hybridize to NR-B, we selected four such clones and analyzed them using Southern hybridization (Fig. 2C, lanes 1–4). Genomic DNAs from ACC-LC5 and normal lymphocytes were digested with either BamHI or with BamHI + NotI, resolved by electrophoresis in agarose gel, transferred to nylon filter and hybridized to the 32P-labeled insert of a NotI linking clone. All four clones clearly demonstrated the presence of an unmethylated NotI recognition site in DNA from normal lymphocytes that was methylated in ACC-LC5 DNA.
Fig 2.
Comparative analysis of small cell lung cancer cell line ACC-LC5. Hybridization patterns of 32P-labeled NRs of normal lymphocyte DNA (A) and ACC-LC5 (B). (C) Southern hybridization of selected NotI clones to normal lymhocyte DNA (N) and ACC-LC5 (L) digested with different restriction enzymes.
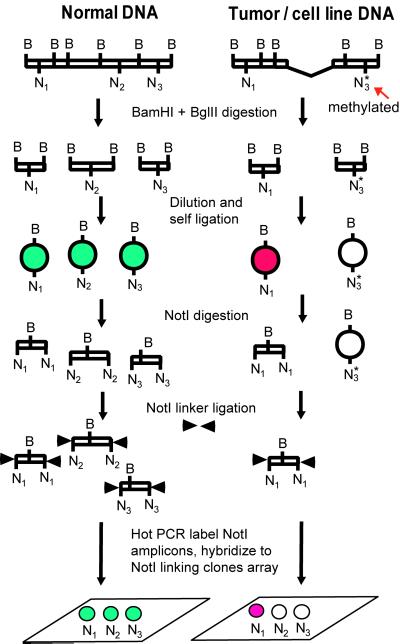
As the next step, we performed similar experiments but using microarrays of DNA from NotI linking clones immobilized to a glass slide. The main idea of this application is shown in Fig. 3. If a particular NotI site is present in the DNA, then the circle will be opened with NotI and labeled. However, if this NotI site is deleted or methylated, then the NR will not contain the corresponding DNA sequences.
Fig 3.
The main idea of using NR for NotI microarrays.
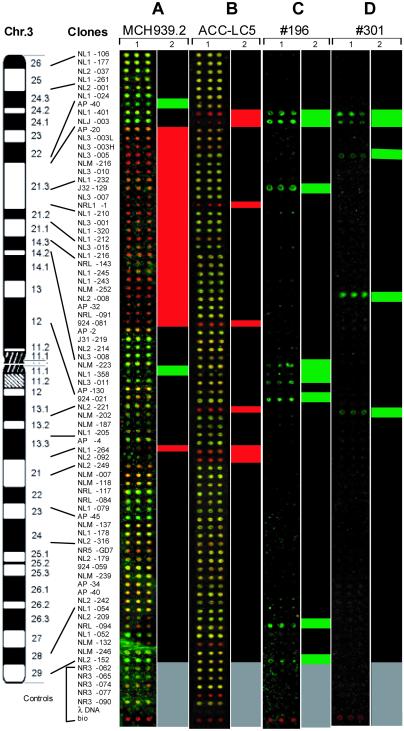
In the first experiment, we used DNA isolated from the human–mouse microcell hybrid cell lines MCH903.1 (which contains the whole of human chromosome 3) and MCH939.2 (chromosome 3 del p14–p22). The NR for MCH903.1 was labeled with Cy5 (red) and the NR for MCH939.3 was labeled with Cy3 (green). Thus, sequences deleted in MCH939.2 should be red. In one experiment, the deletion was precisely mapped (Fig. 4A).
Fig 4.
NotI microarray profiling of deletions/methylation in microcell hybrid MCH 939.2 (A), cell line ACC-LC5 (B), and primary RCC tumors nos. 196 (C) and 301 (D). Representative images of microarrays (1) are ordered according to physical map of chromosome 3. One-dimensional clustering (2) is based on average normalized green/red ratios of fluorescent data (red, R <0.3; green, R >3). For A and B, normal and tested DNA were hybridized together. NR for MCH903.1 (the whole chromosome) was labeled red, and NR for MCH939.2 (3p.14-p22 deletion) was labeled green. Similarly, NR for normal lymphocyte DNA was red, and the small cell lung cancer line ACC-LC5 was labeled green. The red clusters demonstrate a significant overrepresentation of complete chromosome 3 or normal DNA. The green clusters show under-representation of normal DNA. For C and D, one step of NotI-CODE subtraction procedure was performed, and single color hybridization was done. The green clusters demonstrate the significant overrepresentation of normal DNA. Gray color marks controls.
In a second experiment, DNA from ACC-LC5 was again compared with normal lymphocyte DNA (Fig. 4B). If both sequences were present in the NRs, then the combined color was close to yellow, whereas if some clones were deleted in ACC-LC5, then the color of these clones was more red (Fig. 4B). Again, homozygously deleted clones NLJ-003 and NL1–401 were detected unambiguously. Other clones showing a more reddish color probably reflect the fact that in almost 100% of cases of small-cell lung carcinoma, a hemizygous deletion is detected on 3p. Some clones showed the same imbalance as NLJ-003 and NL1–401. This can be explained by methylation of both alleles or deletion of one allele of a NotI site and methylation (or polymorphism) of the other. Indeed, as shown in Fig. 2C, clones NLM-132 and NR3–077 do not contain cleavable NotI sites. In two other cases (AP20 and NRL1–1) that were also completely red, the situation is different. One allele is methylated and the other is deleted (Fig. 4; Table 1; Fig. 2C, lines 5 and 6).
Table 1.
Relative quantitative measurements using the comparative (ΔΔCT) method for normal lymphocyte DNA and ACC-LC5 cell line
| Target/color | Location | NACC-LC5/Nnorm = 2−ΔΔCT | Comments |
|---|---|---|---|
| 924-021/yellow | 3p12.3 | 0.94 (0.83–1.05) | No changes, two copies |
| NRL1-1/red | 3p21.2 | 0.51 (0.41–0.62) | Initial target sequence copy number in ACC-LC5 is two times less than in control (hemizygous deletion); the NRL1-1 site is completely methylated |
| NL3-001/yellow | 3p21.2–21.32 | 1.12 (0.98–1.26) | No changes, two copies |
| NL1-205/yellow | 3p21.2–21.32 | 1.25 (0.75–1.74) | No changes, two copies |
| NLJ-003/red | 3p21.33 | 0.00 | Zero sequence copy number (homozygous deletion) |
To further check the results of this hybridization, TaqMan probes were designed for five NotI linking clones (see Materials and Methods). Quantitative real-time PCR was performed with these primers/probes using the ABI PRISM MODEL 7700 SEQUENCE DETECTOR. Before the experiment was performed with these probes, real-time PCR amplification of the X-chromosome-specific gene PF2K (14) was performed to test whether the method can distinguish between one (male) and two (female) copies per genome. The copy number of PF2K in males (five samples), normalized to a reference (β-actin) and relative to the copy number for PF2K in females (7 samples), was nM/nF = 2−ΔΔCT = 0.5. Thus, the real-time PCR technique allows the successful detection of the difference in X-chromosome copy number between males and females.
The results of quantitative PCR correspond well to the NotI microarray hybridization data (Table 1). It is important to note that the same signal can result from different events. For instance, both homozygously deleted NotI sites (e.g., NLJ-003) and hemizygously deleted and methylated sites (NRL1–1) can give completely red signals (green/red ratio <0.3).
In another control experiment, the ratio of Cy3/Cy5 signals was tested for NotI clones with known copy numbers. As shown in Table 2, the NotI microarrays were sufficiently sensitive to discriminate between normal diploid, hemizygously deleted, and homozygously deleted alleles. In these experiments, 41 arrayed NotI clones from chromosome X were used to detect the ratio between female/female (normal diploid condition) and male/female (equivalent to hemizygously deleted alleles) NRs. For chromosome-X-specific arrays, only NotI clones unmethylated on both chromosomes were used. To detect homozygous deletions, NLJ-003 and NL1–401 were used.
Table 2.
Detection of copy number changes with NotI microarrays
| Arrayed NotI clones, ratio between alleles | NR probe, Cy3/Cy5 | Signal mean, ratio Cy3/Cy5 ± SD |
|---|---|---|
| Chromosome X, specific, normal diploid | XX/XX | 1.00 ± 0.15 |
| Chromosome X, specific, hemizygous deletion | XY/XX | 0.47 ± 0.13 |
| Chromosome 3, specific (NLJ-003, NL1-401), homozygous deletion | ACC-LC5/normal lymphocyte DNA | 0.18 ± 0.13 |
Signal mean, the mean of the intensities of the pixels from all signals.
Two different male/female pairs were used for the comparison.
NotI Microarrays for Genome-Wide Scanning.
In recent years, a wide variety of approaches was offered for genome scanning using different types of microarrays (15–17). However, all of them have clear limitations important for the detection of cancer-associated genes. Thus, array-CGH cannot detect loss of heterozygosity or methylation changes. CpG island microarrays are not suited to study copy number changes; unlike the NotI microarrays, any incomplete digestion will produce an artifactual positive signal; the whole human genome DNA was used for labeling, etc.
The fundamental problems of genome-wide screening using NotI clones are, (i) the size and complexity of the human genome, (ii) the number of repeat sequences, and (iii) the comparatively small sizes of the inserts in NotI clones (on average 6–8 kb). To address these problems, a special procedure was developed to amplify only regions surrounding NotI sites. Other DNA fragments were not amplified. Therefore, only 0.1–0.5% of the total DNA is labeled. Interestingly, sequences surrounding NotI sites contain 10-fold fewer repetitive sequences than the human genome on average (9), and therefore these microarrays are not as sensitive as other methods to the background hybridization caused by repeats. Ribosomal rRNA genes were virtually absent from these NotI flanking sequences.
The NRs can be efficiently used for genomic subtraction, and any enzyme can be used in this procedure for preparing restriction enzyme representations (RRs). By selecting two to three restriction enzymes cutting mainly in CpG islands, this procedure will result in differential cloning of almost all CpG-island-containing DNA fragments. The same RRs can be used for genome screening corresponding microarrays.
Contamination of tumor DNA with normal DNA represents a serious problem for the identification of tumor suppressor genes. Two RCC biopsies containing 30–40% contaminating normal cells were used in a control experiment to check the sensitivity of NotI microarrays to contamination. One step of the NotI-CODE procedure was used before hybridization, and the probe was labeled with only one dye. As shown in Fig. 4, the hybridization clearly identified the two regions most frequently deleted in RCC, 3p21 telomeric (near NLJ-003) and 3p21 centromeric (near NRL1–1). Therefore, the impurity problem that can occur with tumor biopsies can be easily resolved with NotI microarrays. In different types of tumors, aberrant methylation of CpG islands in the promoter region has been observed in many cancer-related genes, resulting in the silencing of their expression. Therefore, by comparing normal and tumor samples, NotI microarrays potentially allow the simultaneous study of genetic and epigenetic factors affecting the same gene.
As a result of our increasing understanding of the role of DNA methylation in carcinogenesis, several new methodologies have been developed to facilitate genome-wide searches for changes in DNA methylation (18–22). Although each of these has its own advantages, none is suited to large-scale screening because all methods are rather inefficient and complicated and can be used for testing only limited number of samples simultaneously (23). For instance, restriction landmark genomic scanning method indeed allows analyzing thousands of NotI sites per experiment (19); however, this approach is rather technically challenging.
The creation and use of microarrays at the genomic level will be very important because it may provide information unavailable at the level of mRNA/cDNA (e.g., methylation or silencing of specific alleles, hemizygous deletions, epigenetic factors, genetic predisposition, work with old samples, etc.). Methylation changes are frequently the earliest events in tumor development and can be detected 1 year before tumor formation is detectable by any other method (18). Furthermore, these microarrays are more sensitive than cDNA microarrays in several ways, because genomic markers are normalized naturally and differences in their copy numbers cannot reach 104 times that is valid for the expression of many genes. Many genes are expressed at levels below 50–100 copies per cell and probably cannot be properly analyzed with cDNA microarrays at all. NotI microarrays have another strong advantage compared with cDNA microarrays, insofar as there is no standard for comparing expression profiles (24). RNA is not a stable molecule; physiological conditions can differ between cancer cells and normal cells, and these conditions can vary very significantly over short time periods, depending on many different factors such as temperature, time of day, and therapeutic regimen. With expression microarrays, it is sometimes difficult to identify the first events and first genetic lesions that lead to the development of cancer.
This is less of a problem with NotI microarrays because genetic lesions (deletions, for example) are irreversible, epigenetic changes (e.g., methylation) are not markedly transient, and normal genomic DNA is a perfect standard for comparison. It is important to mention that NotI microarrays were not designed to replace expression microarrays; on the contrary, they should be used as a complementary approach that can yield additional information.
Using RST microarrays, it is possible to discriminate between deleted and methylated sequences. To achieve this aim, NR should be produced by using DNA that is unmethylated [this can be done by different approaches: limited PCR amplification after the first digestion with restriction enzyme(s), enzymatic demethylation, etc.].
A pilot experiment using NR probes demonstrated the power of the method. We think that NotI microarrays offer a powerful tool with which to study carcinogenesis.
Note Added in Proof.
Chromosome 3-specific NotI microarrays are available now from BD Bioscience CLONTECH.
Acknowledgments
This work was supported by research grants from the Swedish Cancer Society, the Swedish Research Council for Engineering Sciences, the Swedish Foundation for International Cooperation in Research and Higher Education, Ingabritt och Arne Lundbergs Forskningsstiftelse, Pharmacia Corporation, Karolinska Institute, and the Swedish Research Council. It was also partly supported by the Russian National Human Genome Program.
Abbreviations
NR, NotI representation
CODE, cloning of deleted sequences
RCC, renal cell carcinoma
RDA, representational difference analysis
References
- 1.Lisitsyn N., Lisitsyn, N. & Wigler, M. (1993) Science 259, 946-951. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg M., Przybylska, M. & Straus, D. (1994) Proc. Natl. Acad. Sci. USA 91, 6113-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ushijima T., Morimura, K., Hosoya, Y., Okonogi, H., Tatematsu, M., Sugimura, T. & Nagao, M. (1997) Proc. Natl. Acad. Sci. USA 94, 2284-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J., Wang, F., Kashuba, V., Wahlestedt, C. & Zabarovsky, E. R. (2001) Biotechniques 31, 788-793. [DOI] [PubMed] [Google Scholar]
- 5.Bird A. P. (1987) Trends Genet. 3, 342-347. [Google Scholar]
- 6.Kashuba V. I., Gizatullin, R. Z., Protopopov, A. I., Li, J., Vorobieva, N. V., Fedorova, L., Zabarovska, V. I., Muravenko, O. V., Kost-Alimova, M., et al. (1999) Gene 239, 259-271. [DOI] [PubMed] [Google Scholar]
- 7.Zabarovsky E. R., Kashuba, V. I., Kholodnyuk, I. D., Zabarovska, V. I., Stanbridge, E. J. & Klein, G. (1994) Genomics 21, 486-489. [DOI] [PubMed] [Google Scholar]
- 8.Zabarovsky E. R., Gizatullin, R., Podowski, R. M., Zabarovska, V. V., Xie, L., Muravenko, O. V., Kozyrev, S., Petrenko, L., Skobeleva, N., Li, J., et al. (2000) Nucleic Acids Res. 28, 1635-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kutsenko, A. S., Gizatullin, R. Z., Al-Amin, A. N., Wang, F., Kvasha, S. M., Podowski, R. M., Matushkin, Y. G., Gyanchandani, A., Muravenko, O. V., Levitsky, V. G., et. al. (2002) Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 10.Pinkel D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W. L., Chen, C., et al. (1998) Nat. Genet. 20, 207-211. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa S., Kai, M., Tamari, M., Takei, Y., Takeuchi, K., Bandou, H., Yamane, Y., Ogawa, M. & Nakamura, Y. (1997) DNA Res. 4, 35-43. [DOI] [PubMed] [Google Scholar]
- 12.Zabarovsky E. R., Boldog, F., Thompson, T., Scanlon, D., Winberg, G., Marcsek, Z., Erlandsson, R., Stanbridge, E. J., Klein, G. & Sumegi, J. (1990) Nucleic Acids Res. 18, 6319-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diehl F., Grahlmann, S., Beier, M. & Hoheisel, J. D. (2001) Nucleic Acids Res. 29, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boulay J. L., Reuter, J., Ritschard, R., Terracciano, L., Herrmann, R. & Rochlitz, C. (1999) Biotechniques 27, 228-230., 232. [DOI] [PubMed] [Google Scholar]
- 15.Lindblad-Toh K., Tanenbaum, D. M., Daly, M. J., Winchester, E., Lui, W. O., Villapakkam, A., Stanton, S. E., Larsson, C., Hudson, T. J., Johnson, B. E., et al. (2000) Nat. Biotechnol. 18, 1001-1005. [DOI] [PubMed] [Google Scholar]
- 16.Lucito R., West, J., Reiner, A., Alexander, J., Esposito, D., Mishra, B., Powers, S., Norton, L. & Wigler, M. (2000) Genome Res. 10, 1726-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan P. S., Chen, C. M., Shi, H., Rahmatpanah, F., Wei, S. H., Caldwell, C. W. & Huang, T. H. (2001) Cancer Res. 61, 8375-8380. [PubMed] [Google Scholar]
- 18.Palmisano W. A., Divine, K. K., Saccomanno, G., Gilliland, F. D., Baylin, S. B., Herman, J. G. & Belinsky, S. A. (2000) Cancer Res. 60, 5954-5958. [PubMed] [Google Scholar]
- 19.Costello J. F., Fruhwald, M. C., Smiraglia, D. J., Rush, L. J., Robertson, G. P., Gao, X., Wright, F. A., Feramisco, J. D., Peltomaki, P., Lang, J. C., et al. (2000) Nat. Genet. 24, 132-138. [DOI] [PubMed] [Google Scholar]
- 20.Rountree M. R., Bachman, K. E., Herman, J. G. & Baylin, S. B. (2001) Oncogene 20, 3156-3165. [DOI] [PubMed] [Google Scholar]
- 21.Adorjan P., Distler, J., Lipscher, E., Model, F., Muller, J., Pelet, C., Braun, A., Florl, A. R., Gutig, D., Grabs, G., et al. (2002) Nucleic Acids Res. 30, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galm O., Rountree, M. R., Bachman, K. E., Jair, K. W., Baylin, S. B. & Herman, J. G. (2002) Genome Res. 12, 153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugimura T. & Ushijima, T. (2000) Mutat. Res. 462, 235-246. [DOI] [PubMed] [Google Scholar]
- 24.Brazma A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C. A., Causton, H. C., et al. (2001) Nat. Genet. 29, 365-371. [DOI] [PubMed] [Google Scholar]