Abstract
The mechanism by which enzymatic E colicins such as colicin E3 (ColE3) and ColE9 cross the outer membrane, periplasm, and cytoplasmic membrane to reach the cytoplasm and thus kill Escherichia coli cells is unique in prokaryotic biology but is poorly understood. This requires an interaction between TolB in the periplasm and three essential residues, D35, S37, and W39, of a pentapeptide sequence called the TolB box located in the N-terminal translocation domain of the enzymatic E colicins. Here we used site-directed mutagenesis to demonstrate that the TolB box sequence in ColE9 is actually larger than the pentapeptide and extends from residues 34 to 46. The affinity of the TolB box mutants for TolB was determined by surface plasmon resonance to confirm that the loss of biological activity in all except one (N44A) of the extended TolB box mutants correlates with a reduced affinity of binding to TolB. We used a PCR mutagenesis protocol to isolate residues that restored activity to the inactive ColE9 D35A, S37A, and W39A mutants. A serine residue at position 35, a threonine residue at position 37, and phenylalanine or tyrosine residues at position 39 restored biological activity of the mutant ColE9. The average area predicted to be buried upon folding (AABUF) was correlated with the activity of the variants at positions 35, 37, and 39 of the TolB box. All active variants had AABUF profiles that were similar to the wild-type residues at those positions and provided information on the size, stereochemistry, and potential folding pattern of the residues of the TolB Box.
To provide a competitive advantage for nutrient acquisition or colonization of new environments (21, 38), many Escherichia coli cells secrete a ribosomally synthesized bactericidal colicin to kill closely related nonimmune bacteria. The vast majority of colicins exert their lethality by a C-terminal cytotoxic domain that either causes membrane depolarization of the cytoplasmic membrane (the pore-forming colicins) or degradation of RNA or DNA (the enzymatic colicins). Import of the cytotoxic domain requires the cooperation of two other separate structural domains. The first is a centrally located receptor binding domain that targets the colicin to the bacterial outer membrane, and the second is an N-terminal translocation domain that makes contact with several proteins in the outer membrane and periplasmic space of the cell and is believed to act as a signaling mechanism for entry of the cytotoxic domain (4, 48). Colicin translocation can occur via the ton-dependent system, which is used by group B colicins having a TonB box at their N terminus (3), or via the tol-dependent translocation system, which is used by group A colicins such as the enzymatic E colicins (18, 23). Unlike group B colicins which cross the cell envelope by using the proteins TonB, ExbB, and ExbD, group A colicin translocation is more varied and, depending on the colicin, involves a combination of the proteins TolA, TolB, TolR, TolB, and Pal. Translocation of the enzymatic colicins including colicins E9 (ColE9) and E3 (ColE3) requires a specific pentapeptide sequence in the N-terminal translocation region, known initially as the TolA box (12, 31) but which is now known to interact with TolB and is called the TolB box (2, 5). Site-directed mutagenesis of 14 residues within the N-terminal 56 residues of ColE9 only revealed three residues (D35, S37, and W39) where an alanine mutation abolished colicin activity (12). Alanine substitutions of an alternative pentapeptide sequence, DGRGH at residues 5 to 9, and conserved residues Ser 34, Gly 36, Gly 38, Ser 40, Ser 52, and Trp 56 all had some degree of activity that suggested the TolB box consisted of residues 35 to 39. The protein-protein interaction between the T domain of ColE9 and TolB was demonstrated using the yeast two-hybrid system (5), and the interaction was abolished by any of the three alanine mutations in the TolB box that also abolished the biological activity of the colicin.
ColE3 has been suggested to contain a TolR box adjacent to the TolB box and may weakly interact with TolRII (20) but appears to lack a TolA box. No interaction with TolA has been demonstrated with any enzymatic colicin. In contrast, colicin N has a well-defined TolA box and interacts with TolA but not TolB (14), whereas colicin A has a TolA box, a TolB box, and a TolR box and makes contact with TolA, TolB, and TolR (1, 20).
The interaction between the T domain of ColE9 and TolB has been studied using nuclear magnetic resonance (NMR), which confirms that the T domain of ColE9, like that of colicin N (42), E3 (36), and Ia (46), is largely unstructured and flexible (6). The addition of unlabeled TolB to 15N-labeled ColE9 induced significant changes to the peptide NH region of the 1H-15N heteronuclear single-quantum coherence spectrum of the colicin, which affected specific signals to residues A33, S34, D35, W39, S40, S41, E42, N43, and N44. Although these data suggest that the region of ColE9 that interacts with TolB may be larger than the proposed pentapeptide (6), we cannot exclude the possibility that the changes in the NMR spectrum are the result of TolB binding-induced conformational changes in the largely unstructured region flanking the TolB box rather than a direct interaction between TolB and these residues. An extension of the TolB box of colicin A to include the equivalent residues to S40, S41, and E42 in ColE9 has been proposed based upon deletion analysis (2), but there is no published mutational data that confirm the role of any of these flanking residues in the protein-protein interaction with TolB. Here we confirm the presence of an extended TolB box in ColE9 and investigate the possible role of some TolB box residues in the interaction with TolB that provides valuable data to inform models of the translocation mechanism of tol-dependent colicins.
MATERIALS AND METHODS
Plasmids, bacterial strains, and media.
E. coli DH5α (lacZΔM15 recA1) and E. coli JM83 [ara (Δlac-proAB) rpsL Φ80lacZΔM15] cells were used as host strains for cloning and mutagenesis. The ColE9/Im9 complex was expressed from plasmid pNP69 (a derivative of pCS4, described previously [12], containing an SacI site at bp 988) with a C-terminal His-tag on the Im9 protein to facilitate purification (29). TolB containing its N-terminal signal sequence and a C-terminal His-tag was cloned into the complementary NdeI and XhoI sites of pET21a (Novagen), forming the plasmid pRJ376. E. coli BL21(DE3) or E. coli ER2566 were used as host strains for the expression constructs which have a strong, isopropryl-β-d-thiogalactopyranoside-inducible T7 polymerase promoter. All cultures were grown routinely in Luria-Bertani (LB) broth or on plates of LB agar, supplemented when required with ampicillin (100 μg ml−1).
Site-directed mutagenesis.
Site-directed mutagenesis was performed using a two-step PCR method (34). A mutagenic primer was designed with a minimum of six complementary bases on either side of the mutation. This was used in a PCR with a T7 promoter primer to complete the first-stage PCR. The first-stage PCR product was then used in a second-stage PCR with a T7 terminator primer. For the extended TolB box mutations, pNP69 was used as the template in both stages. For the TolB box variant mutants, plasmids pNP56, pCS9, and pNP57 (12), pET21a-based plasmids, expressing inactive ColE9 D35A, S37A, or W39A, respectively, were used as templates so that mutations that restored activity could be identified using the stab test assay.
Large-plate assay of colicin activity.
LB agar plates containing ampicillin (100 μg ml−1) were overlaid with 5 ml of molten 0.7% (wt/vol) agar containing 100 μl of an overnight culture of E. coli JM83 (pET21a). The test proteins were serially diluted in potassium phosphate buffer (50 mM K2HPO4, 50 mM KH2PO4, pH 7), and 2 μl of the diluted proteins was spotted onto the agar and allowed to dry. The plate was incubated for 16 h at 37°C and inspected for zones of inhibition of growth.
Protein purification.
Four milliliters of an overnight LB broth culture of E. coli BL21 cells or E. coli ER2566 cells, transformed with the appropriate plasmid, in LB broth was used to inoculate 400 ml of LB broth, containing 100 μg ml−1 ampicillin. When an optical density at 600 nm (OD600) of 0.6 was reached, the cells were induced by addition of isopropryl-β-d-thiogalactopyranoside to a final concentration of 1 mM. After 2 h of induction, the cells were harvested by centrifugation at 8,000 × g for 10 min.
Colicin wild-type and mutant/Im9 protein complexes were purified using the following method. Cells were lysed by treatment with 5 ml of Bugbuster (Novagen) at room temperature for 30 min. The cell lysate was centrifuged at 20,000 × g for 10 min at 4°C. Proteins were purified using HiTrap chelating high-performance columns (Amersham Biosciences) connected to a BiologicLP high-performance liquid chromatography machine (Bio-Rad). The supernatant was filtered using a 0.2-μm-pore-size syringe filter (Millipore) and applied to the HiTrap column that had been equilibrated with 20 mM Tris-HCl (pH 7.9)-5 mM imidazole-0.5 M NaCl and charged with NiSO4. After application of the supernatant, the column was washed with the equilibrating Tris buffer, and the colicin protein was eluted using a gradient of imidazole (0 to 100%; 0 to 1 M imidazole in 20 mM Tris-HCl, pH 7.9, 5 mM imidazole, 0.5 M NaCl). Proteins were dialyzed in 5 liters of potassium phosphate buffer (50 mM K2HPO4, 50 mM KH2PO4, pH 7) at 4°C for a minimum of 16 h.
ColE9 was purified without its immunity protein using denaturation of the colicin-immunity protein complex by incubation of the complex with guanidine as described previously (45). The free proteins were extensively dialyzed in 5 liters of potassium phosphate buffer (50 mM K2HPO4, 50 mM KH2PO4, pH 7.4), with at least two changes of buffer.
The TolB protein was purified using the following method. Cell pellets were resuspended in NiC buffer (10% [vol/vol] glycerol, 25 mM NaH2PO4, pH 7.4, 50 mM NaCl) containing a complete EDTA-free protease inhibitor cocktail tablet (Roche). The cells were lysed using sonication for 40 min (cycles of 20 s on and 20 s off). The sonicated sample was centrifuged twice at 25,000 × g for 30 min. The supernatant was filtered using a 0.2-μm-pore-size sterile syringe filter (Millipore) and applied to a HiTrap column charged with NiSO4 and equilibrated with 1× phosphate-buffered saline containing 0.5 M NaCl, pH 7.4. Pure protein was eluted using an increasing 1 M imidazole (in 1× phosphate-buffered saline containing 0.5 M NaCl, pH 7.4) gradient ranging from 0 to 100%. Proteins were dialyzed in 5 liters of potassium phosphate buffer (50 mM K2HPO4, 50 mM KH2PO4, pH 7.4) at 4°C for a minimum of 16 h.
Protein concentrations were calculated using Beer's law following absorbance readings at 280 nm.
Luminescence reporter assay.
This assay makes use of an SOS-inducible chromosomal lux operon to detect DNA damage induced by ColE9 in reporter cells (8, 41). All assays were performed in a microtiter plate luminometer (Lucy 1; Anthos Labtech, Salzburg, Austria) at 37°C. The Luminometer, plates, and medium were prewarmed to 37°C to prevent induction of a stress response due to cooling. An overnight culture of E. coli DPD1718, containing a fusion of the E. coli recA promoter region to the Photorhabdus luminescens luxCDABE reporter integrated into the lacZ locus of E. coli DPD1692, was diluted 1:50 in LB broth and grown for 3 h at 37°C in the presence of 30 μg ml−1 chloramphenicol, until the culture reached an OD492 of 0.3 to 0.4. The cells were then diluted 1:2 with LB broth (total volume, 100 μl) in black 96-well optical bottom microtiter plates (Nunc), and 4 μl of the wild-type or mutant colicin was added. Induction of luminescence was followed over a period of 90 min, with readings taken every 600 s. Cell density was also monitored by measuring OD492 values.
The ratios of relative luminescence units (RLU) to OD492 values were used to calculate gamma values (8) at a time point of 50 min using the following equation, where luminescence (L) = RLU/OD492 nm: γ = (Lsample − Lcontrol)/Lcontrol.
SPR.
Surface plasmon resonance (SPR) was performed using a BIAcore X instrument from BIAcore AB (Uppsala, Sweden), operating BIAcore X control software. Operation and maintenance procedures were carried out as detailed in the BIAcore X instrument handbook, using BIAcore AB-certified products. HBS-EP running buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% [vol/vol] surfactant P20), purchased from BIAcore AB, was used throughout. All buffers and solutions used were filtered and degassed using 0.2-μm-pore-size sterile syringe filters (Millipore) or Steritop sterile bottle-top filters (Millipore). Solutions filtered by syringe filter were degassed by centrifugation.
The ligand was immobilized to the matrix of a newly docked CM5 sensor chip, preequilibrated in HBS-EP running buffer (BIAcore AB) at a flow rate of 10 μl min−1 via amine coupling. The carboxymethylated matrix in both flow cells was activated using a mixture of 0.1 M ethyl-N-(3-diethylaminopropyl)carbodiimide (EDC; BIAcore AB) and 0.4 M N-hydroxysuccinimide (NHS; BIAcore AB). The protein sample was diluted in coupling buffer (10 mM sodium acetate, pH 4.5; BIAcore AB) and injected in single-channel mode across flow cell 2 until the required amount of protein was immobilized on the surface. Remaining active esters in both flow cells were then deactivated using 1 M ethanolamine hydrochloride, pH 8.5. The final level of immobilization was determined after two washes with HBS-EP buffer.
Measurement of binding of analytes to the immobilized protein on the CM5 sensor chip was completed at 25°C with HBS-EP running buffer using a flow rate of 30 μl min−1. Proteins to be used as analytes were diluted in HBS-EP buffer to the required concentration, and a 2-min injection of the diluted protein was performed. The surface was then regenerated using a 2-min injection of 10 mM glycine (pH 1.8). The experiment was repeated at least twice for each concentration of protein. Kinetic analysis for binding of analyte to the immobilized ligand was achieved using BIAevaluation software version 3.1. Global analysis was performed employing the Langmuir 1:1 binding isotherm.
RESULTS
Defining the extent of the TolB box of ColE9.
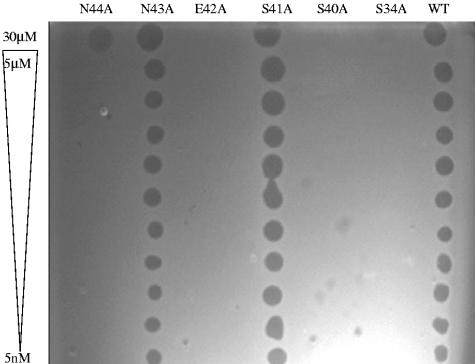
An alignment of the TolB box region of tol-dependent colicins is shown in Fig. 1. Residues in the T domain of ColE9 that have been shown by NMR to be affected by the binding of TolB (6) and lie outside of the defined “TolB box” might be essential for TolB binding. We had previously reported that the ColE9 S34A and S40A mutant protein/Im9 complexes produced very hazy, slow-forming zones of inhibition of indicator E. coli JM83 cells in stab tests (12). We made additional alanine mutations at residues S41, E42, N43, and N44 and then tested all six of the purified, mutant proteins for colicin activity in a large-plate assay (Fig. 2). At a starting concentration of 30 μM, no killing was observed for the ColE9 S34A, S40A, and E42A mutant protein/Im9 complexes, as with the previously described ColE9 D35A, S37A, and W39A TolB box mutants (12). As the indicator cells used in this study were E. coli DH5α, it is possible that the differences in activity of the S34A and S40A mutants compared to our previous findings might be due to differences in the sensitivity of the indicator strains or the longer incubation times used in the stab tests. The relative activity of the ColE9 S41A/Im9 and N43A/Im9 mutants were identical to ColE9/Im9 at all concentrations tested down to 5 nM. The ColE9 N44A/Im9 mutant showed growth inhibition only at a concentration of 30 μM.
FIG. 1.
Alignment of the TolB box region of the tol-dependent colicins, ColE2, ColE3, ColE7, ColE9, and ColA. The pentapeptide TolB box sequences, consisting of residues 35 to 39 in the E colicins and residues 11 to 15 in ColA, are underlined.
FIG. 2.
Determination of the colicin activity of purified ColE9/Im9 and ColE9 S34A, S40A, S41A, E42A, N43A, and N44A mutant protein/Im9 complexes in a large plate assay.
We have recently developed a quantitative DNA damage “reporter” assay that makes use of an SOS-inducible chromosomal lux operon to quantitate the DNA damage induced in the colicin-treated E. coli DPD1718 reporter cells (41). Using a concentration of 0.4 nM, which is within the concentration range that produces a linear response in the reporter assay, it is possible to accurately compare the DNA damage-inducing activity of the ColE9 mutant protein/Im9 complexes, expressed as a gamma value (8), at a fixed time after addition to E. coli DPD1718 cells (41) (Table 1). The results support the large-plate assay data as the ColE9 S34A, S40A, and E42A mutant protein/Im9 complexes showed no significant DNA damage-inducing activity. Interestingly, the ColE9 N44A mutant/Im9 complex retained 3% of the DNA damage-inducing activity of the wild-type ColE9/Im9 complex, which was consistent with the hazy zone seen at a concentration of 30 μM in the large-plate assay (Fig. 2). The ColE9 S41A mutant/Im9 complex retained 26% activity, while the ColE9 N43A mutant/Im9 complex retained 85% activity of the ColE9/Im9 complex in this assay.
TABLE 1.
Properties of extended TolB box mutants
| Mutation | lux (%)a | KD (μM)b |
|---|---|---|
| None | 100 | 13.8 ± 0.3 |
| S34A | 0 | NDc |
| D35A | 0 | ND |
| S37A | 0 | ND |
| W39A | 0 | ND |
| S40A | 0.2 | ND |
| S41A | 26 | 92 ± 2.5d |
| E42A | 0 | ND |
| N43A | 85 | 51.5 ± 1.5 |
| N44A | 3 | 19.4 ± 0.2 |
| W46A | 0.5 | ND |
The lux activity induced by the addition of ColE9/Im9 complex carrying the mutation indicated at 50 min after addition to E. coli DPD1718 cells, expressed as a percentage of the gamma value of that of the ColE9/Im9 complex.
KD values for the interaction of the ColE9/Im9 complex carrying the mutation indicated with TolB were determined from kinetic data obtained from fitting of sensorgrams produced for binding to TolB to the Langmuir 1:1 binding model.
ND, the affinity was too low to be able to determine the KD value.
This represents the lowest affinity determined. The limit of detection is based on the concentration of purified protein and is approximately 100 μM in these experiments.
Using previously described methods (30), we demonstrated that the purified ColE9 S34A, S40A, E42A, and N44A mutant protein/Im9 complexes were able to bind to BtuB receptors and that the “free” ColE9 proteins, in which Im9 has been removed from the complexes, exhibited DNase activity similar to the ColE9 protein (data not shown). This confirms that the DNase and R domains of the four mutant proteins are functional, and thus these four residues are presumably required in some way for translocation of ColE9 and could form part of an extended TolB box as suggested by the NMR data.
Affinity of T domain-TolB interactions by SPR.
To determine the effect of the TolB box mutations on the affinity of the interaction of the ColE9/Im9 mutant complexes with TolB, we used SPR on a Biacore X biosensor. In a typical experiment, 50 nM ligand (TolB) was injected across flow cell 2 of an EDC/NHS-activated CM5 carboxymethylated dextran surface with a contact time of 3 min at a flow rate of 10 μl min−1. This produced a level of immobilization of 300 resonance units (RU), giving a calculated Rmax for ColE9/Im9 binding (assuming a 1:1 binding interaction) of 187.5 RU. The analyte (ColE9/Im9 complex) was injected in solution for 2 min at concentrations of 2, 5, 10, 15, and 20 μM over the TolB chip at a flow rate of 30 μl min−1. The resulting interaction between the ligand and the analyte was recorded in RU, which are directly proportional to the mass accumulation at the surface. After each analyte injection, the sensor chip surface was regenerated with a 1-min pulse of 10 mM glycine, pH 1.8.
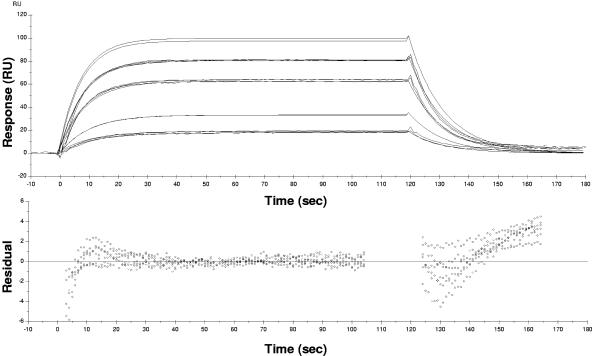
The sensorgrams produced for the range of concentrations of analyte used were corrected by subtraction of the data recorded for flow cell 1 consisting of an equivalent surface without TolB bound to it. Global analysis using BIAevaluation software 3.1 was used to fit the corrected SPR responses to the theoretical 1:1 Langmuir binding model. Corrected sensorgrams and residual plots for the wild-type ColE9/Im9-TolB interaction are shown in Fig. 3. The residual plots highlight deviation of the experimental data from the theoretical fit and are largest at the start and end points of the injections. The kinetics data obtained shows that the association constant per molar per second is 6,130 ± 109 (M−1 s−1), the dissociation constant per second is 0.0847 ± 0.000574 (s−1), the association constant per molar is 72,300 ± 1,374 (M−1) and the dissociation constant per molar (KD) is 13.8 ± 0.26 μM, with a chi-square value of 2.06. A Scatchard plot of the maximum recorded RU/[ColE9/Im9] versus the maximum recorded RU gave an R2 value of 0.97, indicating that the data fit the 1:1 binding model well.
FIG. 3.
TolB interaction with ColE9 determined by SPR. Corrected sensorgrams and residual plots for titration of 2, 5, 10, 15, and 20 μM ColE9/Im9 against TolB at 25°C are shown. The fitted kinetic data for binding are superimposed onto each sensorgram trace. The residual plots are shown underneath and highlight deviation of the experimental data from the theoretical fit.
The binding of the extended TolB box mutant proteins to TolB was compared to that of the wild-type ColE9/Im9 complex in SPR experiments. As expected, binding of the previously reported TolB box mutants, ColE9 D35A/Im9, S37A/Im9, and W39A/Im9 protein complexes to TolB was not detected by SPR (Table 1). Binding of the ColE9 S34A, S40A, and E42A mutant protein-Im9 complexes to TolB was also not detected in this series of SPR experiments. The ColE9 S41A/Im9 and N43A/Im9 mutant proteins gave KD values of 92 μM and 51.5 μM, respectively, for binding to TolB, which agreed both with their relative lux activities of 26% and 85% in the reporter assay (Table 1). The apparent difference in the activities of the ColE9 S41A/Im9 mutant in Fig. 2 and Table 1 is due to the concentration range used in the large-plate assay not being low enough to detect the difference in activity of this mutant protein and ColE9/Im9.
The results with these mutant proteins support the hypothesis of an extended TolB binding region in the T domain of ColE9, in that alanine mutations (S34, S40, and E42) located outside the TolB box pentapeptide region abolished both the biological activity and TolB binding of the mutant colicins. The most surprising result was obtained with the ColE9 N44A mutant protein/Im9 complex which, despite having only 3% of the activity of ColE9/Im9 in the lux reporter assay, bound to TolB with a KD of 19.4 ± 0.22 μM (Table 1). The difference in properties of the ColE9 E42A, N43A, and N44A mutant protein/Im9 complexes perhaps suggests that the C-terminal boundary of the extended TolB box is residue E42. In order to test this hypothesis, we constructed a further alanine mutation of residue W46. This mutant/Im9 protein exhibited 0.5% of the activity of ColE9/Im9 in the lux reporter assay, respectively, while binding to TolB could not be detected. This shows that the C terminus of the TolB box extends at least to residue 46. We did not extend the mutagenesis beyond residue 46 as residues 47, 48, 49, 51, and 53 are all glycine residues that lack any side chains and would be predicted to have little effect on the interactions with TolB based on our studies with the G36A and G38A variants of the TolB box, which remained biologically active and were still able to interact with TolB (5, 12). We had also previously made the mutation S52A that retained significant activity in a stab test assay and did not appear to be part of the TolB box (12). In addition NMR data indicate that TolB binding has little effect on residues beyond N44 (6).
Effect of Im9 on ColE9 binding to TolB in SPR.
In experiments to quantitate protein-protein interactions of colicin N with TolAII-III, it was observed that the KD values for binding were 3 μM for the isolated T domain of colicin N, 8 μM for the T plus R domains, and 18 μM for full-size colicin N (14) The authors suggested that the TolA binding site in colicin N may be masked by other domains of the colicin. In order to test if this also occurred with ColE9, we injected 0.5 to 4 μM of a truncated ColE9, which consisted only of the T plus R domains, across TolB amine coupled to a CM5 chip and determined the KD value of 1.66 ± 0.03 μM, which is significantly different from the value obtained with the ColE9/Im9 complex. When we compared the binding of free ColE9 and the ColE9/Im9 complex to the same TolB chip, there was a significant difference in the KD of binding of the two proteins (1.17 ± 0.025 μM for ColE9 and 13.8 μM for ColE9/Im9). The chi-square value for the ColE9 data was 4.3, and Scatchard analysis of the data gave an R2 of 0.907, which is indicative of a good fit of the data to the binding model. The simplest explanation for these results is that the affinity of ColE9 to TolB immobilized on a CM5 chip is not affected by the presence of the DNase domain but is reduced by the binding of Im9 to the DNase domain of ColE9. The observed results are unlikely to be due to nonspecific binding of the positively charged DNase domain (charge, +7.58; pI 9.56) to the negatively charged dextran surface of the CM5 chip, in the absence of Im9 (charge, −8.6; pI 4.43), as very high binding of ColE9 to the reference flow cell 1 which contains no TolB bound to the CM5 chip would also be observed.
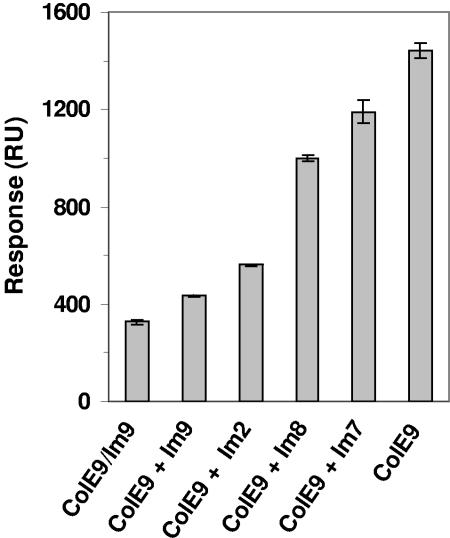
We investigated this phenomenon in a number of ways. During the purification of ColE9 from the ColE9/Im9 complex, denaturation and refolding steps are required to separate Im9 from ColE9. The noncognate immunity proteins Im7, Im8, and Im2 also bind to the DNase domain of ColE9 with KD values of 10−4 M, 10−6 M, and 10−8 M, respectively, compared to the figure of 10−14 M for Im9 (44). After preincubation of 2 μM of each noncognate Im protein with 2 μM of ColE9, the resulting complex was injected across a TolB chip, and the mean response was recorded after 2 min. The results showed that the relative reduction in binding of the resulting ColE9/Im complexes to TolB, compared to ColE9 alone, was related to the KD value for binding of the noncognate Im protein to the DNase domain (Fig. 4) but not in a linear manner because there are several differences in the nature of the complex specificity between cognate and noncognate interactions and also between different noncognate interactions (25, 26). This result is consistent with the hypothesis of an effect of Im binding to the DNase domain that in some way affects binding to TolB.
FIG. 4.
The effect of incubation with noncognate Im proteins on the interaction between free ColE9 and TolB. The bar chart shows a comparison of the mean responses for a 2-min injection of ColE9, ColE9/Im9, and ColE9 preincubated with Im9, Im2, Im8, or Im7 across TolB-amine coupled to a CM5 chip. This experiment was repeated with similar results on three separate occasions enabling a mean to be generated.
No three-dimensional structures of ColE9/Im9 and ColE9 are available to investigate a possible effect of Im9 on the T domain, although the NMR data show that Im9 binding to ColE9 does not affect residues 1 to 83 of the colicin (6). However, the three-dimensional structure of the RNase ColE3/Im3 complex showed that 83% of the Im3 surface area is buried in the ColE3/Im3 complex and that the immunity protein-T domain interface constitutes 40% of the buried surface area (43). We thus investigated the binding of ColE3 and ColE3/Im3 to TolB by SPR to see if a similar effect of the presence of Im3 was observed. The results were qualitatively very similar to those obtained with ColE9/Im9, with KD values for the interaction with TolB of 0.92 ± 0.02 μM for ColE3 and 7.07 ± 0.13 μM for ColE3/Im3.
The results clearly show that the KD of the interaction of the T plus R domain of ColE9 with TolB was very similar to that of ColE9 itself. It is interesting that measuring the affinity of binding for TolB by isothermal titration calorimetry (ITC) revealed ∼1 μM KD values for both ColE9 and ColE9/Im9 (S. Loftus and C. Kleanthous, personal communication), which are very similar to the KD value that we have obtained with free ColE9 by SPR. These results suggest that the specific binding of Im9 to the DNase domain of ColE9 in some as yet unexplained way affects binding of the TolB box region to TolB in an SPR experiment but not in an ITC experiment. Differences between the KD values obtained by ITC and SPR have also been seen in studies of the protein-protein interaction between colicin A and the TolA domain III. This interaction was detected by SPR (9) but not by ITC (14), a result that we have also repeated with the ColA T domain and TolA domain III proteins by using these two methods (data not shown).
Residues that restore activity to TolB box mutants.
The TolB box is contained within an 83-residue sequence that has been shown by NMR to be disordered in solution (6), consistent with crystallographic studies of the related ColE3 in complex with its inhibitor protein Im3 which found that the N-terminal 83 residues were not visible in the electron density map (36). There is no published information on the role of any of the essential TolB box residues (12) in the protein-protein interaction with TolB. Similarly, although it has been reported that it is the C-terminal β-propeller domain of TolB that interacts with the T domain of ColE9 (5), there are no reported mutants in TolB that affect this interaction. In order to obtain information on the possible role of essential TolB box residues in the interaction with TolB, we designed mutagenic primers to randomly mutate the alanine residue of a biologically inactive, ColE9 TolB box mutant to any residue.
Starting with the ColE9 D35A mutant template, a total of 353 clones were screened for activity, of which 3 (0.8%) were active against E. coli JM83 cells in stab tests. These were sequenced, and two contained a different aspartate codon (GAT or GAC) and the other contained a serine codon (AGC) (Table 2). GAT is the codon that occurs in the wild-type ColE9 gene at this position. The titer of the purified ColE9 D35S/Im9 mutant protein complex was identical to that of ColE9/Im9 in a large-plate activity assay (data not shown). Of the inactive clones obtained in this experiment, 45 were tested for expression of full-length ColE9 using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Fourteen of the 22 clones expressing full-length ColE9 and Im9 were sequenced. Six of these contained additional mutations and therefore were discounted. The remaining eight clones gave the results shown in Table 2, which revealed eight different residues that do not restore colicin activity in place of the normal D35 residue in ColE9.
TABLE 2.
Mutations that restore colicin activity in the TolB box
| D35A clones
|
S37A clones
|
W39A clones
|
|||
|---|---|---|---|---|---|
| Activea | Inactiveb | Active | Inactive | Active | Inactive |
| Asp GAT | Asn AAC | Ser TCA (6) | Gln CAA (3) | Trp TGG | Arg CGC (2) |
| Asp GAC | Ile ATT | Ser TCC (2) | Phe TTT | Phe TTT (3) | Ser TCC (2) |
| Ser AGC | Thr ACC | Ser TCT (2) | Pro CCT | Phe TTC (3) | Ser TTC |
| His CAC | Ser TCG | Pro CCC | Tyr TAT (2) | Ala GCG | |
| Leu ATA | Ser AGC | Asp GAC | Tyr TAC (2) | Gln CAA | |
| Phe TTC | Thr ACA (3) | Asn AAT | His CAT | ||
| Arg AGA | Thr ACC | Leu TTG | Ala GCA | ||
| Ala GCA | Thr ACT | Ala GCC | Thr ACA | ||
| Met ATG | Val GTT | ||||
| Ile ATC | |||||
| Ile ATT | |||||
| Ile ATA | |||||
Mutations that can replace residue 35 and restore colicin activity, with the codon found at this position in ColE9 shown in bold.
Mutations that do not restore colicin activity to the ColE9 D35A/Im9 complex. The number in brackets shows how many clones containing that codon were identified.
With the ColE9 S37A template, a total of 290 clones were screened for activity against E. coli JM83 cells. Thirty-two (11%) of these expressed an active colicin, and 15 of these were sequenced, giving the results shown in Table 2. The S37 codon is TCA in ColE9, and six of the active clones sequenced contained this codon, while four other serine codons were also recovered. Threonine can also be tolerated at residue 37 (with three different codons being observed) with no significant difference in biological activity when compared to ColE9 (data not shown). Eleven inactive clones that expressed full-length colicin were sequenced with the results shown in Table 2.
A total of 296 clones were produced after mutagenesis of the ColE9 W39A DNA template. From this number, 21 (7%) were active against E. coli JM83 cells in a stab test. In addition to the tryptophan codon TGG, six phenylalanine mutations were isolated (from both possible codons) and four that encoded tyrosine (from both possible codons) at this position (Table 2). The ColE9 W39F mutant protein showed a colicin activity within one doubling dilution of that of ColE9 (data not shown). The ColE9 A39Y mutant protein produced a hazy zone in the activity assay; therefore, it was impossible to determine an accurate titer. Fourteen of the inactive clones that expressed a full-length ColE9 protein were sequenced (Table 2).
SPR analysis of some of the ColE9/Im9 mutant complexes revealed a KD for binding to TolB of 12 μM for the D35S mutant and 24.3 μM for the S37T mutant compared to the value of 13.8 μM obtained for ColE9/Im9. The results obtained with this strategy have revealed information about alternative residues that can be tolerated within the TolB box sequence of ColE9 that will be discussed in the context of the possible role of residues 35, 37, and 39 in an extended TolB box sequence.
DISCUSSION
Our understanding of protein structure has been changed in recent years by the discovery that many proteins are unfolded, or partly folded, in their native states and only fold into an ordered structure on binding a partner protein (10, 11, 40, 47). Coupling protein folding to a protein-protein interaction could (i) contribute to the specificity of the intermolecular recognition event (37), (ii) enhance the rate of the protein-protein interaction (32, 35), (iii) allow a protein to bind to several different target molecules (11, 22), and (iv) provide large protein-binding surfaces in relatively small proteins (17). The intrinsically disordered N-terminal domain of colicins such as ColA, ColN, ColE3, and ColE9 bind one or more interacting proteins and are a valuable model system to study protein-protein interactions (6, 27, 39).
The KD value obtained from SPR experiments of 13.8 μM for the interaction of ColE9/Im9 with TolB is the first published data on the affinity of enzymatic E colicin/Im protein complex for a Tol protein and is similar to the value of 10.4 μM previously reported for the interaction of colicin A with TolB (14). To our surprise, removal of the Im9 protein from the ColE9/Im9 protein complex, or from ColE9/Im9 mutant protein complexes, resulted in a significant increase in the affinity of binding of the free ColE9 or free ColE9 mutants to TolB in SPR experiments. Data from NMR experiments have suggested little effect on the N-terminal 83 residues of the T domain of ColE9 resulting from Im9 binding (6), but Walker et al. (43) showed a possible effect of the presence of Im3 on the binding of the T domain of ColE3 to TolB from the presence of a significant interface between the T domain of ColE3 and Im3 in the crystal structure of the ColE3/Im3 complex (36). As it is not known if the immunity protein of enzymatic colicins is still attached to the catalytic domain when the TolB box of the translocation domain makes contact with TolB, the influence of Im9 cannot be discounted. Indeed, a reduction in the affinity of the colicin/immunity interaction associated with a noncognate interaction increased the affinity of the TolB box toward TolB (Fig. 4). ITC experiments (S. Loftus and C. Kleanthous, personal communication) investigating the interaction of TolB with free ColE9 have demonstrated very similar KD values to the results produced here. However, no apparent differences in the KD values of the interaction of TolB between free ColE9 or ColE9/Im9 complex were detected in the ITC experiment (S. Loftus and C. Kleanthous, personal communication), indicating that care needs to be taken in the interpretation of SPR experiments involving the interaction of ColE9/Im9 or ColE3/Im3 complexes with TolB.
NMR data indicated that binding of TolB affects residues 34 to 44 of ColE9 (6). This led the authors to suggest that this region may represent an extended TolB box of ColE9 or that binding of TolB to this largely unstructured region of the T domain of ColE9 may cause a conformational change that extends to affect a cluster of adjoining residues. An extension of the TolB box in ColA to include residues S16, S17, and E18 (Fig. 1) had earlier been suggested based upon sequence alignments and deletion analysis (2). The studies reported here provide direct experimental evidence, through mutational and biophysical analysis, that the TolB box of ColE9 is indeed larger than the originally described pentapeptide sequence. The ColE9 S34A, S40A, E42A, N44A, and W46A mutant protein/Im9 complexes are able to bind to the colicin receptor, BtuB, and retain DNase activity. However, each of these mutant protein complexes, with the exception of the N44A mutant, is inactive in vivo due to an inability to interact with TolB, as clearly demonstrated by SPR. The NMR spectra of the D35A and S37A mutations present in a fusion protein consisting of the N-terminal 61 residues of ColE9 connected by an 8-residue linker to the DNase domain (T1-61-DNase) have shown that the major effects of these alanine mutations are limited to the substituted amino acid and its near neighbors (39) and do not perturb the amino acid clusters around the tryptophan residues of the disordered polypeptide, which are the key structural features of the translocation domain.
The data obtained with the ColE9 S41A, N43A, and W46A/Im9 mutants demonstrated reduced affinities of binding to TolB, compared with ColE9/Im9, that are relative to their biological activities in the lux assay, with the W46A/Im9 protein demonstrating the lowest affinity for TolB and relative biological activity (Table 1). The properties of the ColE9 N44A mutant protein/Im9 complex are surprising in that it binds to TolB with similar affinity to the ColE9/Im9 complex, but it exhibits biological activity only at very high concentrations in the large-plate assay (Fig. 2) and has only 3% of the DNA damage-inducing activity of the ColE9/Im9 complex in the lux reporter assay (Table 1). We thus predict that residue N44 is not involved in binding to TolB but may have an alternative, important role in the translocation process.
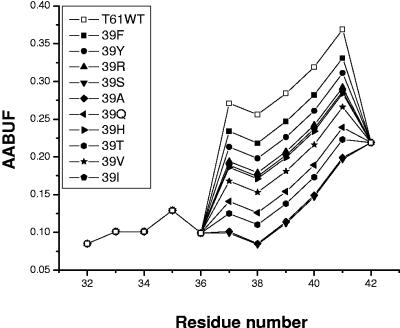
The extended TolB box residues of ColE9 that we have identified may interact directly with TolB residues, although this has not been shown, or they could be essential for maintaining intramolecular interactions that stabilize the structure of the already recognized TolB box to allow this region to form a direct interaction with TolB. The issue of whether an inactivating mutation has an indirect effect on a protein-protein interaction through causing structural changes or a direct effect through leaving a structure unchanged but removing a functional group that interacts with the partner protein is one that is present in all mutational studies. For globular proteins, usually the measurement of some structural indicator, such as the peptide circular dichroism spectra or tryptophan fluorescence spectra, is sufficient to resolve this, but for the T1-61 region of ColE9, neither of these methods provides conformational information because of the intrinsic disorder of the peptide. The intrinsic disorder not only makes characterization of structural features difficult but also makes the conformational properties more susceptible to change induced by mutagenesis than is the case for well-structured globular proteins. This is clearly shown by a combined NMR and mutagenesis study of Rac1 binding to the disordered domain of RhoGD1 (15), which highlighted the role of mutated residues in flexible proteins affecting the transient structure of the free state of RhoGD1 important for binding Rac1. In the absence of heteronuclear NMR data for all of the mutants reported in this paper, we have sought to provide some insight into whether the mutations have materially affected the conformational properties of the free T1-61 region using a predictive approach. This is based on a correlation between the formation of ordered clusters of interacting residues in the TolB box binding region and the size and polarity of amino acid side chains as indicated by their average area buried upon folding (AABUF) (33). As noted in the introduction, 15N NMR relaxation time analyses revealed that the extended TolB box region consists of dynamic clusters of interacting residues, mainly centered around tryptophan residues at positions 39, 46, and 56, and showed that the W39A variant in particular had a profound effect on the local structure of the TolB box and affected the interacting cluster around residues 55 to 60 (39). There is some minor clustering of polar residues at the extreme N terminus of the translocation domain, and therefore the clustering is not entirely hydrophobic in character although the observed correlation with AABUF (33) shows that the local dynamics of the TolB box are determined partly by the size and polarity of the relevant amino acid side chains (39). A detailed analysis of the AABUF calculations of the TolB box region show a strong correlation of the effect of the W39A mutation on the AABUF with the concomitant reduction in clustering around this residue (39). The effect on the AABUF of mutating residue 39 of T1-61-DNase to those residues identified as active and inactive varied by ±52% and is shown in Fig. 5. The inactive alanine variant showed the lowest AABUF compared to the wild-type protein, with the active Phe and Tyr variants more similar to the wild-type protein. In between these variants, however, there is a gradual variation in AABUF for the remaining inactive substitutions. Similar plots of AABUF for the Asp35 and Ser37 active and inactive variants varied by only ±5.5% over all mutations of residue 35 and by ±25.5% for mutations of residue 37 (data not shown). AABUF plots of the active D35S and S37T variants were most similar to the D35 and S37 plots, respectively. Therefore, the AABUF results suggest that Ser, Asn, Thr, and Ala could replace Asp at position 35 and that Thr, Pro, Asp, Asn, and Ala could replace Ser at position 37, with clusters similar to those of the wild-type protein being formed. However, size and polarity are not the only things required to form functional clusters, as our alanine mutagenesis data indicate by revealing that the presence of an Ala, Asn, or Thr residue at position 35 or an Ala, Asn, Asp, or Pro at position 37 results in an inactive colicin (Table 2). We assume that residues 35 and 37 are TolB-contact residues in the complex, positioned appropriately for interaction by the clustering of the free T1-61 region, and that both form polar interactions with TolB (hydrogen bonds or salt bridge in the case of Asp35). In such a case both Pro and Ala will be inactive as they do not have side-chain polarities sufficient for similar intermolecular interactions. Furthermore, Thr and Asn are both more bulky, less flexible, and less likely to form β-turns than Ser and Asp (28), while both Asp and Asn are considerably more polar than Ser (16), prefer to be exposed (19), and have slightly greater radii of gyration (24) than both of the active residues Thr and Ser. The fact that Thr can be tolerated at position 37 and not at position 35 presumably indicates differences in the stereochemical requirements of the residues at each position.
FIG. 5.
Plots of the AABUF for the T1-61-DNase fusion protein and the mutants at position 39. AABUF values were calculated with the ExPASy tool ProtScale (http://us.expasy.org/tools/protscale.html) and normalized from 0 to 10.
Our SPR data demonstrate an important role for residues 34, 40, 42, and 46 in binding to TolB but do not resolve whether these residues interact directly with TolB residues. A model of how this stretch of up to 13 residues might interact with TolB has recently been proposed (39). The 13 residues of the extended TolB box are too small to fold into a globular domain but are predicted to have structure. The amino acids with the highest propensity for forming β-turns are glycine, asparagine, proline, serine, and aspartate (7), and these constitute 10 out of 13 residues in the sequence between 34 and 46. Predictions using the Beta-TurnPrediction server suggest that the TolB binding sequence will consist of two to four β-turns that form a series of turns that runs across the surface of TolB (39). A precedent for this kind of structure has been observed with the protein methanol dehydrogenase that consists of two domains, a 66-kDa eight-bladed β-propeller catalytic domain and an 8.5-kDa domain of unknown function that is essential for activity and is layered across the surface of the β-propeller in an extended fashion, with the N-terminal 30 residues forming a series of turns (13). The observed effects of alanine mutations on the clustering of residues in the largely unstructured T domain of ColE9 implies that there are side-chain interactions between specific residues in the TolB box that may be responsible for maintaining an element of local structure rather than specific interactions with TolB. Determination of the X-ray structure of the T domain of ColE9 bound to TolB will be required to confirm the model of binding to TolB.
Acknowledgments
We are grateful to Philip Bardelang for his assistance with the SPR experiments.
This work was supported by BBSRC Studentships (S.H. and L.E.H.) and a Wellcome Trust Programme grant (M.V.).
REFERENCES
- 1.Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84:413-421. [DOI] [PubMed] [Google Scholar]
- 2.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27:143-157. [DOI] [PubMed] [Google Scholar]
- 3.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 4.Cao, Z., and P. E. Klebba. 2002. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie 84:399-412. [DOI] [PubMed] [Google Scholar]
- 5.Carr, S., C. N. Penfold, V. Bamford, R. James, and A. M. Hemmings. 2000. The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Structure Fold. Des. 8:57-66. [DOI] [PubMed] [Google Scholar]
- 6.Collins, E. S., S. B. Whittaker, K. Tozawa, C. MacDonald, R. Boetzel, C. N. Penfold, A. Reilly, N. J. Clayden, M. J. Osborne, A. M. Hemmings, C. Kleanthous, R. James, and G. R. Moore. 2002. Structural dynamics of the membrane translocation domain of colicin E9 and its interaction with TolB. J. Mol. Biol. 318:787-804. [DOI] [PubMed] [Google Scholar]
- 7.Creighton, T. E. 1993. Proteins: structures and molecular properties, 2nd ed. W. H. Freeman, New York, N.Y.
- 8.Davidov, Y., R. Rozen, D. R. Smulski, T. K. Van Dyk, A. C. Vollmer, D. A. Elsemore, R. A. LaRossa, and S. Belkin. 2000. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat. Res. 466:97-107. [DOI] [PubMed] [Google Scholar]
- 9.Derouiche, R., G. Zeder-Lutz, H. Bénédetti, M. Gavioli, A. Rigal, C. Lazdunski, and R. Lloubès. 1997. Binding of colicins A and E1 to purified TolA domains. Microbiology 143:3185-3192. [DOI] [PubMed] [Google Scholar]
- 10.Dunker, A. K., C. J. Brown, J. D. Lawson, L. M. Iakoucheva, and Z. Obradovic. 2002. Intrinsic disorder and protein function. Biochemistry 41:6573-6582. [DOI] [PubMed] [Google Scholar]
- 11.Dyson, H. J., and P. E. Wright. 2002. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12:54-60. [DOI] [PubMed] [Google Scholar]
- 12.Garinot-Schneider, C., C. N. Penfold, G. R. Moore, C. Kleanthous, and R. James. 1997. Identification of residues in the putative TolA box which are essential for the toxicity of the endonuclease toxin colicin E9. Microbiology 143:2931-2938. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh, M., C. Anthony, K. Harlos, M. G. Goodwin, and C. C. F. Blake. 1995. The refined structure of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens at 1.94 Å. Structure 3:177-187. [DOI] [PubMed] [Google Scholar]
- 14.Gokce, I., E. M. Raggett, Q. Hong, R. Virden, A. Cooper, and J. H. Lakey. 2000. The TolA-recognition site of colicin N. ITC, SPR and stopped-flow fluorescence define a crucial 27-residue segment. J. Mol. Biol. 304:621-632. [DOI] [PubMed] [Google Scholar]
- 15.Golovanov, A. P., D. Hawkins, I. Barsukov, R. Badii, G. Bokoch, L. Lian, and G. Roberts. 2001. Structural consequences of site-directed mutagenesis in flexible protein domains: NMR characterization of the L(55,56)S mutant of RhoGDI. Eur. J. Biochem. 268:2253-2260. [DOI] [PubMed] [Google Scholar]
- 16.Grantham, R. 1974. Amino acid difference formula to help explain protein evolution. Science 185:862-864. [DOI] [PubMed] [Google Scholar]
- 17.Gunasekaran, K., C. J. Tsai, S. Kumar, D. Zanuy, and R. X. Nussinov. 2003. Extended disordered proteins: targeting function with less scaffold. Trends Biochem. Sci. 28:81-85. [DOI] [PubMed] [Google Scholar]
- 18.James, R., C. Kleanthous, and G. R. Moore. 1996. The biology of E colicins: paradigms and paradoxes. Microbiology 142:1569-1580. [DOI] [PubMed] [Google Scholar]
- 19.Janin, J., and S. Wodak. 1978. Conformation of amino acid side-chains in proteins. J. Mol. Biol. 125:357-386. [DOI] [PubMed] [Google Scholar]
- 20.Journet, L., E. Bouveret, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 2001. Import of colicins across the outer membrane of Escherichia coli involves multiple protein interactions in the periplasm. Mol. Microbiol. 42:331-344. [DOI] [PubMed] [Google Scholar]
- 21.Kirkup, B. C., and M. A. Riley. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428:412-414. [DOI] [PubMed] [Google Scholar]
- 22.Kriwacki, R. W., L. Hengst, L. Tennant, S. I. Reed, and P. E. Wright. 1996. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA 93:11504-11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazzaroni, J. C., J. F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84:391-397. [DOI] [PubMed] [Google Scholar]
- 24.Levitt, M. 1976. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 104:59-107. [DOI] [PubMed] [Google Scholar]
- 25.Li, W., S. J. Hamill, A. M. Hemmings, G. R. Moore, R. James, and C. Kleanthous. 1998. Dual recognition and the role of specificity-determining residues in colicin E9 DNase-immunity protein interactions. Biochemistry 37:11771-11779. [DOI] [PubMed] [Google Scholar]
- 26.Li, W., A. H. Keeble, C. Giffard, R. James, G. R. Moore, and C. Kleanthous. 2004. Highly discriminating protein-protein interaction specificities in the context of a conserved binding energy hotspot. J. Mol. Biol. 337:743-759. [DOI] [PubMed] [Google Scholar]
- 27.Macdonald, C. J., K. Tozawa, E. S. Collins, C. N. Penfold, R. James, C. Kleanthous, N. J. Clayden, and G. R. Moore. 2004. Characterisation of a mobile protein-binding epitope in the translocation domain of colicin E9. J. Biomol. NMR 30:81-96. [DOI] [PubMed] [Google Scholar]
- 28.Pellequer, J. L., E. Westhof, and M. H. Van Regenmortel. 1993. Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol. Lett. 36:83-99. [DOI] [PubMed] [Google Scholar]
- 29.Penfold, C. N., C. Garinot-Schneider, A. M. Hemmings, G. R. Moore, C. Kleanthous, and R. James. 2000. A 76-residue polypeptide of colicin E9 confers receptor specificity and inhibits the growth of vitamin B12-dependent Escherichia coli 113/3 cells. Mol. Microbiol. 38:639-649. [DOI] [PubMed] [Google Scholar]
- 30.Penfold, C. N., B. Healy, N. G. Housden, R. Boetzel, M. Vankemmelbeke, G. R. Moore, C. Kleanthous, and R. James. 2004. Flexibility in the receptor-binding domain of the enzymatic colicin E9 is required for toxicity against Escherichia coli cells. J. Bacteriol. 186:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilsl, H., and V. Braun. 1995. Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol. Microbiol. 16:57-67. [DOI] [PubMed] [Google Scholar]
- 32.Pontius, B. W. 1993. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem. Sci. 18:181-186. [DOI] [PubMed] [Google Scholar]
- 33.Rose, G. D., A. R. Geselowitz, G. J. Lesser, R. H. Lee, and M. H. Zehfus. 1985. Hydrophobicity of amino acid residues in globular proteins. Science 229:834-838. [DOI] [PubMed] [Google Scholar]
- 34.Sarkar, G., and S. S. Sommer. 1990. The megaprimer method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 35.Shoemaker, B. A., J. J. Portman, and P. G. Wolynes. 2000. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. USA 97:8868-8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soelaiman, S., K. Jakes, N. Wu, C. Li, and M. Shoham. 2001. Crystal structure of Colicin E3: Implications for cell entry and ribosome inactivation. Mol. Cell 8:1053-1062. [DOI] [PubMed] [Google Scholar]
- 37.Spolar, R. S., and M. T. J. Record. 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science 263:777-784. [DOI] [PubMed] [Google Scholar]
- 38.Tan, Y., and M. A. Riley. 1996. Rapid invasion by colicinogenic Escherichia coli with novel immunity functions. Microbiology 142:2175-2180. [DOI] [PubMed] [Google Scholar]
- 39.Tozawa, K., C. J. MacDonald, C. N. Penfold, R. James, C. Kleanthous, N. J. Clayden, and G. R. Moore. Order in an intrinsically disordered protein: the TolB binding region of colicin E9. Biochemistry, in press. [DOI] [PubMed]
- 40.Uversky, V. N. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11:739-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vankemmelbeke, M., B. Healy, G. R. Moore, C. Kleanthous, C. N. Penfold, and R. James. 2005. Rapid detection of colicin E9 induced DNA damage using Escherichia coli cells carrying SOS promoter::lux fusions. J. Bacteriol. 187:4900-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vetter, I. R., M. W. Parker, A. D. Tucker, J. H. Lakey, F. Pattus, and D. Tsernoglou. 1998. Crystal structure of a colicin N fragment suggests a model for toxicity. Structure 6:863-874. [DOI] [PubMed] [Google Scholar]
- 43.Walker, D., G. R. Moore, R. James, and C. Kleanthous. 2003. Thermodynamic consequences of bipartite immunity protein binding to the ribosomal ribonuclease colicin E3. Biochemistry 42:4161-4171. [DOI] [PubMed] [Google Scholar]
- 44.Wallis, R., K. Y. Leung, A. J. Pommer, H. Videler, G. R. Moore, R. James, and C. Kleanthous. 1995. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the millimolar to femtomolar affinity range. Biochemistry 34:13751-13759. [DOI] [PubMed] [Google Scholar]
- 45.Wallis, R., A. Reilly, A. Rowe, G. R. Moore, R. James, and C. Kleanthous. 1992. In vivo and in vitro characterization of overproduced colicin E9 immunity protein. Eur. J. Biochem. 207:687-695. [DOI] [PubMed] [Google Scholar]
- 46.Wiener, M., D. Freymann, P. Ghosh, and R. M. Stroud. 1997. Crystal structure of colicin Ia. Nature 385:461-464. [DOI] [PubMed] [Google Scholar]
- 47.Wright, P. E., and H. J. Dyson. 1999. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293:321-331. [DOI] [PubMed] [Google Scholar]
- 48.Zakharov, S. D., V. Y. Eroukova, T. I. Rokitskaya, M. V. Zhalnina, O. Sharma, P. J. Loll, H. I. Zgurskaya, Y. N. Antonenko, and W. A. Cramer. 2004. Colicin occlusion of OmpF and TolC channels: outer membrane translocons for colicin import. Biophys. J. 87:3901-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]