Abstract
A new Dictyostelium discoideum cyclase gene was identified that encodes a protein (sGC) with 35% similarity to mammalian soluble adenylyl cyclase (sAC). Gene disruption of sGC has no effect on adenylyl cyclase activity and results in a >10-fold reduction in guanylyl cyclase activity. The scg– null mutants show reduced chemotactic sensitivity and aggregate poorly under stringent conditions. With Mn2+/GTP as substrate, most of the sGC activity is soluble, but with the more physiological Mg2+/GTP the activity is de tected in membranes and stimulated by GTPγS. Unexpectedly, orthologues of sGC and sAC are present in bacteria and vertebrates, but absent from Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana and Saccharomyces cerevisiae.
Keywords: cGMP/chemotaxis/Dictyostelium/evolution/guanylyl cyclase
Introduction
cAMP and cGMP are important signalling molecules in prokaryotes and eukaryotes. In prokaryotes, cAMP regulates gene expression. cGMP is only found in cyanobacteria in relatively high amounts, but its function as an intracellular signalling molecule is not well understood (Ochoa De Alda et al., 2000). In eukaryotes, cAMP and cGMP regulate enzyme activities, channel activities and gene expression, mainly via cAMP- and cGMP-dependent protein kinases (Houslay and Milligan, 1997; Lohmann et al., 1997). A large and complex family of adenylyl cyclases (ACs) and guanylyl cyclases (GCs) is responsible for the synthesis of cAMP and cGMP. At present, five phylogenetically separated classes have been recognized (Danchin, 1993), which are the ACs in enterobacteria (class I), the calmodulin-dependent toxin ACs (class II), the universal group of ACs and GCs (class III) and two recently recognized genetically unrelated ACs in hyperthermophilic archeabacteria (class IV; Sismeiro et al., 1998) and ruminal anaerobic bacteria (class V; Cotta et al., 1998). All five classes of enzymes are present in bacteria, while eukaryotes seem to contain only class III enzymes, which are the subject of this study.
The general topology of class III enzymes is deduced from the three-dimensional structure of mammalian AC (Tesmer et al., 1997; Zhang et al., 1997). Two cyclase domains are associated in an antiparallel manner. This dimer can be formed by two identical cyclase domains or two different domains. In the latter case, these domains can derive from one polypeptide or from two different polypeptides. Additional domains may control the localization and regulation of the cyclases. In higher eukaryotes, four subgroups are recognized (Hurley, 1998): (i) the 12-transmembrane ACs, which are composed of two different cyclase domains and are regulated by G-proteins; (ii) the single-transmembrane GCs, which contain one cyclase domain and dimerize to form a homodimer; these enzymes are stimulated by extracellular peptides, which bind to a receptor domain; (iii) the nitric oxide-sensitive soluble GC, which is a heterodimer of two different proteins with one cyclase domain each; and (iv) the recently discovered mammalian soluble adenylyl cyclase (sAC), which harbours two cyclase domains and shares the highest degree of identity with bacterial ACs (Buck et al., 1999). sAC is regulated by bicarbonate and involved in sperm maturation (Chen et al., 2000).
In the social amoeba Dictyostelium, cGMP is implicated as a second messenger for chemotaxis (Van Haastert and Kuwayama, 1997), while cAMP can act as both first and second messenger (Parent and Devreotes, 1996; Meima and Schaap, 1999b). As first messenger, cAMP induces chemotaxis, cAMP signal relay and gene expression. As second messenger, it controls terminal spore and stalk cell differentiation and spore germination. Three AC genes were identified in Dictyostelium. ACA is similar to mammalian 12-transmembrane ACs and controls cAMP signalling during cell aggregation. ACG has the topology of the single transmembrane cyclases, with one cyclase domain (Pitt et al., 1992); the enzyme produces cAMP, which regulates spore germination. A third adenylyl cyclase activity (ACB) is detectable mainly in the multicellular stage (Meima and Schaap, 1999a). This activity is encoded by the acrA gene, which is most similar to cyanobacterial ACs and controls spore maturation (Soderbom et al., 1999). Not all known effects of cAMP can as yet be attributed to the known ACs, which may suggest the presence of a fourth AC gene. Recently, we identified a Dictyostelium GC gene, gcaA (Roelofs et al., 2001), which encodes a protein with the topology of 12-transmembrane ACs. The two catalytic domains of GCA appear to be interchanged; thus, the second cyclase domain of mammalian AC, which provides most catalytic interactions with ATP, is similar to the first domain of GCA, interacting with GTP. GC enzymes with this topology have also been found in Paramecium and Plasmodium (Linder et al., 1999; Carucci et al., 2000) but, unlike GCA, these proteins have a P-type ATPase at their N-terminus. Cells with an inactivated gcaA gene still possess substantial GC activity, indicating the presence of a second GC in Dictyostelium.
During the last 3 years, a large portion of the Dictyostelium genome has been sequenced; presently, the probability of finding an open reading frame (ORF) of 300 amino acids is ∼80–90%. Using this database, we recognized gene fragments whose deduced amino acid sequence predicts two catalytic domains and a more C-terminal sequence that are similar to mammalian sAC (Buck et al., 1999). The complete amino acid sequence was deduced from genomic and cDNA sequence data. We have inactivated the gene and demonstrate that it encodes a guanylyl cyclase (sGC). Interestingly, databases of several completed genomes suggest that orthologues of Dictyostelium sGC are present in bacteria and mammals, but not in Drosophila, Caenorhabditis elegans, Arabidopsis and Saccharomyces cerevisiae.
Results
Identification of sGC
To identify novel AC or GC candidates, the Dictyostelium genome and cDNA databases were screened with AC and GC consensus sequences (see Materials and methods; Roelofs et al., 2001). This revealed a novel gene that is positioned on the 18 kb contig CHR2.0.71028. As discussed below, the putative protein shares a significant degree of identity with mammalian sAC (Buck et al., 1999), and the Dictyostelium gene was found to encode an sGC; we therefore denote the gene as sgcA and the protein as DdsGC.
The sequence of contig CHR2.0.71028 allows the unambiguous identification of the 3′ end of the sgcA gene, since a distinct ORF that encodes a PIWI transcription factor is positioned downstream of sgcA. The start of sgcA is more difficult to identify, because the deduced amino acid sequence does not reveal an obvious translational start methionine. Mammalian sAC contains a short N-terminal sequence that is followed by two cyclase domains and a long C-terminal sequence. In DdsGC, an in-frame methionine is present ∼140 amino acids before the first catalytic domain, but this methionine is preceded in-frame by 193 amino acids without other in-frame methionines. An upstream ORF contains a perfect start methionine, but potential splice sites would predict an intron of >1 kb, which is extremely long for Dictyostelium. To identify the ORF, we isolated 16 cDNAs and sequenced the three with the largest inserts. This revealed that sgcA consists of at least three introns and four exons (see Figure 1A). The first exon contains a start methionine that conforms well to the Dictyostelium Kozak sequence; the exon encodes 649 amino acids. The first intron is short (157 bp) and AT rich, which is common for Dictyostelium. The second exon is exceptionally short, encoding only 23 amino acids. This exon was observed in all cDNA clones that have the appropriate size; furthermore, its presence is essential as it preserves the reading frame. The second intron is very long (1176 bp) and unusually GC rich for a Dictyostelium intron. The remaining third and fourth exon encode 373 and 1796 amino acids and are separated by a normal 166 bp intron. The total ORF is 8532 bp, and predicts a 2843 amino acid protein with a mol. wt of 315 kDa and a pI of 8.5.
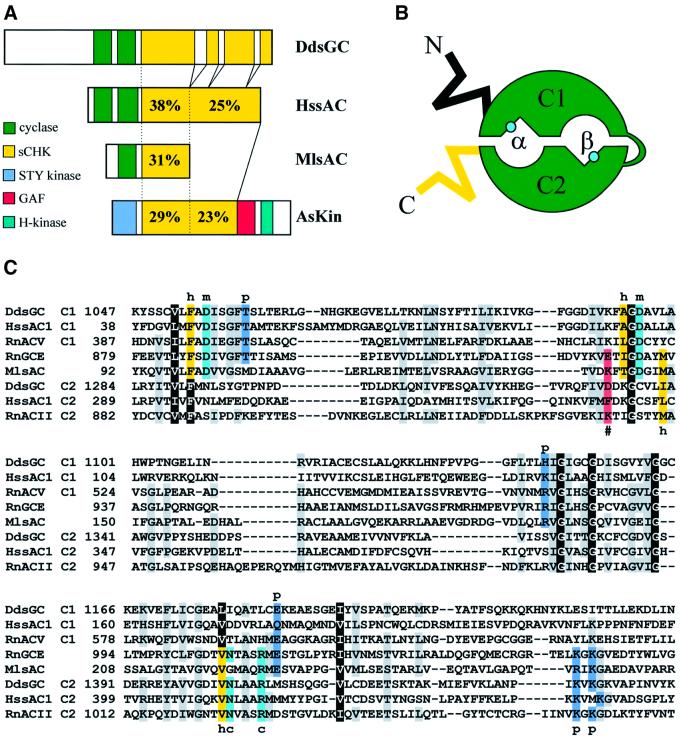
Fig. 1. Comparison of Dictyostelium DdsGC with related sequences. (A) Schematic of the putative domains in DdsGC, mammalian HssAC, bacterial MlsAC and bacterial AsKin, a complex kinase from Anabaena sp. (AAF44635). The C-terminal region of DdsGC (yellow) is denoted as the soluble cyclase kinase homology (sCKH) region; it contains three stretches of repetitive amino acid sequences, which were removed for optimal sequence alignments. STY kinase, serine/threonine/tyrosine protein kinase domain; H kinase, histidine protein kinase. The numbers indicate the percentage similarity to DdsGC. (B) Schematic of the cyclase domains in an antiparallel dimer. The catalytic site is located at the interface of the two domains, but not in the centre. Therefore, there are two potential catalytic pockets, α and β; the α-site is defined as the site that in the X-ray structure is more proximal to the N-terminus of C1, whereas the β-site is closer to the N-terminus of C2. In a homodimer, such as in membrane-bound mammalian GC, the α and β catalytic sites are identical. However, in heterodimers, α and β could be different. The turquoise dot indicates the position of the catalytic arginine (R1029 in ACII). In mammalian AC, ATP is converted in the β-site and the α-site has been mutated to a forskolin-binding site. Sequence alignment (C) indicates that also in Dictyostelium sGC GTP is most probably hydrolysed in the β-site whereas the α-site presumably is inactive. (C) Alignment of the catalytic domains of several mammalian, Dictyostelium and bacterial cyclases, based on the crystal structure of the ACII–ACV dimer (Tesmer et al., 1997). The colour and letter codes below or above the sequences refer to: black, conserved in all sequences; grey, conserved in five or more sequences; yellow (h), hydrophobic pocket for purine binding; red (#, in C2 domain only), determination of adenylyl (K) versus guanylyl (E or D) specificity; turquoise, amino acids essential for the catalytic reaction (c) or metal binding (m); blue (p), phosphate binding.
The sGC protein
The deduced amino acid sequence of DdsGC can be subdivided tentatively into three segments. The N-terminal ∼1000 amino acid region shares no similarity with other proteins in the database. It is rich in a repetitive sequence [poly (Q) and poly (N)], that is found in many Dictyostelium proteins. It also contains 12 repeats of a sequence P(I/L)(S/T)S, without indication for a putative function. Subsequent to this N-terminal segment, DdsGC contains two cyclase domains (amino acids 1051–1410) that share the highest homology with mammalian sAC and bacterial cyclases (see Discussion below).
The cyclase domains of DdsGC are followed by a long 1433 amino acid C-terminal segment that shares a significant degree of amino acid identity with the C-terminal segment of mammalian sAC (Figure 1A). From the many bacterial type III cyclases, two genes were identified in Mycobacterium laepra and Sinorhizobium meliloti that predict C-terminal segments that are similar to the first half of the C-terminal segment of sGC (Figure 1A). Furthermore, the complete C-terminal segment of sGC shares significant identity with the central region of a bacterial protein that also harbours a serine/threonine kinase, a histidine kinase and a GAF domain (Figure 1A). We denote this segment sCKH, the soluble cyclase kinase homology region (see Supplementary data available at The EMBO Journal Online; the alignment of sCKH regions is appended). The only recognizable functional feature of this sCKH region is an AAA-domain with a P-loop motif that is common to many ATP- and GTP-binding proteins (Saraste et al., 1990).
The cyclase catalytic domains
The crystal structure of mammalian AC suggests that AC and GC enzymes are active as a dimer with potentially two ATP- or GTP-binding pockets, α and β, at the dimer interface (Tesmer et al., 1997) (see Figure 1B). Enzymes with one cyclase domain, such as the mammalian GCE or the bacterial MlsAC, form a homodimer and thus will have two identical catalytic centres (Liu et al., 1997; Bieger and Essen, 2001). In the crystal structure of the ACV C1–ACII C2 dimer, ATP is bound in the β-site; the corresponding region of the α-site contains many mutations that would prevent high affinity binding and conversion of ATP or GTP (see also Figure 1C). Although both domains contribute to the binding of the substrate in the β-site, the amino acids involved in the cyclization reaction (notably R1029 in ACII) are located in the C2 domain. Also in sGC, these amino acids are present only in the C2 domain (Figure 1C), indicating that in sGC the substrate is also bound in the β-site. In contrast, GTP appears to be bound and hydrolysed in the α-site of the other Dictyostelium guanylyl cyclase GCA (Roelofs et al., 2001).
In mammalian cyclases, the purine moiety of ATP or GTP is recognized specifically by only a few amino acids. The most pronounced amino acid in AC is a lysine from the domain that also provides the amino acids for catalysis (K938 in the C2 ACII; red in Figure 1C); in GC, this amino acid is a glutamate (E928 in GCE). In the active binding pocket of sGC, the corresponding amino acid is aspartate D1332, suggesting that DdsGC is more likely to encode a GC than an AC.
Phylogeny of the catalytic domains
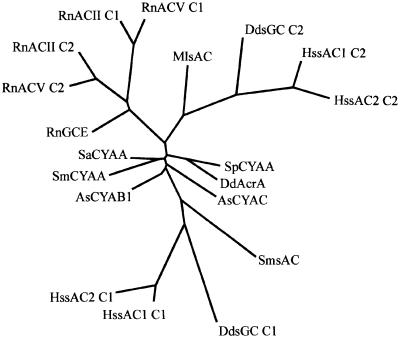
Figure 2 presents a phylogenetic analysis of the cyclase domains of DdsGC, vertebrate sAC, bacterial sAC, mammalian classical AC and GC, and several bacterial AC sequences. The analysis includes a second human soluble adenylyl cyclase (sAC2) that was found in human genome databases; its amino acid sequence could only be partially deduced. [The deduced amino acid sequence of the human contigs AL133404 and AL031778 shows a high degree of identity with sAC (68% in the catalytic region and 55% in the sCKH region, and therefore is denoted as HssAC2. Gene finder programs do not provide a complete ORF; inspection by eye allows the identification of ∼80% of the putative protein sequence.] The phylogenetic tree emphasizes the relatedness between human sAC, Dictyostelium sGC and bacterial cyclases. The C1 and C2 domains of the two human sACs are most related to the C1 and C2 domains of DdsGC, respectively. The two bacterial sACs are positioned on these branches, but closer to the apparent origin, with MlsAC grouping with the C1 domains and SmsAC with the C2 domains. A number of other bacterial cyclases that do not belong to the group of soluble cyclases occupy positions that are intermediate to the C1 and C2 domains. The cyclase domains of all other vertebrate cyclases group together in a separate clade. Two interesting observations can be made. First, the C1 and C2 domains of human sAC and the cyclase domains of other classical human cyclases show approximately equal distances from the apparent root of bacterial cyclases. This may suggest that these three groups of cyclase domains separated very early during evolution. Secondly, no homologues of sGC or sAC can be found in the databases for the completed genomes of Drosophila, C.elegans, Arabidopsis and S.cerevisiae. [Blast searches were carried out using the catalytic or sCKH regions of HssAC, DdsGC, MlsAC and AsKin in DDBJ/EMBL/GenBank (http://www.ncbi.nlm.nih.gov), and several specific databases for Drosophila (http://www.fruitfly.org), C.elegans (http://www.wormbase.org), human (http://www.ncbi.nlm. nih.gov/genome/seq/HsBlast.html and http://publication. celera.com), yeast (http://www.ncbi.nlm.nih.gov), Plas modium (http://www.ncbi.nlm.nih.gov/Malaria/plasmodiumbl.html), Dictyostelium (http://www.sdsc.edu/mpr/dicty/) and Arabidopsis (http://www.arabidopsis.org).]
Fig. 2. Phylogenetic analysis of the cyclase domains. Using the amino acid of the cyclase domains of 12 ACs and two GCs, an alignment was made using Clustal_W, followed by optimizing by eye. The alignment was used as input for phylogenetic analysis using ProtDis and Fitch (Phylip 3.5, J.Felsenstein, 1993). Sequences used: RnACV (Rattus norvegicus: NP_072122), RnACII (Rattus norvegicus: P26769), RnGCE (Rattus norvegicus: P51840), AsCYAB1(Anabaena sp.: BAA13998), AsCYAC (Anabaena sp.: BAA14000), SaCYAA (Stigmatella aurantiaca: P40137), MlsAC (Mycobacterium leprae: CAA19149), SpCYAA (Spirulina platensis: BAA22996), SmCYAA (Sinorhizobium meliloti: P19485), SmsAC (Sinorhizobium meliloti: S60684), DdAcrA (Dictyostelium discoideum: AD50121), HssAC1 (Homo sapiens: NP_060887), HssAC2 (Homo sapiens: AL133404 and AL031778), DdsGC (Dictyostelium discoideum: AF361947).
Developmental expression of sGC
The developmental expression of DdsGC was investigated by northern blot analysis (Figure 3). Two transcripts can be detected of ∼8.2 ± 0.5 and 10.3 ± 0.5 kb, respectively. The larger transcript is ∼3-fold less abundant than the smaller transcript, especially at later stages of development. The presence of two transcripts may indicate different transcription start sites and/or alternative splicing; the isolated cDNA clones do not provide evidence for alternative splicing in the coding region of sGC. Expression of DdsGC is relatively low during growth, increases ∼2.5-fold during aggregation and reaches a maximum at 3–6 h. Expression declines ∼3-fold during the slug stage (9–17 h) to increase slightly during culmination. The expression of the other GC in Dictyostelium, DdGCA, shows approximately the opposite profile, with relatively high expression during growth and the multicellular stage and low expression during aggregation (Roelofs et al., 2001).
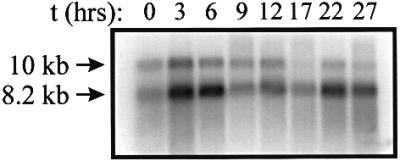
Fig. 3. Developmental expression of DdsGC. Wild-type AX3 cells were allowed to develop for the indicated time periods. Total RNA was isolated, size-separated and probed with the catalytic domains of sGC. The arrows indicate the size of the two transcripts. Ethidium bromide staining indicated equal loading of the samples.
Inactivation of the sgcA gene
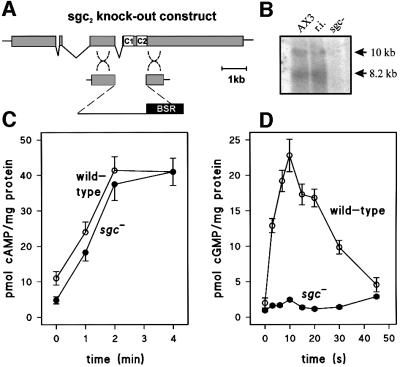
The sgcA gene was inactivated by homologous recombination (Figure 4A) to establish its function. This strategy was preferred over expression of the protein due to the large size of sGC with its long and relatively unknown N- and C-terminal segments; expression of the two His-tagged cyclase domains in Dictyostelium or Escherichia coli did not result in a detectable increase of AC or GC activity in crude Dictyostelium lysates or Ni+-purified bacterial lysates (data not shown). Two independent sgc– knock-out cell lines were obtained with different deletions, yielding identical results. Southern blot analysis demonstrated the disruption of the sgcA gene (data not shown), resulting in the absence of both mRNA species for sGC (Figure 4B).
Fig. 4. Genetic and functional characterization of a DdsGC gene disruptant. (A) Schematic of gene disruption. A second independent sgc– cell lines was obtained with a deletion of the first cyclase domain. (B) Northern blot of wild-type AX3, random integrant (r.i.) and sgc– cells. (C and D) cAMP and cGMP response of wild-type AX2 (open circles) and sgc– (closed circles). Cells were starved for 5 h and stimulated with 10 µM 2′-deoxy-cAMP and 5 mM DTT or with 0.1 µM cAMP for the detection of the cAMP and cGMP response, respectively. At the times indicated, cells were lysed and assayed for cAMP or cGMP, respectively.
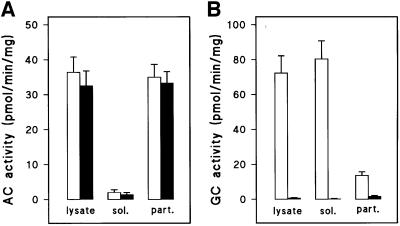
To establish the biochemical identity of sGC, we first tested the effects of sgcA gene disruption on two predominant AC- and GC-mediated responses in aggregating cells: cAMP relay and the cAMP-induced cGMP response. Figure 4C shows that cAMP accumulation, which peaks at 2 min, was unaffected in sgc– cells. In contrast, cGMP accumulation, which occurs within 10 s of cAMP stimulation, was 90% reduced in the sgc– cell line (Figure 4D). sAC, the mammalian orthologue of sGC, is activated by Mn2+ and is present mainly in the cytosol. We measured both AC and GC activities in the soluble and particulate fractions of wild-type and sgc– cells. Figure 5A shows that almost all Mn2+-dependent AC activity was present in the particulate fraction and was equally high in wild-type and sgc– cells. In contrast, Mn2+-dependent GC activity was present predominantly in the soluble fraction and was >95% reduced in sgc– cells (Figure 5B). These data demonstrate that DdsGC encodes a GC, that is mainly cytosolic in the presence of Mn2+ and has no significant AC activity.
Fig. 5. Adenylyl and guanylyl cyclase activity in sgc– cells. AX2 (open bars) and sgc– cells (filled bars) were lysed and separated into a 16 000 g particulate fraction (part.) and a 180 000 g soluble fraction (sol). The fractions were incubated with 2 mM Mn2+ and either 0.5 mM ATP (A) or GTP (B), and assayed for cAMP and cGMP production, respectively.
Phenotype of sgc-null cells
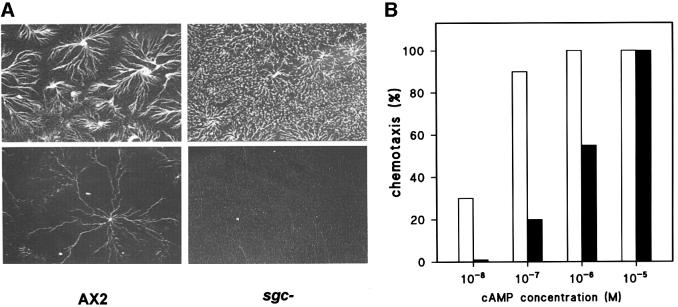
The sgc– mutant aggregated and developed normally into fruiting bodies on standard culture plates. Under standard conditions, the cells are closely packed, and diminished chemotactic sensitivity may not affect cell agglomeration too severely. To test the cells’ ability to aggregate under more stringent conditions, we plated them at decreasing cell densities, submerged in buffer. Figure 6A shows that wild-type cells still aggregated efficiently at 3 × 104 cells/cm2, whereas sgc– cells did not aggregate at all. Aggregation of sgc– cells started at 106 cells/cm2 but was delayed by several hours compared with wild-type cells. The chemotactic response of sgc-null and control cells to cAMP was measured (Figure 6B). Wild-type cells responded to 10–8 and 10–7 M cAMP, which was insufficient for sgc– cells. The sgc– cells started to show a clear chemotactic response to 10–6 M cAMP, indicating that chemotaxis in sgc– required at least 10- to 20-fold higher cAMP concentrations than in wild-type cells.
Fig. 6. Phenotype of the DdsGC gene disruptant. (A) Cell aggregation of wild-type AX2 and sgc– cells in phosphate buffer at two cell densities, 106 and 3 × 104 cells/cm2 (upper and lower panels, respectively). (B) Chemotaxis of wild-type AX3 (open bars) and sgc– cells (filled bars) to different concentrations of cAMP. The percentage of populations that showed a positive response in the small population assay (Konijn, 1970) was measured.
Guanylyl cyclase activity in sgc– cells
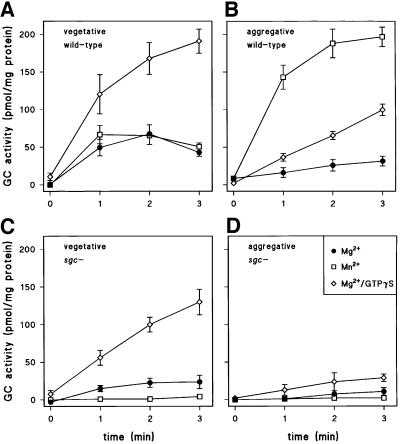
Earlier work showed that the cGMP response requires a heterotrimeric G-protein and that GC activity in cell lysates can be activated by guanosine 5′-O-(3-thiotriphosphate) (GTPγS) (Van Haastert and Kuwayama, 1997). We measured GC activity in lysates of vegetative and 7 h starved cells in the presence and absence of GTPγS. Figure 7A shows that vegetative wild-type cells have approximately equal GC activity when measured in the presence of 2 mM Mg2+ or Mn2+. The Mg2+-dependent GC activity was stimulated 4-fold by GTPγS. In vegetative sgc– cells (Figure 7C), the Mg2+/GTPγS-stimulated activity was reduced by ∼50% if compared with the activity from wild-type cells; interestingly, all Mn2+-dependent GC activity in the sgc– cells is virtually absent (Figure 7C). In 7 h starved wild-type cells, GTPγS induced a 3-fold stimulation of basal Mg2+-dependent GC activity (Figure 7B). Maximal GC activity was detected in the presence of Mn2+, which was 6-fold higher than in the presence of Mg2+ (Figure 7B). In starved sgc– cells, the GTPγS-stimulated activity is 4-fold lower than in wild-type cells, while Mn2+-dependent activity was absent (Figure 7D). These data illustrate the presence of two GC species: an activity, mainly in vegetative cells, that is stimulated by GTPγS, but is not active with Mn2+; and an activity in 7 h starved cells that is activated by both GTPγS and Mn2+. Since the latter activity is lost in sgc– cells, it is most likely to be sGC. The activity in vegetative cells is most likely to be GCA, which is expressed at high levels during growth and decreases during starvation (Roelofs et al., 2001). The residual GCA expression during cell aggregation may also explain the small 10% cGMP response in sgc– cells.
Fig. 7. Guanylyl cyclase in sgc– cells. Wild-type AX2 and sgc– mutant cells were starved for 0 (vegetative cells) or 7 h (aggregative cells). Cells were lysed in the presence or absence of 30 µM GTPγS. The unstimulated lysates were incubated with 0.5 mM GTP and 2 mM MgCl2 or MnCl2, and the GTPγS-stimulated lysates with 0.5 mM GTP and 2 mM MgCl2. At the indicated time intervals, the reactions were terminated and cGMP levels measured by radioimmunoassay.
Characterization of sGC activity
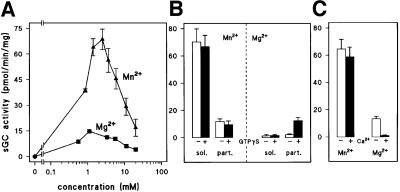
Mammalian cells expressing sAC contain a high background of other ACs, which hinders studies on sAC regulation. Dictyostelium offers the unique possibility to investigate the biochemical properties of this class of soluble enzymes. The experiments were performed in gca– cells to ascertain that only sGC was measured (Roelofs et al., 2001). Figure 8A reveals that sGC is more active with Mn2+ than with Mg2+ as divalent cation. Figure 8B shows that the Mn2+-dependent GC activity could not be stimulated by GTPγS in either the soluble or particulate fraction. A small Mg2+-dependent activity is present in the particulate fraction, which is stimulated 5-fold by GTPγS.
Fig. 8. Characterization of DdsGC enzyme activity in gca– cells. (A) Cells were lysed in the presence of 100 µM GTPγS, and sGC activity was measured in the lysate with 0.5 mM GTP and the indicated concentrations of Mg2+, or Mn2+ in excess of EGTA. (B) Cells were lysed in the presence or absence of 100 µM GTPγS, separated into a 16 000 g soluble (sol.) and particulate (part.) fraction and assayed for sGC activity with 2 mM Mn2+ or Mg2+. (C) Effect of 1 µM free Ca2+ on sGC activity in the lysate measured with Mn2+GTP or Mg2+GTP; the lysate was prepared in the presence of GTPγS.
Submicromolar Ca2+ concentrations inhibit Dictyo stelium GC activity, as is the case for GC in the vertebrate eye (Dizhoor et al., 1995; Van Haastert and Kuwayama, 1997). Figure 8C shows that 1 µM Ca2+ has no effect on Mn2+-dependent sGC activity, whereas it induces >90% inhibition of Mg2+/GTPγS-stimulated sGC activity.
Discussion
The Dictyostelium genome sequencing project has provided a wealth of information on genes that potentially are involved in signal transduction. We identified the Dictyostelium homologue of mammalian sAC. Sequence alignment with both AC and GC domains and functional analysis of the Dictyostelium gene showed that it encodes a guanylyl cyclase (sGC). Similarly to mammalian sAC, sGC shows high activity in the soluble cell fraction with Mn2+/GTP as a substrate. Lower activity can be detected in the particulate fraction. sGC does not contain hydrophobic segments that could result in membrane localization, suggesting that it may form a complex with a membrane-bound protein. In Dictyostelium, Mn2+ is most probably not the physiological bivalent cation of sGC, because the intracellular concentration of Mn2+ (∼10 µM) is too low to support activity, while at the prevailing Mg2+ concentration (∼3.5 mM) significant activity is observed (Padh and Brenner, 1984). The same conclusion was drawn for sAC in mammalian cells (Chen et al., 2000). In the presence of Mg2+, sGC activity was detected mainly in the particulate fraction.
Regulation of ACs and GCs by their natural effectors such as G-proteins or Ca2+ generally is detectable only with Mg2+, while Mn2+ uncovers all intrinsic activity (Tesmer et al., 1999). In agreement with this general observation, Mg2+-dependent sGC activity is stimulated by GTPγS and inhibited by Ca2+ ions, whereas these regulators do not affect Mn2+-dependent sGC activity. The in vitro activation of membrane-associated sGC by GTPγS could represent the activation mechanism in living cells. The large pool of inactive sGC allows for two additional activation mechanisms: A G-protein-mediated signal may modify the soluble enzyme (e.g. covalent modification or the association with other proteins) such that soluble sGC becomes Mg2+ sensitive. Alternatively, a G-protein-mediated signal may result in the translocation of soluble sGC to the membrane, which could result in an Mg2+-sensitive sGC activity.
Inactivation of the sgcA gene demonstrates that the cAMP-induced cGMP response is mediated predominantly by sGC. The sgc– cells show ∼20-fold reduced chemotactic sensitivity towards cAMP and a reduced capacity to aggregate under stringent conditions, which is consistent with the proposed role of cGMP during chemotaxis and cell aggregation. The small ∼10% residual cGMP response in sgc– cells to cAMP is most probably derived from GCA at the aggregation stage. Northern blot analysis reveals that the expression of the sGC gene and of the GCA gene complement each other, with high expression of GCA during growth and late development and highest expression of sGC during aggregation. These expression patterns are also reflected by the enzyme activities. sGC shows an ∼4-fold higher activity during aggregation than during growth, while GCA is 5-fold more active during growth than during aggregation (Figure 7).
The two GCs in Dictyostelium have a very different topology suggesting different modes of regulation. GCA is a 12-transmembrane enzyme, similar to G-protein-coupled ACs, whereas sGC is homologous to bicarbonate-stimulated sAC. Nevertheless, both GCA and sGC are activated in membranes by GTPγS and inhibited by Ca2+ ions. Detailed biochemical characterization and regulation of GCA and sGC can be performed in sgc– and gca– cells, respectively. This may also uncover the mechanism by which GCA is Mn2+ insensitive and sGC is associated with the membrane.
The identification of the two GCs in Dictyostelium, GCA and sGC, which are both homologous to ACs, uncovers unexpected evolutionary traits. From a biochemical point of view, this may not be very surprising, considering the relative ease with which an AC can be converted by site-directed mutagenesis into a GC. No close phylogenetic relationship can be found with mammalian GC, suggesting that GCA and sGC developed independently into a GC. Phylogenetic analysis of hundreds of ACs and GCs suggests that mammalian GC, GCA and sGC developed independently. This analysis also indicates that an AC to GC transition became fixed only a few times during evolution: at the formation of the vertebrate GCs; at the formation of GCA-like GCs in lower eukaryotes such as Dictyostelium, Paramecium and Plasmodium; and at the formation of sGC in Dictyo stelium. Furthermore, it is surprising that Dictyostelium does not have a member of the metazoan GCs, suggesting that in Dictyostelium cGMP synthesis has an independent origin. This might mean that other components of the cGMP pathway, such as phosphodiesterases and cGMP targets, have unique characteristics as well, which may explain the difficulties in identifying these proteins in Dictyostelium.
It is quite fascinating that orthologues of sGC can be detected in mammals and bacteria, but are absent from the completed genome sequences of the invertebrates C.elegans and Drosophila, the yeast S.cerevisiae and the flowering plant Arabidopsis. The eukaryote phylogeny strongly supports an animal/fungi supergroup, with the mycetozoa, such as Dictyostelium, as the immediate sister group (Baldauf et al., 2000). It seems unlikely that all lower animals and fungi have lost the gene, but even more unlikely that in the absence of gene loss, sGC and sAC represent two sequential cases of lateral transfer of an originally bacterial gene. It will be of great interest to trace the evolutionary roots of these enzymes when more complete genome sequences of lower unicellular eukaryotes and lower animals become available.
Materials and methods
Strain and culture conditions
AX3, AX2 (‘wild types’), gca-null (Roelofs et al., 2001) and sgc-null cells (see below) were grown in HG5 medium, which was supplemented with 10 µg/ml blasticidin S for gca– and sgc– cells. Cells were starved for up to 5 h by shaking in 10 mM phosphate buffer pH 6.5 (PB) at a density of 107 cells/ml. For longer starvation periods, cells were deposited on non-nutrient agar plates (1.5% agar in PB) and incubated at 22°C. To study aggregation under submerged conditions, cells were incubated at 22°C in 4 cm Petri dishes with 1 ml of PB at densities ranging from 3 × 103 to 3 × 106 cells/cm2 and photographed at 2 h intervals under dark field illumination. Chemotaxis toward cAMP was measured with the small population assay (Konijn, 1970). Briefly, small populations (0.1 µl containing ∼500 cells) were deposited on hydrophobic agar. After ∼5 h, just before aggregation started, small droplets containing different cAMP concentrations were deposited close to the amoebal populations. The distribution of cells within the small population was observed and scored positive if at least twice as many cells were pressed against the edge of the droplet close to the cAMP concentration if compared with the opposite edge (Konijn, 1970).
Bioinformatics
The Dictyostelium cDNA and genomic databases were screened for putative ACs and GCs with the catalytic domains of bacterial and eukaryotic cyclases. During the early phase of the sequencing project, three putative cyclase domains were recognized. Later, it appeared that one gene with a single cyclase domain (acrA) encoded the adenylyl cyclase ACB. The other two cyclase domains were present in one large contig (contig CHR2.0.71028, that was assembled by the Dictyostelium genome project, genome sequencing centre Jena, Germany; http://genome.imb-jena.de/dictyostelium/). Using cDNA sequence information to confirm potential exon–intron boundaries (see below), the complete ORF of sGC was obtained; this sequence is deposited in DDBJ/EMBL/GenBank (accession No. AF361947). BLAST, SMART and Pfam programs were used to analyse the domain structure of sGC and its sequence similarity to other proteins. A screen of the Dictyostelium database on May 30, 2001 with sequences from all five classes of cyclases did not reveal additional candidate genes for ACs or GCs.
cDNA screens
A 1434 bp DNA fragment encoding part of the catalytic domains of sGC (amino acids 1001–1479) was isolated by RT–PCR and subcloned, yielding pGeasycsCyc. A cDNA library, kindly provided by Dr R.Gomer, was screened with a 571 bp fragment (the 5′ region RT–PCR fragment up to the BglII site). After rescuing 16 λZAP clones as plasmids, three clones with the largest inserts were sequenced. Two clones were long enough to identify all four exons and three introns (see Figure 1A). In Dictyostelium, exons are large, and introns, when present, are small, with an average size of 100–200 bp. One exon of sGC is unusually small (70 bp) while one intron is exceptionally large (1176 bp) and contains many potential splice sites, which prevented gene finder programs from constructing the ORF.
Gene inactivation
Two independent sgc knock-out cell lines were obtained. For construct 1, the bsr selection cassette from pUCBsrΔBamHI (Sutoh, 1993) was cloned into the BglII–ClaI sites of pGeasycsCyc. A linear fragment with the bsr cassette and sGC flanking sequence was obtained by two rounds of PCR. After homologous recombination, this lead to a disruption of the first cyclase domain between amino acids 1192 and 1224. For construct 2, two DNA fragments of sGC comprising amino acids 705–1044 and 1421–1686 were amplified by PCR using oligonucleotides that yielded a 5′ BglI site and a 3′ BamHI site on the first fragment and 5′ BamHI and 3′ EcoRI site on the second fragment. These fragments were cloned in tandem into the BamHI and BamHI–EcoRI sites of pUCBsrΔBamHI. The construct was linearized with BamHI, yielding the plasmid flanked by 5′ and 3′ sGC fragments (see Figure 4A). Homologous recombination with this construct leads to deletion of amino acids 1045–1421, which comprises both catalytic domains. Dictyostelium AX3 and AX2 were transformed by electroporation with 5 µg of DNA constructs 1 and 2, respectively. Blasticidin-resistant clones were screened for homologous recombination by PCR and Southern blot analysis; 80% of the AX3 clones carried a gene disruption (sgc1–) and 90% of the AX2 clones carried sgc2–.
Guanylyl and adenylyl cyclase assays
AX3, AX2, gca-null and sgc-null cells were harvested and starved for 7 h in PB. To measure AC and GC activity, cells were washed and resuspended in ice-cold lysis buffer (1.5 mM EGTA and 250 mM sucrose in 10 mM Tris pH 8.0) and lysed through nucleopore filters (pore size 3 µM) in the absence or presence of 100 µM guanosine 5′-O-(3-thiotriphosphate) (GTPγS). For separation into soluble and particulate fractions, 0.5 ml aliquots of filter lysates were centrifuged for 5 min at 16 000 g and the pellets was resuspended in 0.5 ml of lysis buffer (particulate fraction). In some experiments, the supernatant was cleared further by centrifugation for 5 min at 180 000 g (soluble fraction). Lysates and fractions were incubated at 22°C with 0.5 mM ATP or GTP for AC and GC activity, respectively, in the presence of 10 mM dithiothreitol (DTT), 0.2 mM 3-isobutyl-1-methylxanthine and 2 mM Mn2+ or Mg2+ in lysis buffer (Meima and Schaap, 1999a). Reactions were terminated by the addition of an equal volume of 3.5% perchloric acid, and cAMP or cGMP levels were measured by isotope dilution assays (Snaar-Jagalska and Van Haastert, 1994).
For measuring cGMP responses to cAMP, cells were starved for 5 h, washed and resuspended in PB at 108 cells/ml. Cells were stimulated with 0.1 µM cAMP, reactions were terminated with perchloric acid, and cGMP levels were measured. For measuring the cAMP response, cells were stimulated with 10 µM 2′-deoxy-cAMP and 5 mM DTT, reactions were terminated with perchloric acid, and cAMP levels were measured.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Marten Postma for assistance with the figures, and Helena Snippe for isolation of cDNA clones that allowed the assignment of the ORF. This research was supported by the Netherlands Organization of Scientific Research, and by Wellcome Trust University Award Grant 057137.
References
- Baldauf S.L., Roger,A.J., Wenk-Siefert,I. and Doolittle,W.F. (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science, 290, 972–977. [DOI] [PubMed] [Google Scholar]
- Bieger B. and Essen,L.O. (2001) Structural analysis of adenylate cyclases from Trypanosoma brucei in their monomeric state. EMBO J., 20, 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J., Sinclair,M.L., Schapal,L., Cann,M.J. and Levin,L.R. (1999) Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc. Natl Acad. Sci. USA, 96, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carucci D.J. et al. (2000) Guanylyl cyclase activity associated with putative bifunctional integral membrane proteins in Plasmodium falciparum. J. Biol. Chem., 275, 22147–22156. [DOI] [PubMed] [Google Scholar]
- Chen Y., Cann,M.J., Litvin,T.N., Iourgenko,V., Sinclair,M.L., Levin,L.R. and Buck,J. (2000) Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science, 289, 625–628. [DOI] [PubMed] [Google Scholar]
- Cotta M.A., Whitehead,T.R. and Wheeler,M.B. (1998) Identification of a novel adenylate cyclase in the ruminal anaerobe, Prevotella ruminicola D31d. FEMS Microbiol. Lett., 164, 257–260. [DOI] [PubMed] [Google Scholar]
- Danchin A. (1993) Phylogeny of adenylyl cyclases. Adv. Second Messenger Phosphoprotein Res., 27, 109–162. [PubMed] [Google Scholar]
- Dizhoor A.M., Olshevskaya,E.V., Henzel,W.J., Wong,S.C., Stults,J.T., Ankoudinova,I. and Hurley,J.B. (1995) Cloning, sequencing and expression of a 24-kDa Ca(2+)-binding protein activating photo receptor guanylyl cyclase. J. Biol. Chem., 270, 25200–25206. [DOI] [PubMed] [Google Scholar]
- Houslay M.D. and Milligan,G. (1997) Tailoring cAMP-signalling responses through isoform multiplicity. Trends Biochem. Sci., 22, 217–224. [DOI] [PubMed] [Google Scholar]
- Hurley J.H. (1998) The adenylyl and guanylyl cyclase superfamily. Curr. Opin. Struct. Biol., 8, 770–777. [DOI] [PubMed] [Google Scholar]
- Konijn T.M. (1970) Microbiological assay of cyclic 3′,5′-AMP. Experientia, 26, 367–369. [DOI] [PubMed] [Google Scholar]
- Linder J.U., Engel,P., Reimer,A., Kruger,T., Plattner,H., Schultz,A. and Schultz,J.E. (1999) Guanylyl cyclases with the topology of mammalian adenylyl cyclases and an N-terminal P-type ATPase-like domain in Paramecium, Tetrahymena and Plasmodium. EMBO J., 18, 4222–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ruoho,A.E., Rao,V.D. and Hurley,J.H. (1997) Catalytic mechanism of the adenylyl and guanylyl cyclases: modeling and mutational analysis. Proc. Natl Acad. Sci. USA, 94, 13414–13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann S.M., Vaandrager,A.B., Smolenski,A., Walter,U. and De Jonge,H.R. (1997) Distinct and specific functions of cGMP-dependent protein kinases. Trends Biochem. Sci., 22, 307–312. [DOI] [PubMed] [Google Scholar]
- Meima M. and Schaap,P. (1999a) Fingerprinting of adenylyl cyclase activities during Dictyostelium development indicates a dominant role for adenylyl cyclase B in terminal differentiation. Dev. Biol., 212, 182–190. [DOI] [PubMed] [Google Scholar]
- Meima M. and Schaap,P. (1999b) Dictyostelium development—socializing through cAMP. Semin. Cell Dev. Biol., 10, 567–576. [DOI] [PubMed] [Google Scholar]
- Ochoa De Alda J.A., Ajlani,G. and Houmard,J. (2000) Synechocystis strain PCC 6803 cya2, a prokaryotic gene that encodes a guanylyl cyclase. J. Bacteriol., 182, 3839–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padh H. and Brenner,M. (1984) Studies of the guanylate cyclase of the social amoeba Dictyostelium discoideum. Arch. Biochem. Biophys., 229, 73–80. [DOI] [PubMed] [Google Scholar]
- Parent C.A. and Devreotes,P.N. (1996) Molecular genetics of signal transduction in Dictyostelium. Annu. Rev. Biochem., 65, 411–440. [DOI] [PubMed] [Google Scholar]
- Pitt G.S., Milona,N., Borleis,J., Lin,K.C., Reed,R.R. and Devreotes,P.N. (1992) Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell, 69, 305–315. [DOI] [PubMed] [Google Scholar]
- Roelofs J., Snippe,H., Kleineidam,R.G. and Van Haastert,P.J.M. (2001) Guanylate cyclase in Dictyostelium discoideum with the topology of mammalian adenylate cyclase. Biochem. J., 354, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald,P.R. and Wittinghofer,A. (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci., 15, 430–434. [DOI] [PubMed] [Google Scholar]
- Sismeiro O., Trotot,P., Biville,F., Vivares,C. and Danchin,A. (1998) Aeromonas hydrophila adenylyl cyclase 2: a new class of adenylyl cyclases with thermophilic properties and sequence similarities to proteins from hyperthermophilic archaebacteria. J. Bacteriol., 180, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaar-Jagalska B.E. and Van Haastert,P.J. (1994) G-protein assays in Dictyostelium. Methods Enzymol., 237, 387–408. [DOI] [PubMed] [Google Scholar]
- Soderbom F., Anjard,C., Iranfar,N., Fuller,D. and Loomis,W.F. (1999) An adenylyl cyclase that functions during late development of Dictyostelium. Development, 126, 5463–5471. [DOI] [PubMed] [Google Scholar]
- Sutoh K. (1993) A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid, 30, 150–154. [DOI] [PubMed] [Google Scholar]
- Tesmer J.J., Sunahara,R.K., Gilman,A.G. and Sprang,S.R. (1997) Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsα⋅GTPγS. Science, 278, 1907–1916. [DOI] [PubMed] [Google Scholar]
- Tesmer J.J., Sunahara,R.K., Johnson,R.A., Gosselin,G., Gilman,A.G. and Sprang,S.R. (1999) Two-metal-ion catalysis in adenylyl cyclase. Science, 285, 756–760. [DOI] [PubMed] [Google Scholar]
- van Haastert P.J. and Kuwayama,H. (1997) cGMP as second messenger during Dictyostelium chemotaxis. FEBS Lett., 410, 25–28. [DOI] [PubMed] [Google Scholar]
- Zhang G., Liu,Y., Ruoho,A.E. and Hurley,J.H. (1997) Structure of the adenylyl cyclase catalytic core. Nature, 386, 247–253. [DOI] [PubMed] [Google Scholar]