Abstract
The activity of the enzymes catalyzing the first two steps of sulfate assimilation, ATP sulfurylase and adenosine 5′-phosphosulfate reductase (APR), are confined to bundle sheath cells in several C4 monocot species. With the aim to analyze the molecular basis of this distribution and to determine whether it was a prerequisite or a consequence of the C4 photosynthetic mechanism, we compared the intercellular distribution of the activity and the mRNA of APR in C3, C3-C4, C4-like, and C4 species of the dicot genus Flaveria. Measurements of APR activity, mRNA level, and protein accumulation in six Flaveria species revealed that APR activity, cysteine, and glutathione levels were significantly higher in C4-like and C4 species than in C3 and C3-C4 species. ATP sulfurylase and APR mRNA were present at comparable levels in both mesophyll and bundle sheath cells of C4 species Flaveria trinervia. Immunogold electron microscopy demonstrated the presence of APR protein in chloroplasts of both cell types. These findings, taken together with results from the literature, show that the localization of assimilatory sulfate reduction in the bundle sheath cells is not ubiquitous among C4 plants and therefore is neither a prerequisite nor a consequence of C4 photosynthesis.
Assimilatory sulfate reduction is a pathway used by prokaryotes, fungi, and photosynthetic organisms to convert inorganic sulfate to sulfide, which is further incorporated into carbon skeletons of amino acids to form Cys or homo-Cys (Brunold, 1993). In this pathway, sulfate is first activated by ATP sulfurylase (ATPS) forming adenosine 5′-phosphosulfate (APS). In higher plants, APS is reduced by APS reductase (APR) to sulfite, which is further reduced to the level of sulfide by sulfite reductase (Bick and Leustek, 1998; Suter et al., 2000). APS can also be phosphorylated by APS kinase to phosphoadenosine 5′-phosphosulfate, which is utilized for synthesis of a wide range of sulfated compounds in reactions catalyzed by a variety of sulfotransferases (Varin et al., 1997). Thus, APR is a key step in sulfate assimilation and as such, the enzyme is highly regulated, e.g. by light, sulfur and nitrogen supply, heavy metals, or chilling (Rüegsegger et al., 1990; Neuenschwander et al., 1991; Brunold, 1993; Brunner et al., 1995; Kopriva et al., 1999, 2000).
C4 plants are characterized by an intercellular compartmentation of CO2 and nitrate assimilation between mesophyll and bundle sheath cells (Black, 1973; Moore and Black, 1979). Also, sulfate assimilation was proposed to be restricted to the bundle sheath cells of C4 plants (Gerwick et al., 1980; Schmutz and Brunold, 1984). Several groups reported that 75% to 100% of total leaf ATPS activity in maize (Zea mays) is confined to bundle sheath cells (Gerwick et al., 1980; Passera and Ghisi, 1982; Schmutz and Brunold, 1984). These findings were extended to 17 other C4 species where 95% to 100% of total leaf ATPS activity was found in chloroplasts of bundle sheath cells. Also, the next enzyme in the sulfate reduction pathway, APR, was found exclusively or almost exclusively in bundle sheath cells of maize (Schmutz and Brunold, 1984; Burgener et al., 1998), whereas the activities of the subsequent enzymes of the pathway, sulfite reductase and O-acetyl-Ser-(thiol) lyase, were found in both cell types at comparable levels (Passera and Ghisi, 1982; Burnell, 1984; Schmutz and Brunold, 1985). In accordance, in maize the mRNAs for APR, ATPS, and sulfite reductase accumulated in bundle sheath only, whereas the mRNA for O-acetyl-Ser-(thiol) lyase was also detected in mesophyll cells (Kopriva et al., 2001). Cultivation of maize plants at 12°C resulted in a prominent increase of APR mRNA and activity in bundle sheath cells. In addition, after chilling mRNAs for APR and sulfite reductase, as well as low APR activity, were detected in mesophyll cells. Therefore, it seems that chilling stress is able to affect not only the levels but also the intercellular distribution of mRNAs for enzymes of the sulfate assimilation (Kopriva et al., 2001).
The functional significance of the compartmentation of sulfate assimilation in bundle sheath cells of maize and other C4 species is not yet clear. A possible explanation could be the reduced O2 concentrations in bundle sheath cells compared with mesophyll cells, which might prevent oxidation of the reaction intermediates of sulfate assimilation, SO32− and S2−, or, alternatively, higher availability of Ser, resulting from exclusive localization of Gly decarboxylase and Ser hydroxymethyltransferase in bundle sheath cells (Burgener et al., 1998). As a first step to elucidate the importance of this compartmentation, the question arose of whether the exclusive or almost exclusive localization of the first two enzymes of sulfate assimilation was a prerequisite or a consequence of C4 photosynthesis. To answer this question we turned to the Flaveria genus. The genus Flaveria (Flaveriinae-Asteraceae) is unique because, beside C3 and C4 species, a relatively large number of C3-C4 intermediates exist in this genus (Powell, 1978; Bauwe, 1984; Ku et al., 1991). The levels of C4 enzymes in Flaveria spp. are correlated with the degree of C4 photosynthesis based on the initial products of photosynthesis and CO2 compensation points (Ku et al., 1991; Rosche et al., 1994; Marshall et al., 1996). This means that a continual gradation exists among Flaveria spp. both in the physiology and biochemistry of photosynthesis (Monson and Moore, 1989). Here, we report on activities, mRNA, protein accumulation, and inter- and intracellular localization of ATPS and APR in six species of the genus Flaveria with C3, C3-C4 intermediate, and C4 photosynthesis.
RESULTS
ATPS and APR Activity, Protein Accumulation, and Thiol Levels in Different Flaveria Spp.
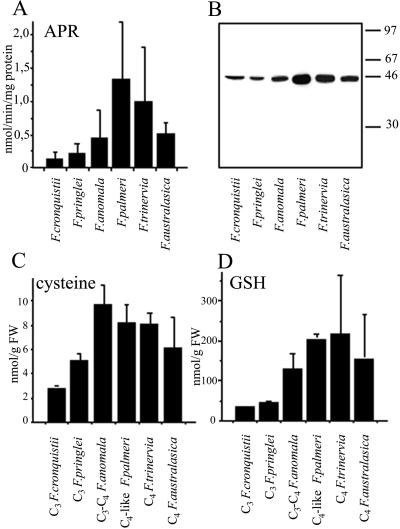
ATPS and APR activities were measured in young fully developed leaves of Flaveria cronquistii (C3), Flaveria pringlei (C3), Flaveria anomala (C3-C4), Flaveria palmeri (C4-like), Flaveria trinervia (C4), and Flaveria australasica (C4). As shown in Figure 1A, the APR activity in C4-like and C4 species F. palmeri, F. trinervia, and F. australasica was significantly higher than in the C3 species. ATPS activities showed a similar pattern, with the highest activity in F. palmeri, but due to great variations between different experiments the differences were not significant (data not shown). The APR protein accumulation was addressed by western analysis with antisera against APR from Arabidopsis. Similar to APR enzyme activities, the highest APR protein amount was detected in F. palmeri (Fig. 1B) and the lowest in the C3 species. Also, Cys and glutathione (GSH) concentrations in young leaves were lower in C3 species then in C4 and C4-like species, analogous to the distributions of APR activity and protein amount (Fig. 1, C and D). There was a strong correlation between the foliar GSH concentrations and measured APR activities among the six Flaveria spp. analyzed (r = 0.906).
Figure 1.
Assimilatory sulfate reduction in young leaves of six Flaveria species with different types of photosynthesis. A, APR activity. The results are presented as mean values + sd from six independent measurements. B, Western-blot analysis of APR. Ten micrograms of leaf proteins was resolved on 12% (w/v) SDS-PAGE gel and transferred onto nitrocellulose membrane. APR was immunologically detected using antisera against recombinant APR2 from Arabidopsis. C, Cys content. The results are presented as mean values + sd from three to six independent measurements. D, GSH content. The results are presented as mean values + sd from three to six independent measurements.
Expression Analysis
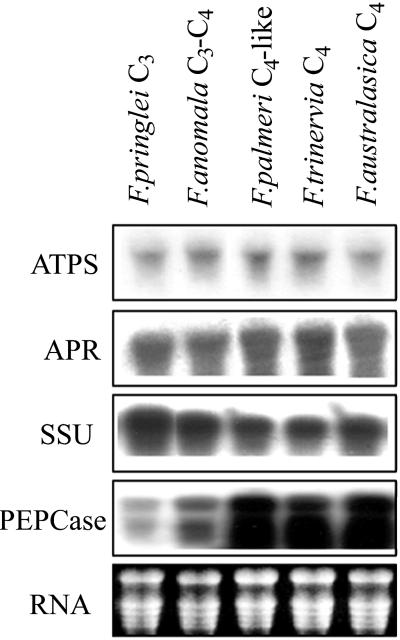
The levels of ATPS and APR mRNA were determined in young, fully developed leaves of five Flaveria spp. by northern blotting. Partial cDNAs for ATPS and APR were cloned from the C4 dicot F. trinervia by reverse transcriptase (RT)-PCR with degenerate primers against conserved domains (Suter et al., 2000). To avoid a bias caused by hybridization with a probe from a C4 species only, cDNA for APR was isolated also from the C3 species F. cronquistii. Both ATPS and APR mRNA levels, quantified with a densitometer, were very similar in all species; only slightly higher levels were observed in C4 and C4-like species than in C3 and C3-C4 species (Fig. 2). The results of hybridizations with an APR probe from C3 and C4 species were essentially identical (data not shown). As controls, the filters were hybridized with cDNA probes coding for Rubisco SSU and PEPCase from F. trinervia. The differences in expression of these genes among the different species corresponded to the expected pattern, i.e. mRNA levels for Rubisco SSU were highest in C3 species and decreased toward the C4 ones, whereas PEPCase mRNA was very abundant in C4-like and C4 species but hardly detectable in C3 species. Therefore, it seems that the variations in ATPS and APR activity (Fig. 1A) in the different species are not regulated solely at the transcriptional level.
Figure 2.
Northern-blot analysis. Total RNA was extracted from young leaves of five Flaveria spp., separated on 1% (w/v) agarose in the presence of formaldehyde, blotted onto Hybond-N nylon membrane, and hybridized with 32P-labeled cDNA fragments of ATPS, APR, Rubisco small subunit (SSU), and phosphoenolpyruvate carboxylase (PEPCase) from F. trinervia. Ethidium bromide-stained RNA is shown as a control of loading and RNA intactness.
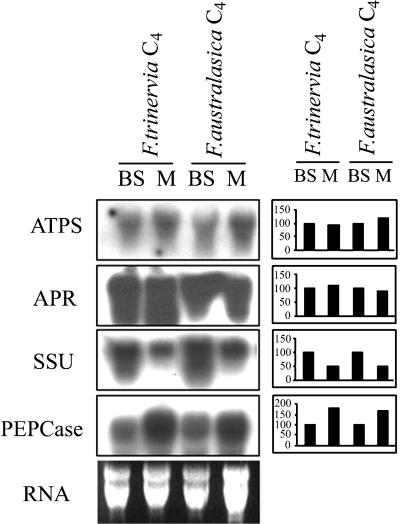
To compare the intercellular distribution of ATPS and APR mRNA in maize and Flaveria spp., we isolated RNA from mesophyll and bundle sheath cells from two C4 species, F. trinervia and F. australasica, and subjected it to northern analysis. Although the RNA preparations from the two cell types were cross contaminated, the quantification of the northern data surprisingly revealed that the mRNAs for ATPS and APR were present in both cell types at about the same levels (Fig. 3A). For both enzymes the relative transcript levels in mesophyll and bundle sheath cells (M/BS) were 1.1 to 1.2, as expected for mRNAs present in both cell types. In contrast, the mRNA of the marker enzyme for bundle sheath cells, Rubisco SSU, was detected predominantly in the RNA prepared from this cell type (M/BS = 0.5). PEPCase mRNA correspondingly was detected at a higher level in the RNA from mesophyll cells (M/BS = 2.1).
Figure 3.
Northern-blot analysis of mesophyll- and bundle sheath-specific RNA. Total RNA was extracted from mesophyll and bundle sheath cells of young leaves of the C4 species F. trinervia and F. australasica, separated on 1% (w/v) agarose in the presence of formaldehyde, blotted onto Hybond-N nylon membrane, and hybridized with 32P-labeled cDNA fragments of ATPS, APR, Rubisco SSU, and PEPCase from F. trinervia. Ethidium bromide-stained RNA is shown as a control of loading and RNA intactness. Right, Results of densitometric quantification of the northern blots. 100%, Represents mRNA level in bundle sheath cells of the particular species.
In Situ RNA Hybridization
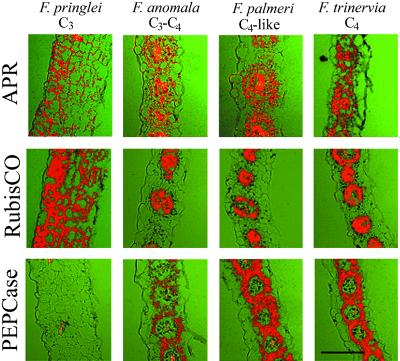
Because the results of northern analysis with bundle sheath and mesophyll RNA in C4 Flaveria differed significantly from maize (Kopriva et al., 2001) we wanted to confirm them using in situ RNA hybridizations with cDNA for APR from F. trinervia. As controls, in situ hybridizations were performed with cDNAs for Rubisco SSU and PEPCase, from F. trinervia. These hybridizations resulted in expected patterns of expression, i.e. bundle sheath-specific localization of Rubisco mRNA and mesophyll-specific expression of PEPCase (Fig. 4) in the C4 and C4-like species. In contrast, in all species analyzed, including C4 and C4-like F. trinervia and F. palmeri, no cell-specific expression pattern was observed and the APR mRNA was localized in both bundle sheath and mesophyll cells at comparable levels (Fig. 4), thus confirming the results of northern analysis.
Figure 4.
Expression of APR, Rubisco SSU, and PEPCase in leaves of four Flaveria spp. Transverse sections (7 μm) of young leaves from F. pringlei (C3), F. anomala (C3-C4), F. palmeri (C4-like), and F. trinervia (C4) were analyzed by in situ hybridization. The sections were hybridized with probes specific for the indicated genes. Bar = 100 μm.
Immunolocalization of APR
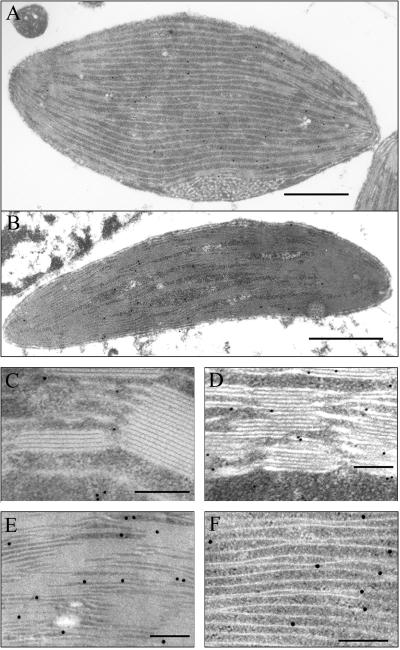
The presence of APR mRNA in the leaf mesophyll cells of F. trinervia does not necessarily mean that the mRNA is translated and the protein is active there. Therefore, antisera against APR2 from Arabidopsis (Kopriva et al., 1999) were used for immunogold electron microscopy on ultrathin leaf sections from three species: F. pringlei, F. anomala, and F. trinervia, to investigate the cellular distribution and spatial localization of this enzyme. APR protein was detected in chloroplasts of F. pringlei and F. anomala and in chloroplasts of both mesophyll and bundle sheath cells of F. trinervia (Fig. 5). In plastids of all three species, APR was localized approximately 30% to stroma and 70% to thylakoid membranes (data not shown). Treatment of the leaf sections with rabbit pre-immune sera did not result in any significant labeling as well as treatment with sera immunoprecipitated with purified recombinant APR. Because in all previous experiments we observed a strong correlation between APR protein accumulation and activity (Kopriva et al., 1999; Koprivova et al., 2000), we conclude that sulfate assimilation takes place in both mesophyll and bundle sheath cells in the C4 dicot F. trinervia.
Figure 5.
Immunolocalization of APR in different Flaveria spp. A, F. trinervia (C4) bundle sheath chloroplast; B, F. trinervia (C4) mesophyll chloroplast; C, F. pringlei (C3) part of chloroplast; D, F. anomala part of mesophyll chloroplast; E, F. trinervia (C4) part of mesophyll chloroplast; F, F. trinervia (C4) part of bundle sheath chloroplast. Bar = 1 μm in A and B; bar = 0.2 μm in C through F.
DISCUSSION
The aim of this study was to determine whether the well-documented bundle sheath-specific localization of sulfate assimilation in C4 plants (Gerwick et al., 1980; Schmutz and Brunold, 1984; Burgener et al., 1998) was a prerequisite or a consequence of C4 photosynthesis. Analogously to the results achieved with maize (Kopriva et al., 2001), we expected that the APR and ATPS mRNA and protein would be localized exclusively in bundle sheath cells in the C4 Flaveria spp. and in all photosynthetic cells in C3 species. From the distribution of ATPS and APR mRNA in C3-C4 intermediate and C4-like species, we thus would obtain information on the significance of the cell-specific localization of sulfate assimilation for C4 photosynthesis. However, both APR mRNA and protein were clearly detected in both types of cells in the C4 species F. trinervia. These results are surprising because in several reports the activity of ATPS and APR were localized exclusively or almost exclusively to bundle sheath cells of various C4 species (Gerwick et al., 1980; Schmutz and Brunold, 1984; Burgener et al., 1998). However, F. trinervia is a dicot and in all previous reports only monocot species were analyzed. We conclude, therefore, that the previously observed bundle sheath-specific localization of sulfate assimilation is not a general feature of C4 photosynthesis. On the other hand, the bundle sheath localization of APR is not a feature of all monocots because in the C3 monocot species Triticum aestivum, APR and ATPS were localized in both mesophyll and bundle sheath cells (Schmutz and Brunold, 1984). In fact, APR and ATPS are not the only enzymes in which cell-type specific expression in C4 dicots differs from C4 monocots. Bundle sheath cells of monocot NADP-malic enzyme C4 species lack photosystem II (Sheen and Bogorad, 1986); however, photosystem II polypeptides were found in both cell types at approximately the same levels in F. trinervia (Ketchner and Sayre, 1992).
The APR activities measured at normal conditions in C3 species vary substantially from 0.3 to 0.8 nmol min−1 mg−1 protein in hybrid poplar (Populus tremula × P. alba; Hartmann et al., 2000) and 5 to 10 nmol min−1 mg−1 protein in Arabidopsis (Kopriva et al., 1999; Koprivova et al., 2000) to 15 to 25 nmol min−1 mg−1 protein in Lemna minor (Neuenschwander et al., 1991). In maize, a C4 species, APR activities of 0.6 to 1.5 nmol min−1 mg−1 protein were determined (Brunner et al., 1995). The APR activities measured in the different Flaveria spp. thus correspond well to those determined in other plant species. The APR activity in C4-like and C4 Flaveria spp. was significantly higher than in C3 and C3-C4 species. On the other hand, ATPS activities in C4 species were not significantly different from C3 ones (Gerwick et al., 1980). Also, the GSH concentrations in Flaveria spp. are in the range measured in other plant species. APR is highly regulated by sulfur demand (Brunold, 1993); therefore, it is not surprising that there was a strong correlation between the foliar GSH concentrations and measured APR activities among the six Flaveria species analyzed. The reason for higher GSH concentrations in C4 Flaveria species than in C3 ones is not clear yet because the plants were grown under identical conditions. It could be speculated, however, that this difference is due to genetic adaptation to conditions of temperature and light stress to which C4 species are more exposed in their natural habitats than the C3 ones.
Isolated chloroplasts are capable of reducing sulfate (Trebst and Schmidt, 1969) and, correspondingly, the ATPS and APR activities were localized in spinach (Spinacia oleracea) chloroplasts (Schmidt, 1976; Fankhauser and Brunold, 1978; Lunn et al., 1990). No APR activity was measured in peroxisomes, mitochondria, or cytosol (Fankhauser and Brunold, 1978), whereas a cytosolic ATPS isoform could be identified in leaves of spinach and Arabidopsis (Lunn et al., 1990; Rotte and Leustek, 2000). Both APR and ATPS also appear to be exclusively localized to proplastids of root cells (Brunold and Suter, 1989). Recombinant APR from Catharanthus roseus was imported into intact pea (Pisum sativum) chloroplasts and correctly processed there (Prior et al., 1999), indicating again plastid localization for APR. In addition, in western analysis of pea chloroplast fractions APR protein was detected in stroma but not in any of the membrane fractions (Prior et al., 1999). In accordance with the former reports, immunogold labeling revealed the APR protein in chloroplasts of all three Flaveria spp. analyzed.
It is surprising that the APR signal was detected prevalently associated with the chloroplast thylakoid membranes. The reduction of APS does not directly rely on photosynthetic products, the electrons are supplied by glutathione (Bick et al., 1998), so that APR itself would not get any advantage by association with the thylakoids. However, the preceding and subsequent enzymes in the pathway, ATPS and sulfite reductase, directly utilize photosynthetic products formed at the thylakoids, i.e. ATP and reduced ferredoxin, respectively. Thus, it is plausible to hypothesize that the enzymes of assimilatory sulfate reduction form a multi-enzyme complex associated with the thylakoid membranes similarly as was proposed for Calvin cycle enzymes (Süss et al., 1993). In addition, the association of the enzymes of sulfate assimilation in a complex would lead to channeling of the reaction intermediates and thus prevent release of highly reactive and cytotoxic sulfite. It was already shown that two last enzymes of this pathway, O-acetyl-Ser(thiol) lyase and Ser acetyltransferase associate in a multienzyme complex (Bogdanova and Hell, 1997; Droux et al., 1998). The second enzymatic function of APR, namely the GSH-dependent reduction of dehydroascorbate (Bick et al., 1998), alternatively might be responsible for the association of APR with thylakoids because vast amounts of dehydroascorbate are produced at the thylakoid membranes during the photoprotective xanthophyll cycle (Demmig-Adams and Adams, 1996). The existence of a specific dehydroascorbate reductase is a matter of controversy (Foyer and Mullineaux, 1997; Morell et al., 1997). The results presented here indicate that APR might be a good candidate.
In conclusion, our findings taken together with results from the literature show that the localization of assimilatory sulfate reduction in the bundle sheath cells is not ubiquitous among C4 plants and is therefore neither a prerequisite nor a consequence of C4 photosynthesis.
MATERIALS AND METHODS
Plant Material
The seeds and/or plants of Flaveria cronquistii, Flaveria pringlei, Flaveria anomala, Flaveria palmeri, Flaveria trinervia, and Flaveria australasica were provided by Prof. P. Westhoff (University of Düsseldorf, Germany). Seeds were sown in soil and plants were grown in a greenhouse at 16-h-light/8-h-dark cycle and a temperature of 25°C ± 3°C.
Enzyme Assays
For extractions, young fully developed leaves of 5- to 6-week-old Flaveria spp. plants were used. Two-hundred milligrams of leaf material was homogenized in 2 mL of 50 mm NaKPO4 buffer (pH 8) supplemented with 30 mm Na2SO3, 0.5 mm 5′-AMP, and 10 mm dithioerythritol (Imhof, 1994), using a glass homogenizator. APR activity was measured in extracts as the production of [35S]sulfite, assayed as acid volatile radioactivity formed in the presence of [35S]APS and dithioerythritol (Brunold and Suter, 1990). The protein concentrations in the extracts were determined according to Bradford (Bradford, 1976) with bovine serum albumin as a standard. The ATPS activity was determined in the same extracts, diluted one-fifth with extraction buffer, by measurement of ATP production from APS and inorganic pyrophosphate using an ATP meter (Schmutz and Brunold, 1982).
Western-Blot Analysis
Aliquots of 10 μg protein from the extracts for APR measurements were subjected to SDS-PAGE and electrotransferred to nitrocellulose filter (Schleicher and Schuell, Dassel, Germany). The blots were analyzed with antisera against recombinant APR2 from Arabidopsis (Kopriva et al., 1999) and developed with the SuperSignal Western Blotting System (Pierce, Lausanne, Switzerland). The western analysis was performed on two independent protein preparations with the same results.
Thiol Measurements
Young leaves were extracted with 0.1 m HCl and the extracts were centrifuged for 30 min at 4°C. The thiols in the supernatant were reduced by bis-(2-mercaptoethylsulfone) (Bernhard et al., 1998) and labeled by monobromobimane (Kranner and Grill, 1996). Total Cys and glutathione were analyzed by reversed-phase HPLC as described by Schupp and Rennenberg (1988) and modified by Rüegsegger and Brunold (1992).
Isolation of RNA and Northern Blotting
Young leaves were pulverized with mortar and pestle in liquid nitrogen and total RNA was isolated by phenol extraction and selective precipitation with LiCl. Mesophyll and bundle sheath specific RNA was isolated from C4 species F. trinervia and F. australasica by a procedure described by Westhoff et al. (1991). Electrophoresis of RNA was performed on formaldehyde-agarose gels at 120 V. RNA was transferred onto Hybond-N nylon membranes (Amersham Pharmacia Biotech, Freiburg, Germany) and hybridized with 32P-labeled cDNA probes for ATPS and APR from F. trinervia. The membranes were washed four times at different concentrations of SSC in 0.1% (w/v) SDS for 20 min, the final washing step being 0.5× SSC, 0.1% (w/v) SDS at 65°C, and exposed to an x-ray film (medical RX, Fuji, Dielsdorf, Switzerland) at −80°C for 2 to 3 d. The autoradiograms were quantified with a densitometer GS-670 (Bio-Rad, Glattbrugg, Switzerland) using the software Molecular Analyst.
Cloning of cDNA for APR and ATPS from F. trinervia
The cDNAs for APR were cloned from F. trinervia and F. cronquistii RNA by RT-PCR with degenerate oligonucleotide primers derived from domains conserved among plant APRs and bacterial phosphoadenosine 5′-phosphosulfate reductases (Suter et al., 2000). The ATPS cDNA fragment was amplified from F. trinervia total RNA by RT-PCR with degenerate primers against conserved domains. The PCR products were cloned into pCR plasmids by the TA cloning kit (Invitrogen, Groningen, The Netherlands) and sequenced on both strands (Microsynth, Balgach, Switzerland).
In Situ RNA Hybridization
For the generation of probes, the cDNA fragments of Rubisco SSU and PEPCase were amplified by RT-PCR from F. trinervia total RNA, cloned into pCR plasmid, and their identity was controlled by sequencing. In situ hybridization experiments were performed on young fully developed leaves of several Flaveria spp. according to the protocol described by Fleming et al. (1993), with modifications described by Reinhardt et al. (1998). After development, the slides were stained in toluidine blue and viewed on an LSM 310 microscope (Carl Zeiss AG, Oberkochen, Germany). Images were taken under bright-field light (shown in false green color) and overlaid with epifluorescence images taken under polarized light exhibiting the silver grain signal (shown in false red color). For each probe, control hybridizations were performed with the corresponding sense probes, with the signals obtained negligible compared to the antisense probes.
Immunogold Localization
One-millimeter2 leaf sections of F. pringlei, F. anomala, and F. trinervia were vacuum infiltrated for a short time with 2% (v/v) formaldehyde and 0.5% (v/v) glutaraldehyde in 50 mm cacodylate buffer (pH 7.2), and kept 2.5 h at room temperature in the same medium. Samples were washed with buffer for 15 min followed by three washes for 15 min with distilled water. Dehydration of samples was done stepwise by increasing the concentration of ethanol and concomitantly lowering the temperature (progressive lowering of temperature) using an automated freeze substitution unit (Leica, Benzheim, Germany). The steps of progressive lowering of temperature substitution were performed as follows: 30% (v/v), 40% (v/v), and 50% (v/v) ethanol for 30 min at 4°C; 60% (v/v) and 75% (v/v) ethanol for 1h at −15°C; and 90% (v/v) ethanol and two times 100% (v/v) ethanol for 1 h at −35°C. The samples were subsequently infiltrated with Lowycryl HM20 resin (Plano GmbH, Marburg, Germany) as follows: 33% (v/v), 50% (v/v), and 66% (v/v) resin in ethanol for 5 h each and then 100% (v/v) resin overnight. Samples were transferred into gelatin capsules, kept there for 3 h in fresh resin, and polymerized at −35°C for 3 d under indirect UV light. The embedded samples were cut into ultrathin sections with a thickness of 70 to 90 nm on an ultramicrotome (Ultra cut F; Reichert) and mounted on copper grids, followed by immunogold labeling with 15 or 10 nm (F. pringlei) gold protein-A as described (Süss et al., 1993), except that the thin sections were slowly agitated during incubation to improve antibody labeling. For controls, APR antiserum was replaced by pre-immunoserum or serum that was incubated with purified recombinant APR. The sections were stained with uranyl acetate and lead citrate prior to examination in a Zeiss CEM 920A transmission electron microscope at 80 kV.
ACKNOWLEDGMENTS
We thank Prof. Dr. Peter Westhoff (Institut für Entwicklungs-und Molekularbiologie der Pflanzen, Universität Düsseldorf) for Flaveria spp. plants and seeds. Further we thank Prof. Dr. Cris Kuhlemeier (Institute of Plant Physiology, University of Bern, Switzerland), Prof. Dr. Heinz Rennenberg (Institut für Forstbotanik und Baumphysiologie, Universität Freiburg, Germany), Dr. Robert Hänsch (Botanisches Institut, Technische Universität Braunschweig, Germany), and Prof. Dr. Peter Westhoff for fruitful discussions and critical reading of the manuscript.
Footnotes
This work was supported by the Swiss National Foundation (grant no. 31–53984.98 to S.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp. 010144.
LITERATURE CITED
- Bauwe H. Photosynthetic enzyme activities and immunofluorescence studies on the localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in leaves of C3, C4, and C3-C4 intermediate species of Flaveria (Asteraceae) Biochem Physiol Pflanzen. 1984;179:253–268. [Google Scholar]
- Bernhard MC, Junker E, Hettinger A, Lauterburg BH. Time course of total cysteine, glutathione and homocysteine in plasma of patients with chronic hepatitis C treated with interferon-a with and without supplementation of N-acetylcysteine. J Hepatol. 1998;28:751–755. doi: 10.1016/s0168-8278(98)80223-7. [DOI] [PubMed] [Google Scholar]
- Bick JA, Aslund F, Chen Y, Leustek T. Glutaredoxin function for the carboxyl-terminal domain of the plant-type 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA. 1998;95:8404–8409. doi: 10.1073/pnas.95.14.8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick JA, Leustek T. Plant sulfur metabolism-the reduction of sulfate to sulfite. Curr Opin Plant Biol. 1998;1:240–244. doi: 10.1016/s1369-5266(98)80111-8. [DOI] [PubMed] [Google Scholar]
- Black CC. Photosynthetic carbon fixation in relation to net CO2 uptake. Annu Rev Plant Physiol. 1973;24:253–286. [Google Scholar]
- Bogdanova N, Hell R. Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J. 1997;11:251–262. doi: 10.1046/j.1365-313x.1997.11020251.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brunner M, Kocsy G, Rüegsegger A, Schmutz D, Brunold C. Effect of chilling on assimilatory sulfate reduction and glutathione synthesis in maize. J Plant Physiol. 1995;146:743–747. [Google Scholar]
- Brunold C. Regulatory interactions between sulfate and nitrate assimilation. In: De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, editors. Sulfur Nutrition and Sulfur Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 61–75. [Google Scholar]
- Brunold C, Suter M. Localization of enzymes of assimilatory sulfate reduction in pea roots. Planta. 1989;179:228–234. doi: 10.1007/BF00393693. [DOI] [PubMed] [Google Scholar]
- Brunold C, Suter M. Adenosine 5′-phosphosulfate sulfotransferase. In: Lea P, editor. Methods in Plant Biochemistry. Vol. 3. London: Academic Press; 1990. pp. 339–343. [Google Scholar]
- Burgener M, Suter M, Jones S, Brunold C. Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiol. 1998;116:1315–1322. doi: 10.1104/pp.116.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN. Sulfate assimilation in C4 plants. Plant Physiol. 1984;75:873–875. doi: 10.1104/pp.75.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW., III The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1996;1:21–26. [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D. Interactions between serine acetyltransferase and O-acetylserine(thiol)lyase in higher plants: structural and kinetic properties of the free and bound enzymes. Eur J Biochem. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- Fankhauser H, Brunold C. Localization of adenosine 5′-phosphosulfate sulfotransferase in spinach leaves. Planta. 1978;143:285–289. doi: 10.1007/BF00392000. [DOI] [PubMed] [Google Scholar]
- Fleming AJ, Mandel T, Roth I, Kuhlemeier C. The patterns of gene expression in the tomato shoot apical meristem. Plant Cell. 1993;5:297–309. doi: 10.1105/tpc.5.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Mullineaux PM. The presence of dehydroascorbate and dehydroascorbate reductase in plant tissues. FEBS Lett. 1997;425:528–529. doi: 10.1016/s0014-5793(98)00281-6. [DOI] [PubMed] [Google Scholar]
- Gerwick BC, Ku SB, Black CC. Initiation of sulfate activation: a variation in C4 photosynthesis plants. Science. 1980;209:513–515. doi: 10.1126/science.209.4455.513. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Mult S, Suter M, Rennenberg H, Herschbach C. Leaf age-dependent differences in sulphur assimilation and allocation in poplar (Populus tremula × P. alba) leaves. J Exp Bot. 2000;51:1077–1088. doi: 10.1093/jexbot/51.347.1077. [DOI] [PubMed] [Google Scholar]
- Imhof M. Die Rollevon Glutathion bei der Resistenz von Arabidopsis thaliana gegen den pathogenen Pilz Pythium paroecandrum. PhD Thesis. Switzerland: Institute of Plant Physiology of the University of Berne; 1994. [Google Scholar]
- Ketchner SL, Sayre RT. Characterization of the expression of the photosystem II-oxygen evolving complex in C4 species of Flaveria. Plant Physiol. 1992;98:1154–1162. doi: 10.1104/pp.98.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Jones S, Koprivova A, Suter M, von Ballmoos P, Brander K, Flückiger J, Brunold C. Influence of chilling stress on the intercellular distribution of assimilatory sulfate reduction and thiols in Zea mays. Plant Biol. 2001;3:24–31. [Google Scholar]
- Kopriva S, Muheim R, Koprivova A, Trachsler N, Catalano C, Suter M, Brunold C. Light regulation of assimilatory sulfate reduction in Arabidopsis thaliana. Plant J. 1999;20:37–44. doi: 10.1046/j.1365-313x.1999.00573.x. [DOI] [PubMed] [Google Scholar]
- Koprivova A, Suter M, Op den Camp R, Brunold C, Kopriva S. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 2000;122:737–746. doi: 10.1104/pp.122.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranner I, Grill D. Determination of glutathione and glutathione disulfide in lichens: a comparison of frequently used methods. Phytochem Anal. 1996;7:24–28. [Google Scholar]
- Ku MSB, Wu JR, Dai ZY, Scott RA, Chu C, Edwards GE. Photosynthetic and photorespiratory characteristics of Flaveria species. Plant Physiol. 1991;96:518–528. doi: 10.1104/pp.96.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Droux M, Martin J, Douce R. Localization of ATP sulfurylase and O-acetylserine(thiol) lyase in spinach leaves. Plant Physiol. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, Stubbs JD, Taylor WC. Two genes encode highly similar chloroplastic NADP-Malic enzymes in Flaveria. Plant Physiol. 1996;111:1251–1261. doi: 10.1104/pp.111.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson RK, Moore BD. On the significance of C3-C4 intermediate photosynthesis to the evolution of C4 photosynthesis. Plant Cell Environ. 1989;12:689–699. [Google Scholar]
- Moore RC, Black CC. Nitrogen assimilation pathways in leaf mesophyll and bundle sheath cells of C4 photosynthesis plants formulated from comparative studies with Digitaria sanguinalis (L.) Scop. Plant Physiol. 1979;64:309–313. doi: 10.1104/pp.64.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell S, Follmann H, De Tulio M, Häberlein I. Dehydroascorbate reduction: the phantom remaining. FEBS Lett. 1997;414:567–570. doi: 10.1016/s0014-5793(98)00282-8. [DOI] [PubMed] [Google Scholar]
- Neuenschwander U, Suter M, Brunold C. Regulation of sulfate assimilation by light and O-acetyl-L-serine in Lemna minor L. Plant Physiol. 1991;97:253–258. doi: 10.1104/pp.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passera C, Ghisi R. ATP sulphurylase and O-acetylserine sulphydrylase in isolated mesophyll protoplasts and bundle sheath strands of S-deprived maize leaves. J Exp Bot. 1982;33:432–438. [Google Scholar]
- Powell AM. Systematics of Flaveria (Flaveriinae-Asteraceae) Ann Miss Bot Garden. 1978;65:590–636. [Google Scholar]
- Prior A, Uhrig JF, Heins L, Wiesmann A, Lillig CH, Stoltze C, Soll J, Schwenn JD. Structural and kinetic properties of adenylyl sulfate reductase from Catharanthus roseus cell cultures. Biochim Biophys Acta. 1999;1430:25–38. doi: 10.1016/s0167-4838(98)00266-0. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998;10:1427–1437. doi: 10.1105/tpc.10.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche E, Streubel M, Westhoff P. Primary structure of the photosynthetic pyruvate orthophosphate dikinase of the C3 plant Flaveria pringlei and expression analysis of pyruvate orthophosphate dikinase sequences in C3, C3-C4 and C4 Flaveria species. Plant Mol Biol. 1994;26:763–769. doi: 10.1007/BF00013761. [DOI] [PubMed] [Google Scholar]
- Rotte C, Leustek T. Differential subcellular localization and expression of ATP sulfurylase and 5′-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiol. 2000;124:715–24. doi: 10.1104/pp.124.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger A, Brunold C. Effect of cadmium on γ-glutamylcysteine synthesis in maize seedlings. Plant Physiol. 1992;99:428–433. doi: 10.1104/pp.99.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger A, Schmutz D, Brunold C. Regulation of glutathione synthesis by cadmium in Pisum sativum L. Plant Physiol. 1990;93:1579–1584. doi: 10.1104/pp.93.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. The adenosine-5′-phosphosulfate sulfotransferase from spinach (Spinacea oleracea L.): stabilization, partial purification and properties. Planta. 1976;130:257–263. doi: 10.1007/BF00387830. [DOI] [PubMed] [Google Scholar]
- Schmutz D, Brunold C. Rapid and simple measurement of ATP sulfurylase activity in crude plant extracts using an ATP meter for bioluminiscence determination. Anal Biochem. 1982;121:151–155. doi: 10.1016/0003-2697(82)90569-3. [DOI] [PubMed] [Google Scholar]
- Schmutz D, Brunold C. Intercellular localization of assimilatory sulfate reduction in leaves of Zea mays and Triticum aestivum. Plant Physiol. 1984;74:866–870. doi: 10.1104/pp.74.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz D, Brunold C. Localization of nitrite and sulfite reductase in bundle sheath and mesophyll cells of maize leaves. Physiol Plant. 1985;64:523–528. [Google Scholar]
- Schupp R, Rennenberg H. Diurnal changes in the glutathione content of spruce needles (Picea abies L.) Plant Sci. 1988;57:113–117. [Google Scholar]
- Sheen J-Y, Bogorad L. Differential expression of six light harvesting chlorophyll a/b binding protein genes in maize leaf cell types. Proc Natl Acad Sci USA. 1986;83:7811–7815. doi: 10.1073/pnas.83.20.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süss K-H, Arkona C, Manteuffel R, Adler K. Calvin cycle multienzyme complexes are bound to chloroplast thylakoid membranes of higher plants in situ. Proc Natl Acad Sci USA. 1993;90:5514–5518. doi: 10.1073/pnas.90.12.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, von Ballmoos P, Kopriva S, Op den Camp R, Schaller J, Kuhlemeier C, Schürmann P, Brunold C. Adenosine 5′-phosphosulfate sulfotransferase and adenosine 5′-phosphosulfate reductase are identical enzymes. J Biol Chem. 2000;275:930–936. doi: 10.1074/jbc.275.2.930. [DOI] [PubMed] [Google Scholar]
- Trebst A, Schmidt A. The mechanism of photosynthetic sulfate reduction by isolated chloroplasts. Biochim Biophys Acta. 1969;180:529–535. doi: 10.1016/0005-2728(69)90031-0. [DOI] [PubMed] [Google Scholar]
- Varin L, Marsolais F, Richard M, Rouleau M. Sulfation and sulfotransferases 6: biochemistry and molecular biology of plant sulfotransferases. FASEB J. 1997;11:517–525. doi: 10.1096/fasebj.11.7.9212075. [DOI] [PubMed] [Google Scholar]
- Westhoff P, Offermann-Steinhard K, Höfer M, Eskins K, Oswald A, Streubel M. Differential accumulation of plastid transcripts encoding photosystem II components in the mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants. Planta. 1991;184:377–388. doi: 10.1007/BF00195340. [DOI] [PubMed] [Google Scholar]