Abstract
Eukaryotic Sm and Sm-like proteins associate with RNA to form the core domain of ribonucleoprotein particles involved in pre-mRNA splicing and other processes. Recently, putative Sm proteins of unknown function have been identified in Archaea. We show by immunoprecipitation experiments that the two Sm proteins present in Archaeoglobus fulgidus (AF-Sm1 and AF-Sm2) associate with RNase P RNA in vivo, suggesting a role in tRNA processing. The AF-Sm1 protein also interacts specifically with oligouridylate in vitro. We have solved the crystal structures of this protein and a complex with RNA. AF-Sm1 forms a seven-membered ring, with the RNA interacting inside the central cavity on one face of the doughnut-shaped complex. The bases are bound via stacking and specific hydrogen bonding contacts in pockets lined by residues highly conserved in archaeal and eukaryotic Sm proteins, while the phosphates remain solvent accessible. A comparison with the structures of human Sm protein dimers reveals closely related monomer folds and intersubunit contacts, indicating that the architecture of the Sm core domain and RNA binding have been conserved during evolution.
Keywords: Archaea/RNA binding/RNase P RNA association/Sm proteins/X-ray structure
Introduction
Eukaryotic Sm and Sm-like (Lsm) proteins are involved in a variety of RNA processing events including pre-mRNA splicing (Mattaj et al., 1993; He and Parker, 2000; Pannone and Wolin, 2000), telomere replication (Seto et al., 1999), histone mRNA processing and mRNA degradation (Boeck et al., 1998; Bouveret et al., 2000; Tharun et al., 2000). Members of this protein family share a common bipartite sequence motif, known as the Sm domain, consisting of two conserved segments separated by a region of variable length and sequence (Hermann et al., 1995; Séraphin, 1995). They assemble in at least three different complexes each containing seven distinct members of the Sm or Lsm protein family (Achsel et al., 1999; Mayes et al., 1999; Salgado-Garrido et al., 1999; Bouveret et al., 2000; Tharun et al., 2000). These complexes interact with various RNAs including the spliceosomal U small nuclear (sn) RNAs, pre-RNase P RNA, telomerase RNA, viral RNA and other RNAs. In the eukaryotic spliceosomal U1, U2, U4/U6 and U5 small nuclear ribonucleoprotein particles (snRNPs), the seven canonical Sm proteins bind to the so-called Sm site, a uridine-rich single-stranded region of the U snRNA (Branlant et al., 1982), forming the Sm core RNP particle or Sm core domain. In vitro experiments indicate that a native-like Sm core domain can be assembled using a nonameric Sm site oligonucleotide (Raker et al., 1999). Electron micrographs of negatively stained human canonical Sm core domains and a recent cryo-electron microscopic study of the human U1 snRNP show doughnut-shaped ring structures with a diameter of ∼70–80 Å (Kastner et al., 1990; Stark et al., 2001). Particles of similar size and shape were also observed for the RNA-free complexes formed by human Lsm proteins, which were shown to interact with the 3′-terminal U tract of U6 snRNA (Achsel et al., 1999). Based on the crystal structures of the human dimeric D3B and D1D2 Sm protein complexes, Nagai and co-workers have proposed a heptameric ring model for the human core domain consistent with the available biochemical and electron microscopic (EM) observations (Kambach et al., 1999a). According to their model, the Sm site RNA is binding inside the doughnut-shaped ring structure involving conserved residues near loops 3 and 5 of the Sm proteins.
Recently, database sequence searches have revealed the presence of Sm-related proteins of unknown function in Archaea (Salgado-Garrido et al., 1999). These Sm-related proteins share the Sm domain with the eukaryotic Sm and Lsm proteins, but in general do not contain any C-terminal extensions present in some of the eukaryotic proteins. In contrast to eukaryotes, archaeal genomes encode a maximum of one (in the Pyrococcus family) or two Sm-related proteins, which appear to belong to two subfamilies we will refer to as Sm1 and Sm2 (Salgado-Garrido et al., 1999). Within the Sm1 family, there is high sequence homology (up to 60% sequence identity), while the identity level is only ∼30% or less between Sm1 and Sm2 proteins from the same organism (Figure 1A). As part of a structural and functional analysis of Sm-related proteins in Archaea, we report here the crystal structures of the Archaeoglobus fulgidus AF-Sm1 protein and its complex with a uridine oligonucleotide, providing the first high-resolution picture of an Sm core domain. Our results indicate that its architecture and the mode of RNA binding have been conserved during evolution, and suggest how specific binding to the U-rich Sm site occurs in the human Sm core. We further show by immunoprecipitation and bandshift experiments that the A.fulgidus Sm1 and Sm2 proteins associate with RNase P RNA in vivo, and bind RNA in vitro.
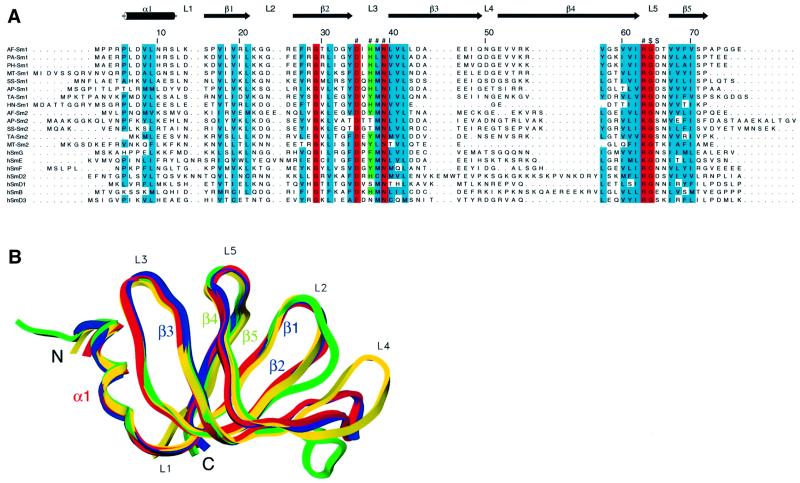
Fig. 1. The Sm fold is conserved between Archaea and eukaryotes. (A) Sequence alignment of archaeal Sm proteins with the human canonical Sm (hSm) proteins G, E, F, D2, D1, B and D3. Shown on the top is the secondary structure assignment in AF-Sm1 as determined by X-ray analysis. Loop regions are labelled L1–L5, β-strands β1–β5; α1 denotes the N-terminal α-helix. β-strands 1, 2 and 3 constitute the first, and strands 4 and 5 the second half of the bipartite Sm domain. Residues fully (N39) or almost fully conserved throughout the Sm and Lsm protein family are marked in red, with highly conserved residues in blue. Residues forming the uracil-binding pocket are labelled ‘#’ or ‘$’ (if they interact through their main chain amide groups); His37, in stacking contact with the base, and the corresponding tyrosine or phenylalanine residues in human Sm proteins, are shown in green. The names of the archaeal proteins indicate the species (AF, Archaeoglobus fulgidus; PA, Pyrococcus abyssi; PH, Pyrococcus horikoshi; MT, Methanobacterium thermoautotrophicum; SS, Sulfolobus solfataricus; AP, Aeropyrum pernix; TA, Thermoplasma acidophilum; HN, Halobacterium sp. NRC-1) and the subfamily (Sm1- or Sm2-type). (B) Superposition of the Cα traces of the archaeal PA-Sm1 (our unpub lished results), AF-Sm1 and AF-Sm2 proteins with human D3 (Kambach et al., 1999a) shown in red, blue, green and yellow, respectively. Differences are restricted to the N- and C-termini as well as the loop regions, in particular loop 4.
Results and discussion
The structure of the AF-Sm1 monomer: the Sm fold is conserved between Archaea and eukaryotes
Recombinant AF-Sm1 protein was expressed in Escherichia coli and diffraction quality crystals were grown using the vapour diffusion technique (see Materials and methods for details). The structure of the AF-Sm1 protein was solved by molecular replacement using the coordinates of the A.fulgidus AF-Sm2 and the Pyrococcus abyssii PA-Sm1 proteins (our unpublished results). The resulting model, consisting of 28 copies of the AF-Sm1 protein arranged in four seven-membered rings, was refined at 2.5 Å to a final R-factor of 20.7% (Table I). The AF-Sm1 monomer has a barrel-type structure consisting of an N-terminal α-helix followed by a strongly bent five-stranded β-sheet (Figure 1B). A closely related fold was found in the crystal structures of the human dimeric D3/B and D1/D2 Sm complexes recently reported by Kambach et al. (1999a). Superposition of the archaeal and human Sm monomers shows that structural differences, presumably at least partly induced by crystal contacts, occur mainly at the N- and C-termini as well as in loop 4, which has a variable length in the human Sm proteins (Figure 1A). Within the two conserved sequence motifs of the Sm domain consisting of β-strands 1–3 and 4–5, respectively, the monomer folds closely superimpose (with r.m.s.ds of ∼0.6 Å for the Cα atoms). The comparison of the structures clearly shows that the Sm fold is conserved between Archaea and eukaryotes.
Table I. Data collection and refinement statistics.
| AF-Sm1 | AF-Sm1 RNA | |
|---|---|---|
| Wavelength (Å) | 0.842 | 0.964 |
| Resolution (Å) | 2.50 | 2.75 |
| Total no. of reflections | 255 898 | 70 211 |
| No. of unique reflections | 63 291 | 28 746 |
| Rsym (%) | 7.2 (35.3) | 5.0 (35.4) |
| Completeness (%) | 99.3 (99.9) | 97.6 (97.5) |
| <I/σI> | 14.1 (3.5) | 11.2 (2.0) |
| Rcrystal (%) (no. of reflections) | 20.7 (60126) | 22.5 (27253) |
| Rfree (%) (no. of reflections) | 26.4 (3164) | 30.0 (1469) |
| No. of protein atoms | 15 463 | 7811 |
| No. of solvent atoms | 126 | 57 |
| No. of RNA atoms | – | 182 |
| R.m.s.d. bond distance (Å) | 0.008 | 0.008 |
| R.m.s.d. bond angle (°) | 1.50 | 1.55 |
| <B> protein | 48.5 | 82.0 |
Rsym = Σhkl|I – <I>|/ΣhklI, where I is the observed intensity for a reflection of index hkl and <I> is the mean intensity. Rsym, completeness and <I/σI> for the highest resolution shell are given in parentheses. The highest resolution shells are 2.50–2.60 Å for the native and 2.75–2.90 Å for the complex structure, respectively.
AF-Sm1 forms a heptameric ring structure
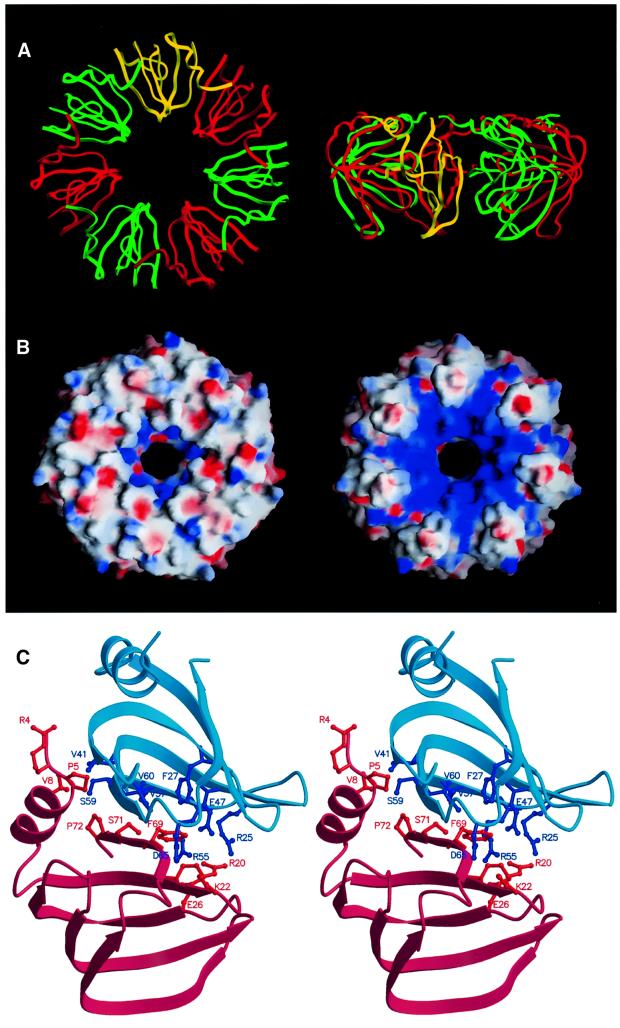
The AF-Sm1 protein forms a seven-membered, doughnut-shaped ring, ∼65 Å in diameter, ∼30 Å thick, with a central cavity ∼13 Å across (Figure 2A and B). Residues in loops 2, 3 and 5 are exposed at the inner surface of the ring, while both the N- and C-termini are located at the outer surface. The two faces of the ring-like structure are distinctly different with regard to shape and charge distribution. While the face containing the N-terminal helix is relatively smooth, there are distinct grooves separating the subunits on the other face mainly caused by the protruding loop 4. These grooves are positively charged (containing residues R20, R25, R55 and K56) and, together with lysine residues projecting into the hole (K22 in loop 2), give rise to a positively charged region surrounding the central cavity on this side of the ring (Figure 2B). Hydrogen bonding between β-strands 4 and 5 of neighbouring subunits gives rise to the formation of a continuous β-sheet in the ring-like structure. Additional salt bridges and predominantly hydrophobic contacts involving residues in the N-terminal helix as well as β-strands 4 and 5 (Figure 2C) result in an extremely stable complex, which even resists denaturating conditions (data not shown). Ultracentrifuge and EM studies show that the Sm1 proteins from A.fulgidus and P.abyssii form stable complexes in solution under a wide range of conditions, while the oligomerization of the A.fulgidus Sm2 protein is strongly dependent on the pH and the presence of RNA (data not shown). In this respect, the archaeal Sm1 proteins behave like Lsm proteins, which were shown to form ring-shaped structures in the absence of RNA (Achsel et al., 1999), while the AF-Sm2 protein resembles the canonical Sm proteins requiring RNA for stable core complex formation (Kambach et al., 1999b).
Fig. 2. Structure of the AF-Sm1 heptamer. (A) Ribbon representation of the AF-Sm1 heptamer (top and side view). For clarity, the monomers are drawn alternately in red and green, and one monomer is depicted in yellow. (B) Electrostatic surface charge potential showing the two faces of the seven-membered ring. Shown on the left is the side binding the RNA and containing the N-terminal helix (corresponding to the top view shown in A). It is relatively flat, while the other side exhibits pronounced positively charged grooves emanating from the centre (as indicated by the blue colour). The figure was produced with GRASP (Nicholls et al., 1991). (C) Dimer contacts in the AF-Sm1 heptamer. The molecules are shown as ribbons of different colours for the pair of interacting molecules. Side chains involved in contacts (<4.0 Å), mainly located on β-strands 4 and 5 as well as the N-terminal helix, are represented in ball-and-stick mode. The figure was produced with MOLSCRIPT (Kraulis, 1991).
The intersubunit contacts described above for the archaeal AF-Sm1 complex are similar to the dimer interfaces seen in the X-ray structures of the human D3/B and D1/D2 Sm complexes (Kambach et al., 1999a) and consistent with mutagenesis studies (Camasses et al., 1998). The structure of the homo-heptameric archaeal AF-Sm1 complex strengthens the model for the human Sm core domain (Kambach et al., 1999a) derived on the basis of the dimer structures and supports the presence of a single copy of each canonical Sm B/D1/D2/D3/E/F/G per complex. Consistently, doughnut-shaped ring structures with outer diameters of ∼70–80 Å have been observed in EM studies of human canonical Sm core RNPs, human U1 snRNP and RNA-free Lsm complexes (Kastner et al., 1990; Achsel et al., 1999; Stark et al., 2001). We conclude that not only the Sm fold but also the architecture of the Sm core domain have been conserved during evolution and are similar in Archaea and eukaryotes.
RNA binding to AF-Sm1: structure of a primitive RNP core domain
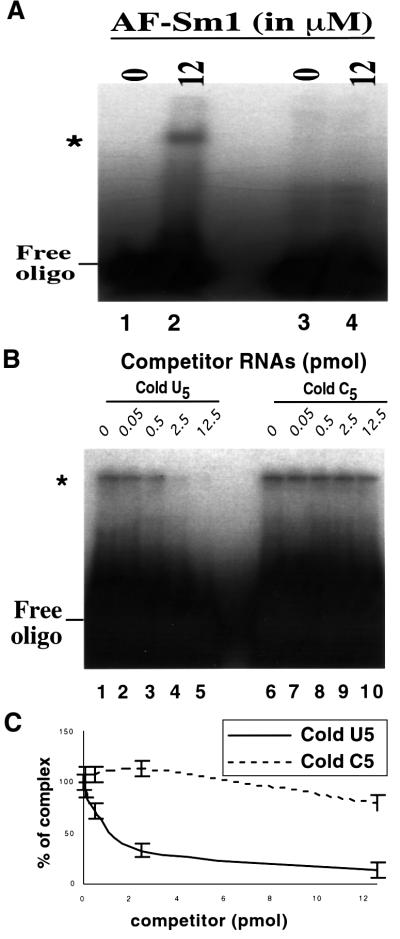
As eukaryotic Sm and Lsm proteins interact with U-rich RNA sequences containing stretches of 4–6 uridines (Branlant et al., 1982; Heinrichs et al., 1992; Achsel et al., 1999; Raker et al., 1999), we tested whether archaeal Sm proteins also bind to such sequences. Gel shift experiments demonstrated that AF-Sm1 interacts weakly with a synthetic Sm site but more strongly with a U5 oligonucleotide (data not shown). Binding to U5 is specific as no interaction was detected with a C5 RNA. This result was confirmed by competition experiments (Figure 3).
Fig. 3. In vitro binding of oligo(U) to AF-Sm1. (A) Direct binding assay by gel shift. Radiolabelled RNA was incubated with or without AF-Sm1 and complexes were resolved following native gel electro phoresis. U5, but not C5, produces a bandshift. A 75 fmol concentration of 32P-labelled oligouridine (lanes 1 and 2) or oligocytidine (lanes 3 and 4) was incubated alone (lanes 1 and 3) or with 1 µg of purified AF-Sm1 (lanes 2 and 4). (B) Competition experiments demonstrate the specificity of the interaction. Various concentrations of cold oligonucleotides (indicated above each lane) were incubated with the 32P-labelled oligouridine (U5) probe and 12 µM AF-Sm1. Complex formation was assayed following native gel electrophoresis. The data demonstrate successful competition by U5 (lanes 1–5) and not by C5 (lanes 6–10). This reveals specific binding of AF-Sm1 to U5. (C) Quantification of competition experiments. Cold U5 displaces AF-Sm1 from 32P-labelled U5 (solid line, n = 2). In comparison, cold C5 does not displace it except when it is present in great excess (dashed line, n = 2). Error bars are derived from two independent experiments. The slower migrating band (marked by *) corresponds to the protein–RNA complex while the faster migrating band corresponds to the free probe.
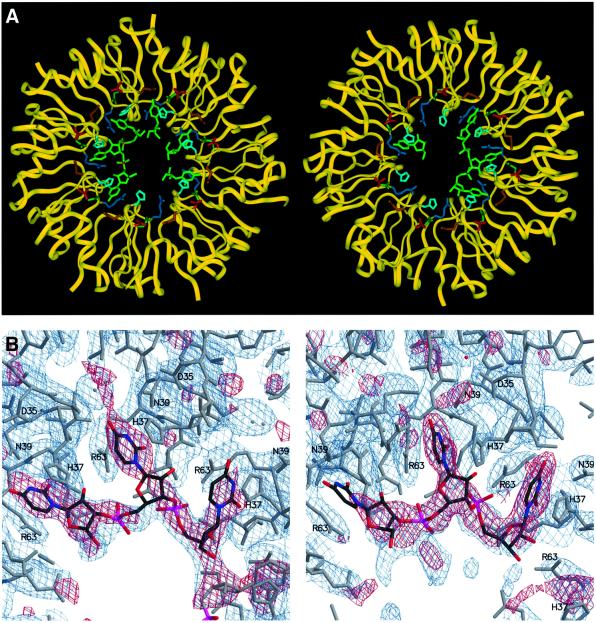
Co-crystallization of the AF-Sm1 protein with a U5 oligonucleotide yielded crystals of an RNA complex diffracting to 2.75 Å, whose structure was determined by molecular replacement using the AF-Sm1 heptamer as the search model (see Materials and methods). In both heptameric rings present in the asymmetric unit of the co-crystals, the oligonucleotide is bound in the central hole on the relatively smooth and, notably, less positively charged face of the ring (Figure 4A and B). With the exception of the orientation of a few residues interacting directly with the RNA and at the N- and C-termini, no significant conformational changes are induced by the binding of RNA. In line with the biochemical observations, the AF-Sm1 heptamer obviously represents a rigid, pre-formed RNA-binding unit.
Fig. 4. The AF-Sm1–U5 complex. (A) The two AF-Sm1 heptamers in the asymmetric unit are shown in a ribbon plot representation (yellow), and the bound oligonucleotides and residues forming the uracil-binding pocket in stick mode, with the oligo(U) coloured green, Asp35 red, His37 light blue, Met38 gold, Asn39 dark green and Arg63 blue. Continuous electron density is present for a trinucleotide as well as for three isolated uridines in the first ring (shown on the left), and for a trinucleotide and a single uridine in the second ring. (B) Portions of the 2Fo – Fc (blue) and Fo – Fc (red) omit maps not including the RNA, contoured at 1σ and 2.6σ, respectively (ring 1 on the left). Atom-type colouring is used for the oligo(U), while the protein is shown in grey. Residues interacting with the bases are labelled.
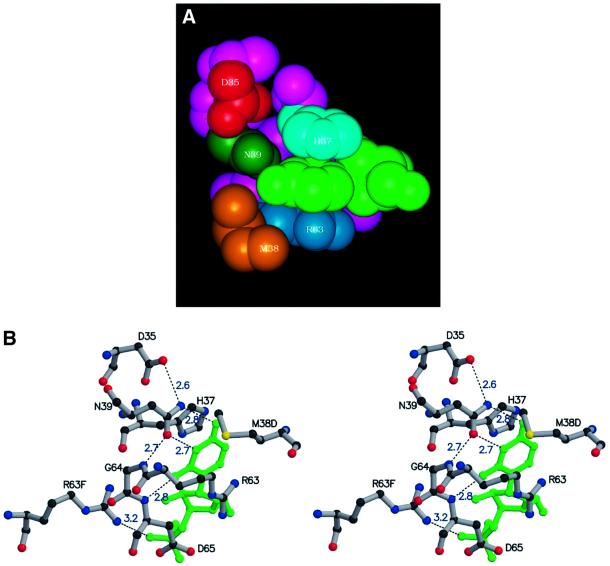
The RNA bases are firmly bound within pockets made up of residues His37, Asn39 and Arg63, located in loops 3 and 5 of the Sm monomer, which are exposed in the interior of the AF-Sm1 heptamer. The uracil ring is sandwiched between the histidine and the arginine side chains and forms hydrogen bonds to the universally conserved asparagine located at the bottom of the pocket. In what one could call the ‘fully bound state’ Asn39 is hydrogen bonded to both the O4 and N3 positions of the uracil, and in addition the O2 atom is in close contact with the main chain amide group of Asp65 (Figure 5A and B). This hydrogen bonding pattern is specific for U, since the orientation of the Asn39 side chain is fixed by contacts with the carboxylate group of Asp35 and the main chain amide group of Gly64 (Figure 5B). In addition, the side chain of Met38 from a neighbouring subunit forms the bottom of the binding pocket, close to the O4 position. This binding mode provides optimal interactions with uracil bases, even though other bases can also be accommodated, albeit with lower affinity. Binding of thymine bases in single-stranded DNA will be strongly discriminated against because of steric clashes of the 5-methyl group with the main chain carbonyl of Ile36 from a neighbouring subunit. Arg63, in addition to being involved in a stacking-type interaction with the base, in most cases forms a salt bridge to the solvent-accessible 5′-phosphate of the same nucleotide. The combination of hydrophobic and electrostatic contacts is likely to result in stable complexes, in line with observations on eukaryotic Sm and Lsm complexes (Lührmann et al., 1990; Raker et al., 1999; Stark et al., 2001). Combined stacking and polar interactions involving bases in single-stranded regions of RNA have been found in a number of RNA–protein complexes (Jones et al., 2001).
Fig. 5. The base-binding pocket. van der Waals (A) and ball-and-stick stereo (B) representation of the residues forming the uracil-binding pocket. The colour code in (A) is as in Figure 4A. The uracil is sandwiched between His37 and Arg63 and forms specific hydrogen bonds with Asn39 and the backbone NH group of Asp65 (distances in Å are indicated). Note that M38D and R63F belong to neighbouring protein subunits.
A notable feature of our X-ray analysis is the fact that we do not observe continuous density for more than three nucleotides of the bound U5 oligonucleotide and that in several binding pockets density is present only for an isolated uridine (Figure 4). The non-uniform binding of RNA in the two rings cannot be explained by a distortion of the heptamers induced by crystal packing forces. In neither of the two rings did we observe any significant deviation from 7-fold rotational symmetry. Obviously there is some disorder in the crystals and, apparently, more than one U5 molecule, only partially visible in the density, is bound in the heptameric rings. In one case, we see alternative orientations for a nucleotide (Figure 4B). The presence of isolated uridines may also be indicative of a partial averaging in the crystal, i.e. multiple binding modes of the RNA in the heptamers. We think, however, that this feature is also a consequence of the architecture of the core domain and reflects the disparity of the repeat distances in the RNA and the Sm heptamer. This allows maximally three consecutive nucleotides to bind in a similar fashion to neighbouring Sm subunits, while a fourth one would have to bind in a distinctly different manner, or may instead bind across or thread through the inner hole of the complex. A more irregular binding of the RNA with respect to the Sm proteins and threading of the RNA through the hole of the ring has been proposed to occur in eukaryotic Sm and Lsm complexes (Kambach et al., 1999a; Stark et al., 2001; Urlaub et al., 2001). Further structural studies are necessary to verify these predictions.
While the resolution of our X-ray analysis (2.75 Å) does not allow the sugar conformation of an isolated nucleotide to be defined unambiguously, it is clear that the electron density for the two trinucleotides is compatible only with a C2′-endo conformation of the riboses. It appears that the architecture of the Sm core requires the C2′-endo conformation, which is not normally found in RNA, as it increases the P–P distances and thereby enables the binding of consecutive bases in neighbouring binding pockets. Interestingly, in the UGU7 complex of the sex-lethal protein, most of the riboses also display C2′-endo sugar conformations (Handa et al., 1999).
Implications for RNA binding in the eukaryotic spliceosomal Sm core domain
The biochemical and electron microscopic data available on the binding of RNA to the Sm core domain of spliceosomal snRNPs suggest that, as in the AF-Sm1–U5 complex, a U-rich, single-stranded region of the RNA, i.e. the Sm site, binds in the inner surface of a ring-like structure (Raker et al., 1999; Stark et al., 2001; Urlaub et al., 2001). Based on the D3/B and D1/D2 Sm dimer structures and the model proposed for the human Sm core, Kambach et al. (1999a) predicted that residues Asn39, Asp35, Gly74 and Arg73, which are conserved in the human Sm core proteins, might be involved in the binding of Us within the Sm site. Indeed, most of the residues involved in RNA binding to AF-Sm1 are either fully or highly conserved in archaeal as well as eukaryotic Sm proteins (Figure 1A). In one of the seven human canonical Sm proteins (B), all residues forming the binding pocket in AF-Sm1 (marked by ‘#’ and ‘$’ in Figure 1A) are conserved, and in another (D2) only Met38 at the bottom of the pocket is replaced by a cysteine. In proteins F and G, His37 is replaced by another aromatic residue (phenylalanine or tyrosine, respectively), allowing for even stronger stacking interactions. In these four proteins, and possibly also in protein E, in which Arg63 is replaced by a lysine, we can expect the binding of the uracil bases to be the same or highly similar to what we see in the archaeal protein. This assumption is fully consistent with experiments showing that spliceosomal snRNP cores can be assembled with a U9 oligonucleotide, thereby demonstrating the high affinity binding of a single-stranded U tract to the human Sm proteins (Raker et al., 1999). Further experimental evidence for a closely related mode of binding comes from a recently published UV cross-linking study showing that His37 of the human Sm protein B/B′ and the peptide Phe37–Met38–Asn39 of the Sm G protein contact uridines in the third and first positions, respectively, of the U4 Sm site oligonucleotide (5′-AAU1UU3UUGA-3′) in a minimal Sm core RNP (Urlaub et al., 2001). These data are compatible with our stuctural results. A clearly different mode of binding is to be expected for the D1 and D3 proteins, where no stacking with an aromatic side chain is possible, since position 37 is occupied by a serine or asparagine, respectively. Consistently, no cross-linking was found to these proteins in this study. The fact that no cross-link to protein D2 was detected could either be due to the M38C substitution or, more probably, might be related to the order in which the Sm proteins are contacted by the RNA. A more detailed picture of these interactions needs to await the determination of the crystal structure of the human core domain. However, the structure of the archaeal AF-Sm1–RNA complex provides compelling arguments for the assumption that the architecture of the core domain and the specific binding of Sm proteins to U-rich sequences are very similar in eukaryotic and archaeal RNPs.
Association of the A.fulgidus Sm proteins with RNase P RNA
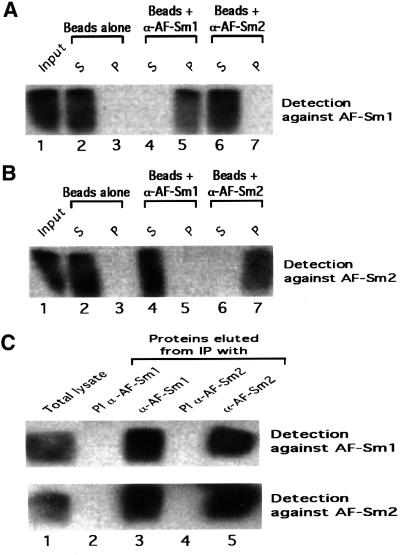
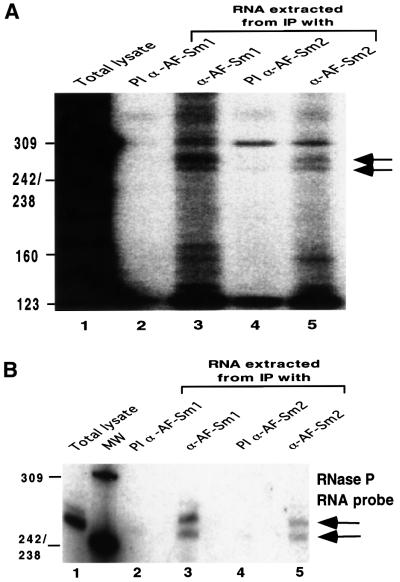
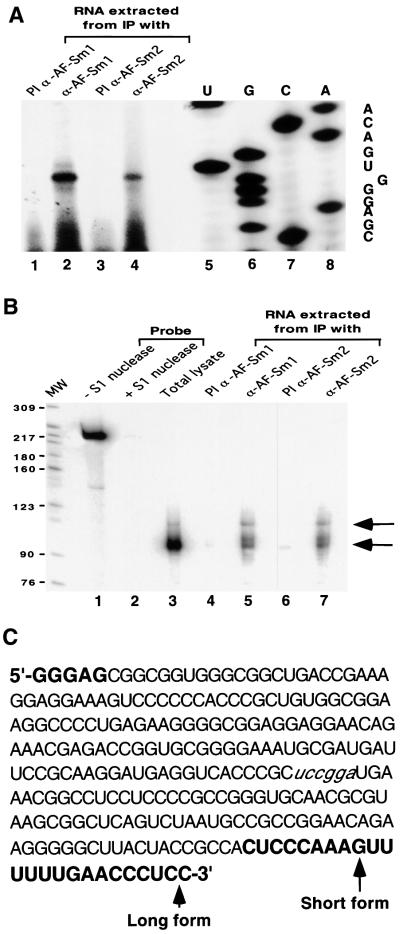
While the crystallographic data and bandshift experiments demonstrate that archaeal Sm proteins interact with RNA in vitro, they give no clue concerning the function of these factors in vivo. To investigate this question, we have raised antibodies specific for either the Sm1 or the Sm2 protein from A.fulgidus. Control western blotting experiments with recombinant proteins confirmed that the sera were specific for each protein, with no cross-reactivity (Figure 6A and B). Western blotting of proteins present in a total cell extract indicates that both the AF-Sm1 and AF-Sm2 proteins are expressed in vivo (Figure 6C, lane 1). Interestingly, AF-Sm1 was detected specifically in fractions immunoprecipitated by the AF-Sm1 and AF-Sm2 antibodies, and vice versa (Figure 6C), indicating an association of the two proteins in vivo. Protein co-precipitation was resistant to RNase pre-treatment, suggesting, but not proving, that this interaction may not be RNA mediated (data not shown). To test for the presence of RNAs in the pellet, we used pCp labelling (Uhlenbeck and Gumport, 1982). Two bands ∼250 nucleotides long were detected reproducibly in the fractions obtained from immunoprecipitation with antibodies against AF-Sm1 and AF-Sm2 and were absent in the respective pre-immune fractions (Figure 7A). Consistent with results obtained in related studies of archaeal RNP complexes (Omer et al., 2000), only a limited amount of material could be recovered, preventing direct RNA sequencing. However, the size of these RNA species suggested that they could represent RNase P RNA, which is predicted to be 248 nucleotides long in A.fulgidus (Brown, 1999). Northern blotting using several probes complementary to the RNase P RNA demonstrated that both species are derived from the RNase P RNA locus (Figure 7B and data not shown). Primer extension was used to map the exact 5′ end (Figure 8A). A single band was obtained in both the total and immunoprecipitated RNA fractions, indicating that the two RNA species share a common 5′ end. S1 nuclease mapping indicated, however, that the two species differ at their 3′ ends (Figure 8B), suggesting that they represent different forms (e.g. mature and precursor) of the A.fulgidus RNase P RNA. It is noteworthy that the 3′-extended species was precipitated more efficiently than the shorter one (Figures 7B and 8B), suggesting an effect of the 3′ U-rich extension (Figure 8C) on the interaction with archaeal Sm proteins. In this behaviour the A.fulgidus proteins resemble the human Lsm proteins, which were shown to interact with an oligo(U) stretch at the 3′ end of U6 snRNA (Achsel et al., 1999). While we have not shown that the interaction of AF-Sm1 and AF-Sm2 with the RNase P RNA is direct, this is likely to be the case given the direct interaction of eukaryotic Sm proteins with RNA and our observation that purified recombinant AF-Sm1 and AF-Sm2 are able to interact with short RNA oligonucleotides in bandshift experiments (Figure 3 and data not shown). Further studies will be required to test whether the interaction of Sm-related proteins with the RNase P RNA is conserved in other archaeal species and whether it is mediated by other proteins.
Fig. 6. Sm proteins from A.fulgidus are expressed in vivo and co-precipitate. (A) Antibodies directed against AF-Sm2 do not cross-react with AF-Sm1. A 0.5 µg aliquot of recombinant AF-Sm1 (Input, lane 1) was incubated with protein A-coated agarose beads that had been previously coupled to antibodies directed against AF-Sm1 (α-AF-Sm1, lanes 4 and 5), AF-Sm2 (α-AF-Sm2, lanes 6 and 7) or, as control, no antibodies (lanes 2 and 3). The AF-Sm1 protein present in the pellet (P, lanes 3, 5 and 7) and supernatant (S, lanes 2, 4 and 6) fractions of each immunoprecipitation reaction was detected by western blotting following gel electrophoresis using α-AF-Sm1 antibodies. Smearing of the AF-Sm1 protein results from incomplete denaturation in standard gel conditions (data not shown). No cross-reaction of α-AF-Sm2 antibodies with AF-Sm1 was observed. (B) Antibodies directed against AF-Sm1 do not cross-react with AF-Sm2. The procedure described in (A) was repeated using 0.5 µg of AF-Sm2 and detection with α-AF-Sm2 antibodies. Smearing of the AF-Sm2 protein results from incomplete denaturation in standard gel conditions (data not shown). No cross-reaction of α-AF-Sm1 antibodies with AF-Sm2 was observed. (C) Total lysate from A.fulgidus cells was prepared as indicated in Materials and methods and used in immunoprecipitation reactions (IP) with α-AF-Sm1 (lane 3), α-AF-Sm2 (lane 5) and the corresponding pre-immune serum (PI α-AF-Sm1 and PI α-AF-Sm2, lanes 2 and 4, respectively). An aliquot of the total lysate before precipitation was loaded in lane 1. The presence of AF-Sm1 and AF-Sm2 in the various fractions was detected by western blotting with antibodies against AF-Sm1 or AF-Sm2.
Fig. 7. RNase P RNA is co-immunoprecipitated by Sm proteins. (A) RNA samples were extracted from immunoprecipitated fractions and analysed by pCp labelling as described in Materials and methods. Arrows indicate two bands found reproducibly in the immuno precipitated fraction obtained with the antibodies against AF-Sm1 or AF-Sm2. (B) Northern blotting with an RNase P RNA-specific probe was carried out with the RNA obtained as described in Materials and methods. The same bands were detected in independent experiments using the same as well as two other RNase P RNA probes.
Fig. 8. RNase P RNA 5′ and 3′ end mapping. (A) RNA samples were prepared as previously described and analysed by primer extension with ST14 (see Materials and methods). A sequence ladder was obtained with the same primer and the cloned RNase P RNA gene as a template to map the 5′ end. (B) The two RNA species differ at their 3′ ends. RNA samples were analysed by S1 nuclease mapping with an overlapping 3′ end fragment. The 3′ end determination was done according to the molecular size of the band. (C) The RNase P RNA sequence obtained from 5′ and 3′ end determination. Sequences in bold letters correspond to nucleotides not present in the sequence provided at the RNase P database web site. Nucleotides in italic correspond to the 3′ end of the probe used for the S1 mapping. The 5′ end corresponds to the end determined by primer extension. Short and long 3′ end forms correspond to those determined by S1 mapping.
Concluding remarks
In providing the first X-ray structure of an Sm protein core domain bound to RNA, our data show that archaeal Sm1-type proteins assemble in seven-membered ring structures, likely to resemble closely the eukaryotic Sm core. Our results therefore strengthen the model proposed by Kambach et al. (1999a) for the human Sm core. The binding of an oligo(U) in the AF-Sm1 protein complex suggests how eukaryotic Sm proteins interact specifically with the uridine-rich Sm site of the U snRNAs present in snRNPs (Branlant et al., 1982; Raker et al., 1999) or how some Lsm proteins specifically recognize the oligo(U) sequence present at the U6 snRNA 3′ end (Achsel et al., 1999). Our data also demonstrate that the A.fulgidus Sm proteins interact with two RNase P RNA species, one of which is likely to be a precursor. In yeast, several Lsm proteins have been shown to interact with the RNase P precursor RNA that similarly contains an oligo(U) sequence at its 3′ end (Salgado-Garrido et al., 1999). This suggests that archaeal and eukaryotic Sm proteins may play a direct or indirect role in tRNA processing. This is also consistent with the evolutionary conservation of one or more function(s) of Sm-related proteins between eukaryotes and Archaea. Taken together, our data suggest that archaeal Sm protein complexes may represent a primitive form of the Sm core domain of eukaryotic RNPs. The presence of primitive snRNPs and snoRNPs (Omer et al., 2000) in Archaea would be consistent with these organisms being closely related to the precursor of eukaryotic nuclei (Puhler et al., 1989).
Materials and methods
Cloning, expression, antibody preparation and purification of AF-Sm1 and AF-Sm2
Archaeoglobus fulgidus strains were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Germany). The AF-Sm1 and AF-Sm2 open reading frames were obtained by PCR from A.fulgidus DNA with the oligonucleotides SM1F, ATAATTCCATGGCACCAAGACCATTGGATGTGCTAAACAGGTCGGTAGGCCTTCTCAAAGTTTTGAGG, and SM1B, ATAATAGGTACCTCACTCACCACCCGGAGCTGGTGAAACGAAAACAACCGTG, for AF-Sm1, and SM2F, ATAATTCCATGGTGCTTCC AAATCAGATGGTAAAGTCAATGGTGG, and SM2B, ATAATAGGTACCTCATTCTTCTTGCGGCTGGATTAGAACGACGTTATTACC, for AF-Sm2, digested with NcoI and KpnI, and cloned into a modified pET24d expression vector. The proteins were overexpressed in E.coli BL21(DE3) cells as glutathione S-transferase (GST) fusion proteins containing an additional histidine tag. After a first purification step using an Ni2+–agarose column, eluted proteins were used to raise antibodies. Approximately 500 µg of His-GST–TEV-tag proteins were injected into a rabbit every 4 weeks. Sera were collected every 2 weeks after each boost. For crystallization, the His-GST–TEV-tag was removed by cleavage with TEV protease. Dialysed protein solutions were then heated to 86°C for 15 min to yield homogeneous Sm proteins after removing the denatured host proteins by centrifugation. The proteins were concentrated to ∼11 mg/ml for crystallization.
Crystallization and data collection
Diffraction quality crystals of the AF-Sm1 protein were obtained at 20°C by the hanging drop vapour diffusion method. The crystallization drops consisted of 1 µl of an 11 mg/ml protein solution and a 1 µl reservoir containing 12% PEG6000 as precipitating agent and 100 mM sodium citrate buffer at pH 4.3. Prism-shaped crystals with the maximal size of 0.2 × 0.1 × 0.05 mm3 grew within a month. Their space group was P21, with unit cell dimensions a = 110.4 Å, b = 64.5 Å, c = 129.8 Å, β = 92.1°. Small RNA complex crystals of 0.05 × 0.05 × 0.03 mm3 were obtained by co-crystallizing the protein with the U5 oligonucleotide at a molar ratio of 1:1 at pH 4.4 using otherwise identical conditions. The space group was P21 with unit cell dimensions a = 69.9 Å, b = 130.4 Å, c = 70.0 Å, β = 115.4°. Prior to data collection, the crystals were coated with a silicone oil film and were frozen in liquid nitrogen. Diffraction data were collected using beamline BW7b at the Hamburg Outstation at DESY and the microfocus beamline (ID13) at the ESRF for the 2.5 Å data of the native protein and the 2.75 Å data of the protein–RNA complex, respectively.
Structure determination and refinement
Data were processed using XDS (Kabsch, 1993) and subsequently converted to CNS (Brünger et al., 1998) and CCP4 (1994) format. The protein structure was solved with the molecular replacement program MOLREP (Vagin and Teplyakov, 1997) using a heptameric ring of PA-Sm1 as the search model (our unpublished results). The resulting model, which contains four seven-membered rings in the asymmetric unit, was refined using CNS reserving 5% of the reflections for the calculation of the free R-factor. In an initial step, the sequence was corrected to the AF-Sm1 sequence and rigid body refinement was carried out defining each monomer as a rigid body, followed by simulated annealing. Several cycles of geometry- and group-based B-factor refinement applying weak non-crystallographic restraints in the first cycles, together with model building, were done before water molecules were added to complete the model. The model building was done in O, version 7 (Jones et al., 1991). The structure of the RNA complex, consisting of two heptameric rings in the asym metric unit, was also solved by MOLREP using the coordinates of the refined AF-Sm1 heptamer as a model. Refinement was accomplished using CNS by applying non-crystallographic restraints for the invariable parts of the monomer. The refinement protocol was almost identical to that of the AF-Sm1 protein. Uridine phosphates with 75% occupancy were built into the electron density map and the nucleotides were covalently linked wherever it was stereochemically possible and indicated by the difference density. The final model consists of two heptameric rings and a total of 10 nucleotides (a trinucleotide and three single nucleosides bound in one ring, a second trinucleotide and a single nucleoside in the other ring). The final models of both the native and RNA-bound AF-Sm1 structures show good stereochemistry as judged by the program PROCHECK (Laskowski et al., 1993). The coordinates of the two structures have been deposited with the Protein Data Bank (PDB codes 1I4K and 1I5L).
Total lysate preparation
Cells obtained from DSMZ were resuspended in 200 µl of buffer B [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM dithio threitol (DTT)] and sonicated six times for 15 s at 4°C. The solution was centrifuged for 20 min at 14 000 g. The resulting supernatant was centrifuged for 30 min at 40 000 g. Supernatant was diluted once in buffer C (20 mM Tris–HCl pH 7.5, 0.1% Tween-20) to obtain the total lysate. Total lysate was submitted to immunoprecipitation and/or RNA extraction.
Immunoprecipitation using anti-AF-Sm1 and anti-AF-Sm2 antibodies
Protein A–Sepharose was swollen in IPP buffer (20 mM Tris–HCl pH 7.5, 75 mM NaCl, 2.5% glycerol, 0.5 mM DTT, 0.05% Tween-20), washed three times in 900 µl and then resuspended in 300 µl of IPP buffer. A 100 µl aliquot of antibodies was added. Beads were rotated for 2 h at 4°C. The antibody-coupled beads were washed four times with IPP buffer and then resuspended in 300 µl. A 100 µl aliquot of total cell lysate was added subsequently and the complete mixture was rotated at 4°C for 2 h. Four washes of 20 min with 900 µl of IPP were carried out after incubation. Beads were recovered by centrifugation.
RNA preparation
Total lysate or beads from immunoprecipitation were treated with proteinase K (20 mg/ml) for 45 min at 50°C in 200 µl of PK buffer (100 mM Tris–HCl pH 7.5, 12.5 mM EDTA, 150 mM NaCl, 1% SDS). The solution was centrifuged to pellet the protein A–Sepharose and the resulting supernatant extracted twice with phenol–chloroform– isoamylalcohol. The aqueous phase containing RNA was precipitated with a half volume of 7.5 M ammonium acetate and 2.5 vols of cold 100% ethanol. After incubation for 1 h at –20°C, the pellet was recovered by centrifugation, washed twice with cold 80% ethanol, dried under vacuum and resuspended in water.
Western blot analysis
Samples were mixed with 3× SDS loading buffer (50 mM Tris–HCl pH 6.8, 10% glycerol, 0.05% bromophenol blue, 2% SDS), boiled for 5 min at 95°C and electrophoresed on 18% SDS–polyacrylamide gels according to Laemmli’s method. Proteins were transferred to Protran nitrocellulose transfer membrane (Schleicher & Schuell) in 25 mM Tris–HCl, 40 mM glycine, 20% methanol, 0.05% SDS for 45 min at 200 mA with a trans-blot semi-dry transfer cell (Bio-Rad). The membrane was blocked for 1 h with 20 ml of blocking solution [5% fat milk in T-PBS (0.2% Tween-20, 100 mM Tris–HCl pH 7.5, 0.9% NaCl] and subsequently washed twice in T-PBS. Primary antibodies were then added in 10 ml of blocking solution, incubated for 1 h at room temperature and washed four times for 20 min. The membrane was incubated for 30 min with the secondary antibodies (horseradish peroxidase-coupled IgG antibodies; 1:5000 dilution, Bio-lab). Proteins were detected with an ECL kit (Amersham Pharmacia Biotech) as recommended by the manufacturer.
PCp labelling
The standard procedure was used. Briefly, 20 pmol of cytidine 3′-phosphate (Cp) were incubated with 7 mM DTT, 100 mM Tris–HCl pH 8.0, 10 mM MgCl2, 10 µl of [γ-32P]ATP (100 Ci/ml, >5000 Ci/mmol) and 10 U of T4 polynucleotide kinase (10 U/µl; Biolabs) for 45 min at 37°C. RNAs were incubated overnight on ice at 4°C with labelled pCp in the presence of 25 µM ATP, 13.3 mM DTT, 10% dimethylsulfoxide, 50 mM Tris–HCl pH 7.5, 15 mM MgCl2 and 1 µl of RNA ligase (15 U/µl; Amersham Pharmacia Biotech). Labelled RNAs were loaded on 8% polyacrylamide–7 M urea and run at 25 W for 2 h. The gel was dried and exposed with a screen at –80°C.
Northern blot analysis
RNAs were extracted as described above and were loaded onto 8% polyacrylamide–7 M urea gels and run at 25 W until the xylene cyanol dye migrated to the bottom of the gel. The RNAs were transferred to a Hybond-N membrane for nucleic acids (Amersham Life Science) by electroblotting and subsequently cross-linked to the membrane by UV irradiation. A chemically synthesized 23mer deoxyoligonucleotide RPSE, TGACCTCATCCTTGCGGAATCAT, complementary to residues 125–148 of the putative RNase P RNA from A.fulgidus according to the RNase P database (http://www.mbio.ncsu.edu/RNaseP/home.html), was labelled with 32P at its 5′ end as previously described for pCp labelling. The membrane was pre-hybridized at 65°C for 1 h in a buffer containing 5× Denhardt’s solution [1 g of bovine serum albumin (BSA), 1 g of polyvinylpyrrolidone, 1 g of Ficoll 400, 100 ml of H2O], 6× SSC, 0.5% SDS and 100 µg/ml salmon sperm DNA per ml. Hybridization with 32P-labelled probe (1 × 107 c.p.m.) was carried out for 1 h at 65°C and cooled down overnight to 40°C in the same buffer. The membrane was washed for 20 min with 2× SSC, 0.1% SDS and for 20 min with 0.1× SSC, 0.5% SDS at 45°C prior to autoradiography.
Primer extension
The primer used was a chemically synthesized 21mer deoxyoligonucleotide ST14, GTGGGGGGACTTTCCTCCTTT, complementary to residues 28–49 of the putative RNase P RNA. It was labelled with 32P at its 5′ end and purified on a G10 column (Chroma spin, Clontech Laboratories) according to the manufacturer’s instructions. Immuno precipitated RNAs were incubated with 0.5 µl of primer (∼4 × 105 c.p.m.) in 6 µl of buffer containing 50 mM Tris–HCl pH 8.0, 40 mM KCl, 0.5 mM EDTA. The solution was incubated for 2 min at 65°C and 1 h at 52°C. A 0.5 µl aliquot of AMV reverse transcriptase (20 U/l, Stratagene) was added in the presence of 2.5 mM dNTP, 200 mM MgCl2, 20 mM DTT and 125 µg/ml actinomycin D. Primer was extended at 45°C for 30 min. The reaction was stopped by adding 10 µl of loading dye containing deionized formamide, 10 mM NaOH, 1 mM EDTA, bromophenol blue and xylene cyanol. The product was loaded onto an 8% polyacrylamide–7 M urea gel and detected by autoradiography as described above.
S1 mapping
The standard procedure was used. Briefly, a 3′-end-labeled probe was prepared by cleaving with BspEI, a plasmid carrying the RNase P RNA coding sequence and ∼100 nucleotides of flanking sequences. The BspEI site was filled-in with the Klenow fragment of DNA polymerase I in the presence of [α-32P]dCTP. After cleavage with PstI, the labelled single-stranded probe was purified by gel electrophoresis. Total or immunoprecipitated RNA(s) were mixed with the DNA probe dissolved in 30 µl of 500 mM PIPES pH 6.6, 1 mM EDTA, 0.4 M NaCl and 80% deionized formamide. After denaturation for 5 min at 75°C, the mixture was hybridized at 56°C overnight. Hybrids were diluted to 300 µl of 30 mM ZnSO4, 0.2 M NaCl, 0.1 M sodium acetate pH 4.5, 2 mg/ml salmon sperm DNA and 3000 U/ml of S1 nuclease (400 U/µl; Roche) and digested for 45 min at 37°C. Products were recovered by ethanol precipitation, fractionated on denaturing gels and detected by autoradiography.
Gel shift experiment
Oligonucleotides, from Xeragon, were 32P labelled at their 5′ end as previously described and purified on a 10% polyacrylamide–7 M urea gel. After overnight elution, oligonucleotides were recovered by n-butanol precipitation according to the procedure described by Cathala and Brunel (1990). A 1 µl aliquot of purified oligonucleotides (75 fmol) was mixed with 6 µl of shift buffer (66 mM HEPES–KOH pH 7.9, 166 mM KCl, 16.6 mM MgCl2, 16.6% glycerol and 0.66 mM EDTA) and 1 µl of BSA (10 mg/ml; Boehringer). A 1 µg aliquot of purified AF-Sm1 was added and incubation was carried out at 65°C for 45 min. For competition experiments, 50 fmol of radioactively labelled U5 were mixed with water (no competition) or with 1 µl of cold oligonucleotide (U5 or C5) at 0.05, 0.5, 2.5 or 12.5 pmol/µl prior to incubation. Samples were mixed with 10 µl of shift loading dye, containing 16% glycerol, 10 mg/ml heparin and marker dyes, and 10 µl were loaded on an 8% polyacrylamide–0.5× TBE gel. After 30 min migration, the electrophoresis was stopped and the gel was dried prior to autoradiography.
Acknowledgments
Acknowledgements
We thank Hiang Dreher, Martin Dreher and Stephen Weeks for help with the production of antibodies and the cloning, purification and crystallization of the AF-Sm2 and the PA-Sm1 proteins, and Luc Moulinier for help with building the PA-Sm1 search model. We gratefully acknowledge the help of the staff of the EMBL outstations in Hamburg and Grenoble with data collection. C.M. was supported by an EMBO long-term fellowship and S.T. by the French government.
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R., Lapeyre,B., Brown,C.E. and Sachs,A.B. (1998) Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol., 18, 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E., Rigaut,G., Shevchenko,A., Wilm,M. and Séraphin,B. (2000) An Sm-like protein complex that participates in mRNA degradation. EMBO J., 19, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol,A., Ebel,J.P., Lazar,E., Haendler,B. and Jacob,M. (1982) U2 RNA shares a structural domain with U1, U4 and U5 RNAs. EMBO J., 1, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W. (1999) The ribonuclease P database. Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Camasses A., Bragado,N.E., Martin,R., Séraphin,B. and Bordonné,R. (1998) Interactions within the yeast Sm core complex: from proteins to amino acids. Mol. Cell. Biol., 18, 1956–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathala G. and Brunel,C. (1990) Use of n-butanol for efficient recovery of minute amounts of small RNA fragments and branched nucleotides from dilute solutions. Nucleic Acids Res., 18, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computer Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for the recognition of the tra mRNA precursor by the sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- He W. and Parker,R. (2000) Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol., 12, 346–350. [DOI] [PubMed] [Google Scholar]
- Heinrichs V., Hackl,W. and Lührmann,R. (1992) Direct binding of small nuclear ribonucleoprotein G to the Sm site of small nuclear RNA. Ultraviolet light cross-linking of protein G to the AUU stretch within the Sm site (AAUUUGUGG) of U1 small nuclear ribonucleoprotein reconstituted in vitro. J. Mol. Biol., 227, 15–28. [DOI] [PubMed] [Google Scholar]
- Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Hornig,H., Brahms,H. and Lührmann,R. (1995) snRNP Sm proteins share two evolutionary conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Daley,D.A.T., Luscombe,N.M., Berman,H.M. and Thornton,J.M. (2001) Protein–RNA interactions: a structural analysis. Nucleic Acids Res., 29, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker,V.A., Lührmann,R., Li,J. and Nagai,K. (1999a) Crystal structure of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 96, 375–387. [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke,S. and Nagai,K. (1999b) Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol., 9, 222–230. [DOI] [PubMed] [Google Scholar]
- Kastner B., Bach,M. and Lührmann,R. (1990) Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc. Natl Acad. Sci. USA, 87, 1710–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lührmann R., Kastner,B. and Bach,M. (1990) Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta, 1087, 265–292. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W., Tollervey,D. and Séraphin,B. (1993) Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J., 7, 47–53. [DOI] [PubMed] [Google Scholar]
- Mayes A.E., Verdone,L., Legrain,P. and Beggs,J.D. (1999) Characteriza tion of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J., 18, 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls K.A., Sharp,B. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- Pannone B.K. and Wolin,S.L. (2000) Sm-like proteins wRING the neck of mRNA. Curr. Biol., 10, R478–R481. [DOI] [PubMed] [Google Scholar]
- Puhler G., Leffers,H., Gropp,F., Palm,P., Klenk,H.P., Lottspeich,F., Garrett,R.A. and Zillig,W. (1989) Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc. Natl Acad. Sci. USA, 86, 4569–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Hartmuth,K., Kastner,B. and Lührmann,R. (1999) Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol., 19, 6554–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson,E., Kandels-Lewis,S. and Séraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B. (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J., 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A.G., Zaug,A.J., Sobel,S.G., Wolin,S.L. and Cech,T.R. (1999) Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature, 401, 177–180. [DOI] [PubMed] [Google Scholar]
- Stark H., Dube,P., Lührmann,R. and Kastner,B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature, 409, 539–542. [DOI] [PubMed] [Google Scholar]
- Tharun S., He,W., Mayes,A.E., Lennertz,P., Beggs,J.D. and Parker,R. (2000) Yeast Sm-like proteins function in mRNA decapping and decay. Nature, 404, 515–518. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O.C. and Gumport,R.I. (1982) T4 RNA ligase. In Boyer,P.D. (ed.), The Enzymes. Academic Press, Inc., New York, NY, Vol. 15B, pp. 31–58.
- Urlaub H., Raker,V.A., Kostka,S. and Lührmann,R. (2001) Sm protein– Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J., 20, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A. and Teplyakov,A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr., 30, 1022–1025. [Google Scholar]