Abstract
In a transgenic rice line, a β-glucuronidase reporter gene under the control of the rice tungro bacilliform virus promoter became gradually methylated, and gene activity was lost concomitantly. Methylation was observed only in the homozygous offspring and was initially restricted to the promoter region and accompanied by loss of expression in the vascular bundle tissue only. This expression pattern was similar to that of a promoter with a deletion of a vascular bundle expression element. The gene activity could be reestablished by treatment with 5-azacytidine. Methylation per se did not inhibit the binding to the promoter region of protein factors which also bound to the unmethylated sequence. Instead, promoter methylation enabled the alternative binding of a protein with specificity for sequence and methylation. In further generations of homozygous offspring the methylation spread into the transcribed region and gene activity was completely repressed also in nonvascular cells. The results indicate that different stages are involved in DNA methylation-correlated gene inactivation, and that at least one of them may involve the attraction of a sequence and methylation-specific DNA-binding protein.
Activity of a gene in a chromosomal context is determined by the interaction of general and specific transcription factors with a gene-specific set of cis-acting sequences in the context of chromatin structure. Chromatin structure can inactivate genes or even chromosomes despite the presence of activating transcription factors. Silent chromatin is often associated with the presence of deacetylated and specifically methylated histones (1, 2) and with cytosine-methylated DNA (3). The silent state can be developmentally controlled or completely stable and even inheritable (4, 5). Alterations of chromatin structure require a complex chromatin-remodeling machinery (1, 6). It is still unclear how many mechanisms exist to single-out specific genes or chromosome regions for packaging into repressive chromatin and what contribution the individual modifications of chromatin have on the regulation of gene expression.
Transcriptional gene silencing in plants is often associated with DNA methylation (7, 8) and also includes chromatin-remodeling steps (6, 9). It can be induced by the presence of double-stranded (ds) RNA, which covers transcription control regions (10–14). Alternatively, DNA–DNA interactions between homologous regions may be involved (15–18). Methylation patterns can be exchanged between homologous DNAs in a recombination-like process (19). DNA regions present as dsRNA become specifically and densely methylated at all cytosines (10, 20, 21) and repeat induced methylation usually also coincides with the repeated regions (22). Whether DNA methylation is the first step in the process or a later stabilization of gene silencing is still unclear. It has been shown that DNA methylation can induce histone modifications, but the inverse relation has also been observed (2, 3, 23, 24). Transcriptional silencing in plants is often released after treatment with methylation inhibitors (8) or in mutants with defects in specific DNA methylases (25–27), but reactivation in the absence of notable demethylation has also been described (28). In contrast, interference with histone deacetylation had strong developmental effects in plants (29) but only few cases of reactivation of a silenced gene have been reported (30, 31).
A difficulty of the analysis of intermediate steps in transcriptional gene silencing lies in its unpredictable occurrence and/or in the speed of its establishment after introduction of a locus with known transsilencing activity.
We describe here a case of silencing in rice, which reproducibly began with the inactivation of a tissue-specificity element in the promoter of rice tungro bacilliform virus (RTBV) in homozygous, transgenic plants. It was correlated with increased methylation of the respective promoter region and binding of a protein to this methylated region. Tissue specificity of silencing and the concomitant DNA methylation pattern was stable in one plant line, whereas in a twin ling line, which was serendipitously regenerated from the same transformation event, methylation spread into the transcribed region and silencing became more complete.
Experimental Procedures
Rice Transformation.
The generation of the transgenic rice lines (cv. Taipei 309) by particle bombardment and the DNA constructs have been described (32).
Cultivation of Seedlings on 5-Azacytidine (5-AC).
Rice seeds were dehusked and surface-sterilized by rinsing for 1 min in 70% ethanol and subsequent incubation in 6% Ca(ClO)2/0.01% Triton X-100 for 1 h, followed by three washes in sterile water. The seeds were germinated under light at 25°C on MS medium (33) supplemented with 30 g/liter sucrose and with or without 30 μM 5-AC (Fluka) and solidified with 4 g/liter Gelrite. Expression of the β-glucuronidase (GUS) gene from the seedlings was tested 6–7 days after germination.
GUS Assays and RNA Analysis.
Histochemical and fluorimetrical GUS assays and RNA analysis by RNase A/T1 protection was performed as described (32).
DNA Gel Blot Analysis.
DNA was extracted from rice leaves by using the Nucleon DNA extraction kit (Amersham Pharmacia) and was digested with the appropriate restriction enzymes. DNA fragments were separated in 1–1.25% agarose gels, transferred to nylon membranes (Amersham Pharmacia) and cross-linked with 0.12 MJ by a UV Stratalinker 1800 (Stratagene). PCR-amplified, digoxigenin-labeled (Roche Molecular Biochemicals) probes from the coding region of GUS (300 bp), the coding region of aph4 (240 bp), or the RTBV-dps (280 bp) were used for hybridization. Hybridization, washing, and detection were performed according to the supplier's instructions (Roche Molecular Biochemicals).
5-Methylcytosine Mapping.
The bisulfite reaction was basically performed as described (34). In brief, 3 μg of genomic DNA in 30 μl of water were denatured by the addition of 3 μl of 2 M NaOH and incubation at room temperature for 10 min. Added to the DNA was 270 μl of 5 M bisulfite solution (2.5 M sodium metabisulfite (Merck)/125 mM hydroquinone); the tube was quickly vortexed and incubated at 50°C for 4 h. The DNA was recovered with 10 μl of QIAEX II (Qiagen, Chatsworth, CA) solution and was eluted in 100 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). The DNA was desulfonated by the addition of 3 μl of 10 M NaOH, mixing and incubation at 37°C for 15 min, and was precipitated by the addition of 150 μl of 5 M ammonium acetate (pH 7.0) and 500 μl of ethanol and incubation for 30 min at −20°C. The DNA was pelleted by centrifugation for 15 min at 4°C, washed with 70% ethanol, and dissolved in 20 μl of TE buffer. The PCR mixture contained 500 pg of each degenerated forward primer (5′-TGCTCTAGACAATCATAA(G/A)TACAAACATATTACAC-3′ or 5′-AGGGTGTAGGATAAATATAAGAG-3′) and backward primers (5′-CCGCTCGAGA(C/T)AT(C/T)AATAGAAGATGAATGG-3′ or 5′-ACACAAT(G/A)TCCT(G/A)CACCACATC-3′), 0.2 mM dNTP, 5 μl of 10× reaction buffer (Qiagen), 10 μl of Q-solution (Qiagen), 2.5 μl of modified DNA, and 2.5 units of Taq-polymerase and H2O were mixed to a final volume of 50 μl. The PCR reaction was performed by denaturing at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 2 min. After 40 cycles the PCR products were examined on a 1% agarose gel, purified on PCR-product purification columns (Roche Molecular Biochemicals), and sequenced by using the forward primer described above. Control reactions were performed by mixing purified plasmid DNA with DNA from untransformed rice plants to monitor complete conversion of unmethylated cytosines under our assay conditions.
Synthetic Oligonucleotides.
Methylated oligonucleotides were synthesized by Microsynth (Balgach, Switzerland). All synthetic oligonucleotides were purified on a 15% denaturing polyacrylamide gel. For generating double-stranded oligonucleotides used for labeled probes and unlabeled competitors, equimolar amounts of the complementary strands were denatured in the presence of 50 mM NaCl at 90°C for 10 min and slowly cooled to room temperature.
Electrophoretic Mobility Shift Assay (EMSA).
Nuclear extracts from cell suspensions of Oryza sativa line Oc were prepared as described (35). Methylated or nonmethylated double-stranded oligonucleotides corresponding to sequences from −169 or −165 to −100, with a 5′-protruding end on the antisense strand, were labeled with [α-32P]dCTP with the Klenow fragment of DNA polymerase. The labeled probe was purified on a 5% native polyacrylamide gel. Typical mixtures (15 μl) for in vitro binding reactions contained 5 μg of poly(dI-dC), 10 μg of nuclear extracts, and 15,000 cpm of labeled DNA probe (around 0.04 pM DNA) in 10 mM Hepes (pH 7.6)/8 mM MgCl2/1 mM DTT/4 mM spermidine/5% (vol/vol) glycerol. Variable amounts of competitors were included. The reaction mixture was preincubated for 10 min at room temperature before the addition of the labeled DNA probe, and incubated for a further 20 min at room temperature after the addition of the labeled DNA probe and the competitors.
Results
An Identical Expression Pattern Can Be Caused by Epigenetic Promoter Modification and by Deletion of a Promoter Specificity Element.
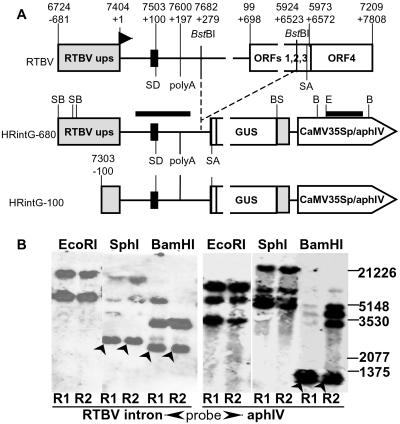
A series of transgenic rice plants had been produced by particle bombardment to analyze the RTBV promoter (Fig. 1A) including its upstream and downstream enhancer elements (32). About two thirds of the lines expressed the GUS transgene in T0 generations and the selfed offspring normally—i.e., mainly in vascular tissue and epidermis of young leaves (HRintG-680; ref. 32). However, the remaining lines showed abnormal or extremely low transgene expression. Two of the abnormal lines (R1, R2) are described here. Southern blot analyses revealed a largely identical integration pattern (Fig. 1B), indicating that the lines were derived from the same primary transformation event and contain two to four cosegregating transgene copies. The quantitative differences with the methylation-sensitive enzyme BamHI suggest differences in initial methylation status. Both lines expressed the reporter gene in the T0 generation normally (Fig. 2A). About one quarter of the T1 offspring showed no GUS expression because of transgene loss by segregation, and one half showed the same phenotype as the parent plant (Fig. 2A). The remaining quarter expressed very little GUS or none at all in the vascular tissue, while maintaining expression in the epidermis (Fig. 2B). These plants turned out to be homozygous for the transgene, whereas the normally expressing plants were hemizygous.
Fig 1.
Schematic map of the RTBV promoter constructs used for the generation of transgenic rice plants and Southern analyses of obtained lines. (A) The structure of the RTBV genome is shown in a linearized version on top, with the RTBV upstream promoter sequences (RTBV ups) as a gray box, the RNA leader sequence as a thin line, and the other ORFs as boxes. The positions of the splice donor (SD) and splice acceptor (SA) (77), the first short ORF in the leader (black box), and the RTBV polyadenylation signal (polyA) are marked. Sequence coordinates and positions with respect to the transcription start site (marked by a bent arrow at +1) are indicated, and the BstBI restriction sites used to generate the recombined intron (32) are shown. The GUS expression constructs mentioned in the text are shown below. GUS is fused to the RTBV ORF4, and translation of the spliced mRNA starts with the short ORF in the leader. In HRintG-680, the position of SphI (S), BamHI (B), and EcoRI (E) and the positions of probes (black bars) used for Southern analyses are shown. (B) Southern blot analysis of hemizygous R1 and R2 plants. Bands corresponding to predictable fragments are marked by arrowheads.
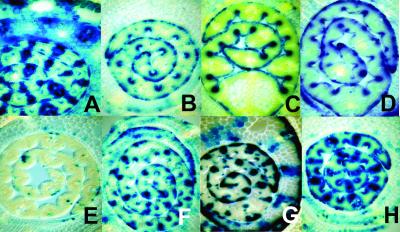
Fig 2.
Histological GUS staining of cross-sections of rice plantlets. The older leaf sheaths enclose the blade of the youngest leaf. (A) Line R1, hemizygous (T2 generation): normal GUS expression of HRintG in vascular bundles and epidermal cells of the leaf blade and in vascular bundles and parenchyma cells of the leaf sheaths. (B) Line R1, homozygous (T2): silenced expression in the vascular bundles, expression exclusively in epidermal cells. (C) Line HRintG-100 containing a GUS gene under control of a deletion variant of the RTBV promoter, which is only active in the epidermis. (D) Line R2, homozygous (T3): expression exclusively in the epidermal cells. (E) Line R1, homozygous (T3): almost completely silenced expression. (F) F1 generation plant of a sexual cross between homozygous R2 and a nontransgenic plant. (G) F2 generation plant (hemizygous) of the sexual cross in F. (H) Line R1, homozygous (T2): restored expression in the vascular bundles after germination on 30 μM 5-AC.
GUS expression only in epidermal cells in the first generation of homozygous R1 and R2 plants resembled the pattern (Fig. 2C) obtained in hemizygous and homozygous plants harboring a similar transgene (HRintG-100; Fig. 1A) with a deletion of the enhancer region of the RTBV promoter upstream of position −100 (32). This resemblance suggests that in R1 and R2 the same vascular tissue-specific enhancer region is affected by an epigenetic effect mimicking the deletion mutation.
Inheritance of the Transgene Activity Status.
Independent of the generation number of the hemizygous, normally expressing parents, both R1 and R2 showed the reproducible loss of gene activity in the vascular cells in the first homozygous generation. This expression pattern was stable in further homozygous generations of the R2 line (Fig. 2D); however, selfing of abnormally expressing, homozygous R1 plants led to a further reduction of GUS expression affecting also the epidermal tissue in the next generation (Fig. 2E). The reproducibility of the development of the transgene expression patterns in the two lines suggests that they are mediated by a stable trigger set early after the primary transformation event during the regeneration of the plants.
The abnormal expression pattern was maintained only in homozygous plants. Backcrossing of homozygous R2 plants of the T3 generation with nontransformed rice resulted in F1 plants with occasional reappearance of expression in the vascular tissue (Fig. 2F) and further normalization of the expression pattern in the F2 generation (Fig. 2G).
Correlation with C-Methylation.
When the transgenic rice seeds were germinated in the presence of the methylation inhibitor 5-AC, GUS activity of the seedlings was partially restored, whereas the expression of nonsilenced lines was little affected (Fig. 2H; see Fig. 6C).
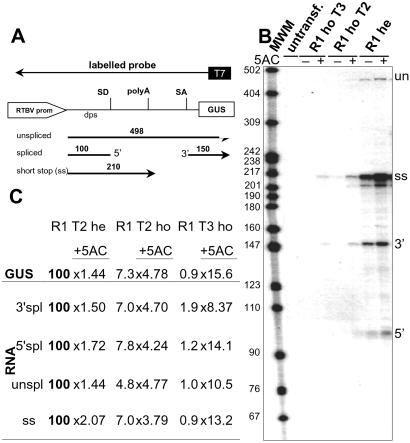
Fig 6.
Analysis of transgene-derived RNAs by RNase protection. (A) Schematic presentation of the expected protection pattern. dps, downstream promoter sequence; SD, splice donor site; SA, splice acceptor site; 5′, protected 5′ exon of the spliced RNA; 3′, protected 3′ exon of the spliced RNA. The region covered by the antisense probe used for the assay in B is shown schematically. Length of expected fragments is indicated. (B) RNase protection assay with total RNA isolated from the leaves of freshly germinated seedlings of untransformed and R1 rice plants; fragments are designated as in A. 5-AC, seedlings grown in the absence (−) or presence (+) of 30 μM 5-AC; MWM, molecular weight marker. (C) Quantification of RNA fragments from B and correlation to the respective GUS activity of the plants. Values are relative to the normally expressing hemizygous line R1 (= 100), and the stimulation factor for incubation with 30 μM 5-AC is given.
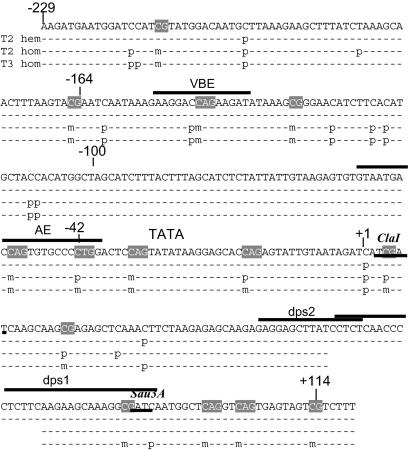
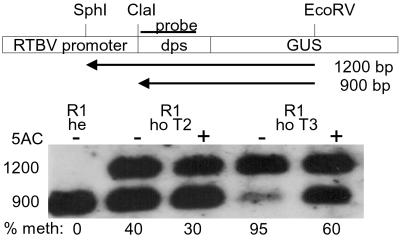
5′-Methylcytosine mapping by sequencing after sodium bisulfite treatment (34) was used on hemizygous and homozygous plants of T2 and T3 generation R1 plants. Whereas only a few cytosines were methylated in the hemizygous plants, partially and fully methylated sites were detected in the homozygous lines. Six sites were fully methylated in the homozygous plants (Fig. 3)—i.e., CG motifs at −165 and −130 and CAG motifs at −212, −146, −52, and −34. Several additional sites were fully methylated and new ones became partially methylated in the next homozygous generation. The site at position +5, which was partially methylated in the T2 generation and fully methylated in the T3 generation, coincides with a methylation-sensitive ClaI restriction site (Fig. 4). By restriction analysis the ClaI site was shown to remain unmethylated in the hemizygote, partially methylated in the T2 homozygote, and nearly fully methylated in the T3 homozygote. Methylation of the ClaI site was partially alleviated by 5-AC treatment (Fig. 4). The results of the analyses by ClaI restriction and by direct sequencing of pools of PCR fragments were compatible, indicating that the sequenced fragment pool was representative for the plant DNA.
Fig 3.
5-Methylcytosine mapping of RTBV [dps] promoter from position −229 to +114 in line R1. Completely (m) or partially (p) methylated cytosines are indicated. hem, hemizygous; hom, homozygous. The gap around +70 indicates a region for which no clear sequencing data could be obtained. Sites that were also analyzed by restriction analysis and promoter elements that bind proteins under our experimental conditions are indicated [VBE (40); AE, activator element (41); dps1 and 2, downstream promoter sequence (35, 61)].
Fig 4.
Methylation analysis of the ClaI site at the 5′ end of the transcribed region. A schematic presentation of the fragments obtained by ClaI, EcoRV, and SphI digestion with methylated and nonmethylated ClaI sites is shown (dps, downstream promoter sequences) together with the resulting Southern blot. 5-AC, seedlings germinated in the presence (+) or absence (−) of 5-AC; he, hemizygous; ho, homozygous; % meth, relative amounts of methylated DNA.
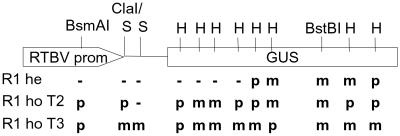
Restriction analysis with several different enzymes revealed methylation in the 3′ region of the GUS gene already in the hemizygous parent plant (Fig. 5). The methylated region was extended toward the 5′ end of the GUS gene in plants with reduced transgene activity. It is unclear whether the extension of the methylated region contributes to the loss of gene activity; however, the initial methylation in the 3′ end had apparently no influence on expression.
Fig 5.
Summary of restriction analyses of normally expressing and silenced lines. he, hemizygous; ho, homozygous; S, Sau3AI; H, HpaII; m, methylated; -, not methylated; p, partially methylated.
Analysis of Transgene-Derived RNAs.
Because of the presence of a transcription termination/polyadenylation signal closely downstream of the RTBV promoter (Fig. 1A), the transgene normally produced three different RNAs: the spliced mRNA, its unspliced precursor, and a short-stop RNA (Fig. 6; refs. 32 and 36). In plants with reduced transgene expression, the levels of all three RNA species were reduced similarly and mRNA levels correlated very well with the observed GUS activity (Fig. 6 B and C). Treatment with 5-AC restored the levels of all three RNA species to the same degree as GUS expression (Fig. 6C), but never to the level of the hemizygous T1 plants. Because it has been reported that the presence of double-stranded RNA can lead to the methylation of the corresponding DNA sequence (10–14, 20, 21), we used a probe that would allow detection of sense transcripts from the RTBV promoter region in the very sensitive RNase A/T1 protection assay. Such transcripts were indeed found in many other lines with abnormal expression (results not shown), but not in the line described here (Fig. 6B). In the RNase protection assays, we usually obtained also a signal corresponding to fragments of 15–25 nt in length (not shown). This signal was not related to any abnormality of expression and therefore unlikely represents short aberrant RNAs as they are described for some cases of gene silencing (37).
RTBV-Promoter Methylation Alters the Protein-Binding Pattern.
DNA methylation can directly interfere with the binding of some transcription factors (38, 39) or can lead to binding of proteins to a DNA sequence (2, 3). Some RTBV promoter regions affected by methylation in the homozygous plants overlap with previously identified protein-binding sites (Fig. 3; ref. 40). An activator element located between positions −35 and −70 was identified as a putative core-promoter element that binds at least two proteins (44). A vascular bundle-expression element (VBE) was located between positions −100 and −169 (32, 42). Nuclear proteins binding to the VBE region were named “vascular bundle expression element-binding proteins.” Protein binding was also reported for a region at about position −90 and for the region around the transcription start site (43), but this binding was not detectable with our assays (not shown).
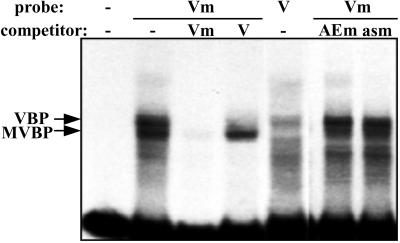
We synthesized DNA fragments of the activator element and the VBE region as C-methylated and unmethylated versions and compared protein-binding patterns by an EMSA. No alteration of the binding pattern was observed when methylated and nonmethylated DNA fragments covering the −35 to −70 region were compared (not shown). Methylation of the −100 to −169 VBE region, which includes two of the CpG and one of the CpApG motifs shown to be fully methylated in the T1 homozygous generation of our transgenic plants (Fig. 3), did not interfere with vascular bundle expression element-binding protein binding. However, a new methylation-specific gel shift was observed (Fig. 7), and the corresponding protein was named “methylated vascular bundle element-binding protein” (MVBP). MVBP binding did not lead to a supershift, showing that the two types of protein bind independently to the methylated substrate. The binding of MVBP was neither competed by the unmethylated fragment (Fig. 7) nor by other DNA fragments containing methylated CpG and CpNpG motifs, as shown here for a fragment containing two as1 elements from the CaMV 35S promoter and for a fragment covering the region from −35 to −70 of the RTBV promoter (Fig. 7). It therefore seems unlikely that MVBP would be simply a nonspecific methylC-binding protein like proteins MeCP1 and 2 and MBD1 and 2, which bind to DNA with either single or multiple CpG-methylated DNA and are involved in gene repression (2, 3). Our further analysis confirmed that MVBP is a sequence and methylation-specific DNA-binding protein detectable also in extracts from rice plants (40).
Fig 7.
Binding of nuclear proteins to methylated and unmethylated vascular bundle-specific enhancer region. DNA fragments covering the region −169 to −100 of the RTBV promoter (TAAGTACGAATCAATAAAGAAGAAGGACCAGAAGATATAAAGCGGGAACATCTTCACATGCTACCACATGGCT) were used either as is or methylated at their two CG and one CNG sites in EMSAs. Competitions were performed with 500-fold excess of unlabeled DNA fragments. V, VBE; Vm, methylated VBE; AEm, methylated sequence around the RTBV activator element GTAAGAGTGTGTAATGACCAGTGTGCCCCTGGACTC; asm, methylated sequence around the CaMV as1 element (CCACTGACGTAAGGGATGACGCACAATC). VBP, vascular bundle element-binding protein.
Discussion
Transcriptional transgene silencing in plants usually leads to a stable shut-down of gene activity throughout the plant. Here we show that it can also affect the specificity of a promoter. An epigenetic alteration of the RTBV promoter in transgenic rice plants mimicked the deletion of a known specificity element, which is required for promoter activity in the vascular bundle (32). Loss of expression was correlated with methylation of the respective promoter region, mainly at CpG and CpNpG sites, and inhibition of methylation restored the original expression pattern, suggesting methylation as a cause for promoter inactivation in the vascular tissue. Direct interference of a methyl group with protein binding has been described for several transcription factors in animals (38, 44–51) and plants (39, 52, 53). Interference can be absolute or depend on the degree of methylation or the context of a binding site (45, 46, 50, 54, 55), and it can result in methylation-dependent silencing (56). However, in gel-shift assays we found that all proteins binding to the nonmethylated VB) and probably at least in part necessary for promoter activity (32, 40–43) also can bind to the methylated VBE. In addition, a different binding activity was observed. The protein (MVBP) displayed specificity for the VBE region and the degree of its methylation; therefore, it is different from the variety of unspecific methylC-binding proteins known to direct transcriptional repressors or corepressors to methylated DNA in other eukaryotes (3) or to mask binding sites for other factors as has been shown for a vitellogenin (56) and a retinoblastoma promoter (48).
Proteins with sequence and methylation specificity have been detected in mammalian cells, where proteins of the MDBP/RFX family are involved in the regulation of several mammalian genes (57), Kaiso is a methylation-dependent transcriptional repressor (58), and a protein with specificity for CmC(A/T)GG is most likely involved in gene silencing in mature B cell lymphomas (59). For plants, the DNA binding of the Ac transposase at some of its binding sites was enhanced by hemimethylation (60).
Methylation of other RTBV promoter regions, including the activator element upstream of the TATA box (41, 43), had no detectable effect on protein binding. The specific loss of promoter activity in the vascular bundle is most likely due to a specific interference with VBE binding, because any effect on chromatin structure should lead to a more general repression. The remaining activity in the epidermal cells was as strong as in HRintG-100 plants. The lack of EMSA supershifts indicated that binding of MVBP and other VBE-binding proteins is mutually exclusive. MVBP could therefore interfere with the binding of the other proteins, but a general repressive effect, which is not active in epidermal cells, is also possible.
In the lines described here, silencing occurred reproducibly in the first homozygous generation. The transgene locus contains at least one intact copy of the transgene but also some rearranged copies, and is expressed well in hemizygous plants despite the presence of some methylation in the 3′ region of the GUS transgene. Additional methylation in vascular bundle-silenced plants was restricted to RTBV promoter regions but was apparently uniform in all present copies. The trigger must either involve a threshold-dependent factor (57) or allelic interactions (16–18). In our RNase protection assays, no evidence could be obtained for the presence of a promoter-covering RNA, which could, as a double-stranded RNA molecule, be involved in RNA-triggered DNA methylation (11). Also the methylation at mainly CpG and CpNpG sites is different from that induced by dsRNA, where usually a high proportion of all of the other cytosines also become methylated (10, 20, 21). However, at present, this mechanism cannot be excluded.
The methylated VBE region comprises CpNpG and CpG target sites for chromomethylases and Met-1, respectively (25, 27, 68, 69), and methylation of both is required to affect protein binding (40). De novo methylation of these sites could be achieved by simultaneous targeting of different methylases or by a novel methylase. Alternatively, region-specific histone modifications could be the first step or be triggered by an initial DNA methylation and then induce (further) DNA methylation (2, 23, 24). Vascular expression normalized within two generations after crossing with untransformed plants, suggesting that maintenance methylation is not completely efficient. Similar observations have been made with other silenced or paramutated genes after removal of the original inactivating locus (70, 71). In other cases, methylation remained stable or reversions were very rare (31, 72, 73).
In homozygous plants of line R2, the initially established methylation and expression pattern was stable in further generations, whereas in R1 an extension of the methylated region and a complete shut-down of promoter activity in epidermal cells was also observed. This repression could be caused either by another direct interference with factor binding at the initiator (43) or further downstream regions (61), by interference with early transcription elongation, or by chromatin remodeling (62–66). The transgene integration patterns indicated that lines R1 and R2 were twins, regenerated from the same primary transformed cell. Apparently, processes early during or after transformation can mark transgene sequences for a certain epigenetic fate. Similar observations have been reported for cre/lox-mediated integration of transgenes into defined sites in tobacco (31). Recombination and rearrangements within the many DNA molecules introduced into cells by all methods of direct gene transfer could certainly produce structures that could trigger silencing processes which could then be maintained also in the absence of the original trigger (74). This silencing process could occur in the transformed cell but also later in progeny cells, as long as free DNA were still present. How such a mark could be set and later converted into stable suppression of a gene remains unclear. Lines R1 and R2 may differ in DNA methylation in other regions than analyzed here, as indicated by analyses with methylation-sensitive restriction enzymes.
It seems that DNA methylation might interfere with expression in a variety of ways, depending on which binding factors are attracted or excluded by methylation. Therefore, the initial effects may depend on the particular enhancer region and possibly also on the cell type. In the case described here, methylation of a short promoter region suffices to alter the tissue specificity; in other cases, methylation of even single cytosines can be effective (56, 75). It seems possible that such regional methylation could allow for a stable control of promoter specificity in contrast to the general on/off control usually associated with gene silencing. For the latter, a higher density of methylation may be required (76) to attract proteins like MeCP1, which only binds to DNA with several methyl cytosines (3) and may induce a repressive chromatin structure by histone modifications to interfere with all transcription initiation or elongation.
Acknowledgments
We thank Katrin Konya and Sabine Klarer for the help with maintenance of plants, Sandra Corsten for help with protection assays, C. Sautter for help with microscopy, and N. Hernández-Schärer and O. Mittelsten Scheid for critical reading of the manuscript. J.F. thanks W. Gruissem for his support. We gratefully acknowledge the financial support by the Swiss National Foundation of Sciences and the Swiss Agency for Development and Cooperation (DEZA/SDC).
Abbreviations
ds, double-stranded
GUS, β-glucuronidase
5-AC, 5-azacytidine
MVBP, methylated vascular bundle element-binding protein
VBE, vascular bundle-expression element
EMSA, electrophoretic mobility shift assay
RTBV, rice tungro bacilliform virus
References
- 1.Narlikar G. J., Fan, H.-Y. & Kingston, R. E. (2002) Cell 108, 475-487. [DOI] [PubMed] [Google Scholar]
- 2.Richards E. J. & Elgin, S. C. R. (2002) Cell 108, 489-500. [DOI] [PubMed] [Google Scholar]
- 3.Bird A. (2002) Genes Dev. 16, 6-21. [DOI] [PubMed] [Google Scholar]
- 4.Jones P. A. & Takai, D. (2001) Science 293, 1068-1070. [DOI] [PubMed] [Google Scholar]
- 5.Martienssen R. A. & Colot, V. (2001) Science 293, 1070-1074. [DOI] [PubMed] [Google Scholar]
- 6.Verbsky M. L. & Richards, E. J. (2001) Curr. Opin. Plant Biol. 4, 494-500. [DOI] [PubMed] [Google Scholar]
- 7.Meyer P. (2000) Plant Mol. Biol. 43, 221-234. [DOI] [PubMed] [Google Scholar]
- 8.Paszkowski J. & Whitham, S. A. (2001) Curr. Opin. Plant Biol. 4, 123-129. [DOI] [PubMed] [Google Scholar]
- 9.Jeddeloh J. A., Stokes, T. L. & Richards, E. J. (1999) Nat. Genet. 22, 94-97. [DOI] [PubMed] [Google Scholar]
- 10.Wassenegger M. (2000) Plant Mol. Biol. 43, 203-220. [DOI] [PubMed] [Google Scholar]
- 11.Mette M. F., Aufsatz, W., van der Winden, J., Matzke, M. A. & Matzke, A. J. M. (2001) EMBO J. 19, 5194-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M.-B. & Waterhouse, P. M. (2001) Curr. Opin. Plant Biol. 5, 146-150. [DOI] [PubMed] [Google Scholar]
- 13.Matzke M., Matzke, A. J. M. & Kooter, J. M. (2001) Science 293, 1080-1973. [DOI] [PubMed] [Google Scholar]
- 14.Thomas C. L., Jones, L., Baulcombe, D. C. & Maule, A. J. (2001) Plant J. 25, 417-426. [DOI] [PubMed] [Google Scholar]
- 15.Wu C. & Morris, J. R. (1999) Curr. Opin. Genet. Dev. 9, 237-246. [DOI] [PubMed] [Google Scholar]
- 16.Selker E. U. (1999) Cell 97, 157-160. [DOI] [PubMed] [Google Scholar]
- 17.Matzke M., Mette, M. F., Jakowitsch, J., Kanno, T., Moscone, E. A., van der Winden, J. & Matzke, A. J. M. (2001) Genetics 158, 451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rassoulzadegan M., Magliano, M. & Cuzin, F. (2002) EMBO J. 21, 440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colot V., Maloisel, L. & Rossignol, J. L. (1996) Cell 86, 855-864. [DOI] [PubMed] [Google Scholar]
- 20.Pelissier T. & Wassenegger, M. (2000) RNA 6, 55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M.-B., Wesley, S. V., Finnegan, E. J., Smith, N. A. & Waterhouse, P. M. (2001) RNA 7, 16-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luff B., Pawlowski, L. & Bender, J. (1999) Mol. Cell 3, 505-511. [DOI] [PubMed] [Google Scholar]
- 23.Jackson J. P., Lindroth, A. M., Cao, X. & Jacobson, S. E. (2002) Nature (London) 416, 556-560. [DOI] [PubMed] [Google Scholar]
- 24.Tamaru H. & Selker, E. U. (2001) Nature (London) 414, 277-283. [DOI] [PubMed] [Google Scholar]
- 25.Kishimoto N., Sakai, H., Jackson, J., Jacobsen, S. E., Meyerowitz, E. M., Dennis, E. S. & Finnegan, E. J. (2001) Plant Mol. Biol. 46, 171-183. [DOI] [PubMed] [Google Scholar]
- 26.Bartee L. & Bender, J. (2001) Nucleic Acids Res. 29, 2127-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartee L., Malagnac, F. & Bender, J. (2001) Genes Dev. 15, 1753-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amedeo P., Habu, Y., Afsar, K., Mittelsten Scheid, O. & Paszkowski, J. (2000) Nature (London) 405, 203-206. [DOI] [PubMed] [Google Scholar]
- 29.Tian L. & Chen, Z. J. (2001) Proc. Natl. Acad. Sci. USA 98, 200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z. J. & Pikaard, C. S. (1997) Genes Dev. 11, 2124-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day C. D., Lee, E., Kobayashi, J., Hoplappa, L. D., Albert, H. & Ow, D. W. (2000) Genes Dev. 14, 2869-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klöti A., Henrich, C., Bieri, S., He, X., Chen, G., Burkhardt, P. K., Wünn, J., Lucca, P., Hohn, T., Potrykus, I. & Fütterer, J. (1999) Plant Mol. Biol 40, 249-266. [DOI] [PubMed] [Google Scholar]
- 33.Murashige T. & Skoog, F. (1962) Physiol. Plant 15, 473-497. [Google Scholar]
- 34.Raizis A. M., Schmitt, F. & Jost, J.-P. (1995) Anal. Biochem. 226, 161-166. [DOI] [PubMed] [Google Scholar]
- 35.Chen G., Rothnie, H. M., He, X., Hohn, T. & Fütterer, J. (1996) J. Virol. 70, 8411-8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fütterer J., Rothnie, H. M., Hohn, T. & Potrykus, I. (1997) J. Virol. 71, 7984-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamilton A. J. & Baulcombe, D. C. (1999) Science 286, 950-952. [DOI] [PubMed] [Google Scholar]
- 38.Iguchi-Ariga S. M. M. & Schaffner, W. (1989) Genes Dev. 3, 612-619. [DOI] [PubMed] [Google Scholar]
- 39.Inamdar N. M., Ehrlich, K. C. & Ehrlich, M. (1991) Plant Mol. Biol. 17, 111-123. [DOI] [PubMed] [Google Scholar]
- 40.He X., Fütterer, J. & Hohn, T. (2001) J. Biol. Chem. 276, 2644-2651. [DOI] [PubMed] [Google Scholar]
- 41.He X., Hohn, T. & Fütterer, J. (2000) J. Biol. Chem. 275, 11799-11808. [DOI] [PubMed] [Google Scholar]
- 42.Yin Y., Chen, L. & Beachy, R. N. (1997) Plant J. 12, 1179-1188. [DOI] [PubMed] [Google Scholar]
- 43.Yin Y. & Beachy, R. N. (1995) Plant J. 7, 969-980. [DOI] [PubMed] [Google Scholar]
- 44.Prendergast G. C. & Ziff, E. B. (1991) Science 251, 186-189. [DOI] [PubMed] [Google Scholar]
- 45.Gaston K. & Fried, M. (1995) Gene 157, 257-259. [DOI] [PubMed] [Google Scholar]
- 46.Radke F., Hug, M., Georgiev, O., Matsuo, K. & Schaffner, W. (1996) Biol. Chem. Hoppe Seyler 377, 47-56. [DOI] [PubMed] [Google Scholar]
- 47.Thain A., Jenkins, O., Clarke, A. R. & Gaston, K. (1996) J. Virol. 70, 7233-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Fiore B., Palena, A., Felsani, A., Palitti, F., Caruso, M. & Lavia, P. (1999) Nucleic Acids Res. 27, 2852-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bell A. C. & Felsenfeld, G. (2000) Nature (London) 405, 482-485. [DOI] [PubMed] [Google Scholar]
- 50.Campanero M. R., Armstrong, M. I. & Flemington, E. K. (2000) Proc. Natl. Acad. Sci. USA 97, 6481-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hark A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M. & Tilghman, S. M. (2000) Nature (London) 405, 486-489. [DOI] [PubMed] [Google Scholar]
- 52.Gälweiler L., Conlan, R. S., Mader, P., Palme, K. & Moore, I. (2000) Plant J. 23, 143-157. [DOI] [PubMed] [Google Scholar]
- 53.Sturaro M. & Viotti, A. (2001) Plant Mol. Biol. 46, 549-560. [DOI] [PubMed] [Google Scholar]
- 54.Kunze R. & Starlinger, P. (1989) EMBO J. 8, 3177-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye F. & Signer, E. R. (1996) Proc. Natl. Acad. Sci. USA 93, 10881-10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark S. J., Harrison, J. & Molloy, P. L. (1997) Gene 195, 67-71. [DOI] [PubMed] [Google Scholar]
- 56.Jost J. P., Saluz, H. P. & Pawlak, A. (1991) Nucleic Acids Res. 19, 5771-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santoro R. & Grummt, I. (2001) Mol. Cell 8, 719-725. [DOI] [PubMed] [Google Scholar]
- 57.Sengupta P. K., Ehrlich, M. & Smith, B. D. (1999) J. Biol. Chem. 274, 36649-36655. [DOI] [PubMed] [Google Scholar]
- 58.Prokhortchouk A., Hendrich, B., Jörgensen, H., Ruzov, A., Wilm, M., Georgiev, G., Bird, A. & Prokhortchouk, E. (2001) Genes Dev. 15, 1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malone C. S., Miner, M. D., Doerr, J. R., Jackson, J. P., Jacobson, S. E., Wall, R. & Teitell, M. (2001) Proc. Natl. Acad. Sci. USA 98, 10404-10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ros F. & Kunze, R. (2001) Genetics 157, 1723-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pröls F. & Meyer, P. (1992) Plant J. 2, 465-475. [DOI] [PubMed] [Google Scholar]
- 61.He X., Fütterer, J. & Hohn, T. (2002) Nucleic Acids Res. 30, 497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kass S. U., Goddard, J. P. & Adams, R. L. P. (1994) Mol. Cell. Biol. 13, 7372-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Blokland R., Ten Lohuis, M. & Meyer, P. (1997) Mol. Gen. Genet. 257, 1-13. [DOI] [PubMed] [Google Scholar]
- 66.Sekinger E. A. & Gross, D. S. (2001) Cell 105, 403-414. [DOI] [PubMed] [Google Scholar]
- 67.Meins F. & Kunz, C. (1995) Curr. Top. Microbiol. Immunol. 197, 105-120. [DOI] [PubMed] [Google Scholar]
- 68.Papa C. M., Springer, N. M., Muszynski, M. G., Meeley, R. & Kaeppler, S. (2001) Plant Cell 13, 1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindroth A. M., Cao, X., Jackson, J. P., Zilberman, D., McCallum, C. M., Henikoff, S. & Jacobson, S. E. (2001) Science 292, 2077-2080. [DOI] [PubMed] [Google Scholar]
- 70.Matzke M. A. & Matzke, A. J. M. (1991) Plant Mol. Biol. 16, 821-830. [DOI] [PubMed] [Google Scholar]
- 71.Hollick J., Dorfweiler, J. & Chandler, V. (1997) Trends Genet. 13, 302-307. [DOI] [PubMed] [Google Scholar]
- 72.Cubas P., Vincent, C. & Coen, E. (1999) Nature (London) 401, 157-161. [DOI] [PubMed] [Google Scholar]
- 73.Jacobsen S. E. (1999) Curr. Biol. 9, R617-R619. [DOI] [PubMed] [Google Scholar]
- 74.Jones L., Ratcliff, F. & Baulcombe, D. C. (2001) Curr. Biol. 11, 745-757. [DOI] [PubMed] [Google Scholar]
- 75.Houchins K., O'Dell, M., Flavell, R. B. & Gustafson, J. P. (1997) Mol. Gen. Genet. 255, 294-301. [DOI] [PubMed] [Google Scholar]
- 76.Hsieh C.-L. (1994) Mol. Cell. Biol. 14, 5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fütterer J., Potrykus, I., Valles Brau, M. P., Dasgupta, I., Hull, R. & Hohn, T. (1994) Virology 198, 663-670. [DOI] [PubMed] [Google Scholar]