Abstract
The Cdc6 DNA replication initiation factor is targeted for ubiquitin-mediated proteolysis by the E3 ubiquitin ligase SCFCDC4 from the end of G1 phase until mitosis in the budding yeast Saccharomyces cerevisiae. Here we describe a dominant-negative CDC6 mutant that, when overexpressed, arrests the cell cycle by inhibiting cyclin-dependent kinases (CDKs) and, thus, prevents passage through mitosis. This mutant protein inhibits CDKs more efficiently than wild-type Cdc6, in part because it is completely refractory to SCFCDC4-mediated proteolysis late in the cell cycle and consequently accumulates to high levels. The mutation responsible for this phenotype destroys a putative CDK phosphorylation site near the middle of the Cdc6 primary amino acid sequence. We show that this site lies within a novel Cdc4-interacting domain distinct from a Cdc4-interacting site identified previously near the N-terminus of the protein. We show that both sites can target Cdc6 for proteolysis in late G1/early S phase whilst only the newly identified site can target Cdc6 for proteolysis during mitosis.
Keywords: Cdc4/Cdc6/cell cycle/proteolysis/replication
Introduction
Precise and tightly controlled DNA replication is an essential feature of the eukaryotic cell cycle. The regulated assembly of pre-replicative complexes (pre-RCs) at origins of DNA replication during the G1 phase is a critical step in this process. In budding yeast the main cyclin (Cln)-dependent kinase (CDK), Cdc28, triggers the initiation of DNA replication from origins containing pre-RCs and, at the same time, prevents the assembly of new pre-RCs (Diffley, 1996, 2001; Dutta and Bell, 1997; Donaldson and Blow, 1999; Pasero and Schwob, 2000). Thus, pre-RCs can assemble only early in G1 phase when Cdc28 kinase activity is low.
The Cdc6 protein is required for the assembly of pre-RCs (Cocker et al., 1996; Detweiler and Li, 1997; Tanaka et al., 1997) and Cdc6 levels are tightly regulated during the cell cycle by CDK activity (Piatti et al., 1995; Drury et al., 1997, 2000; Elsasser et al., 1999; Calzada et al., 2000). The CDC6 gene is transcribed periodically in early G1 phase (Zhou and Jong, 1990; Zwerschke et al., 1994; Piatti et al., 1995; McInerny et al., 1997). In addition to this transcriptional regulation, evidence has emerged of an intricate system for ensuring the effective elimination of Cdc6 throughout the cell cycle. We have previously defined three distinct modes of Cdc6 proteolysis based on cis- and trans-acting genetic requirements (Drury et al., 2000). Cdc6 proteolysis during pre-Start G1 (mode 1) is moderately rapid (t1/2 ∼15 min), is independent of the two best-characterized cell cycle-regulated pathways for ubiquitin-mediated proteolysis, the SCF and the APC/cyclosome, and does not require any of the eight consensus Cdc28 phosphorylation sites in Cdc6.
Near the end of G1 phase, Cdc6 proteolysis becomes extremely rapid (t1/2 < 2 min). This rapid ‘mode 2’ proteolysis requires SCFCDC4. Mode 2 degradation occurs only after the activation of the G1 cyclin-dependent kinase. Moreover, it requires at least some subset of CDK consensus phosphorylation sites in Cdc6, since a mutant in which all eight consensus sites have been mutated is resistant to mode 2 proteolysis. Whilst it requires G1 cyclins, mode 2 proteolysis does not require the S-phase-specific cyclins Clb5 or Clb6, suggesting that it is direct phosphorylation of Cdc6 by Cln-Cdc28 that targets it for proteolysis.
Later in the cell cycle, after activation of the B type cyclins, Cdc6 proteolysis is significantly slower than it is during S phase (t1/2 ∼15 min). This ‘mode 3’ proteolysis, however, is still entirely dependent upon SCFCDC4 and also requires some subset of CDK consensus sites. In our previous work, modes 2 and 3 were distinguished primarily on the basis of proteolysis rate. Here we show that Cdc6 possesses two distinct SCFCDC4-interaction domains, one, previously identified, near the N-terminus of the protein and a second novel domain located near the middle of the protein. Interestingly, this second domain contains two CDK consensus sites and mutation of either site stabilizes the protein during G2/M. Mode 2 proteolysis works through both SCFCDC4 sites but primarily through the first, whereas mode 3 is entirely dependent upon the second. This is a novel mechanism for modulating the rate of SCFCDC4-dependent proteolysis during the cell cycle.
Results
CDC6-d2 overexpression prevents mitosis
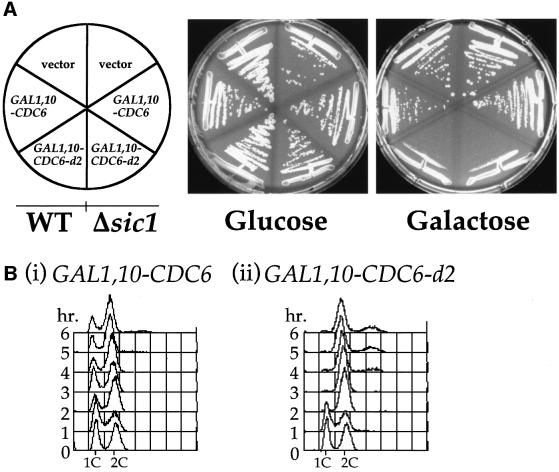
To better understand the function and regulation of Cdc6 in DNA replication we isolated and characterized dominant-negative mutant alleles of CDC6 (Perkins and Diffley, 1998). In this screen, a plasmid containing the CDC6 gene under the control of the GAL1,10 promoter was randomly mutagenized by passage through a mutator Escherichia coli strain. This library of mutants was transformed into yeast and plated on glucose-containing plates. Transformants were then replica plated on to galactose-containing plates to allow expression of the mutant proteins. From 20 000 transformants, two candidates were identified that grew on glucose- but not galactose-containing medium. One of these, CDC6-d1, which has previously been described in detail, prevented normal S phase progression by interfering with pre-RC assembly (Perkins and Diffley, 1998). The second, CDC6-d2, is described in this paper. Figure 1A shows that yeast carrying either the wild-type GAL1,10–CDC6 plasmid or the vector alone formed colonies on both glucose- and galactose-containing plates. However, yeast carrying the GAL1,10–CDC6-d2 plasmid grew well on glucose but showed severely retarded growth on galactose-containing plates.
Fig. 1. (A) W303-1a or W303-1aΔsic1 were transformed with pRS426 (Christianson et al., 1992), pRS426 GAL1,10–CDC6 or pRS426 GAL1,10–CDC6-d2 and grown on glucose (no expression from the GAL1,10 promoter) or galactose to induce expression of the CDC6 gene. Expression of CDC6-d2 but not CDC6 resulted in an inhibition to growth. As can be seen on the right-hand half of each plate this effect was not dependent on SIC1. (B) W303-1a with an integrated copy of either (i) GAL1,10–CDC6 or (ii) GAL1,10–CDC6-d2 was grown in YPRaf until mid-log phase. At this point the cells were transferred into YPGal and allowed to continue cycling. Samples were taken every hour and processed for flow cytometry.
To begin to determine the mechanism of growth suppression, we asked whether Cdc6-d2 overexpression arrested cells at a specific stage of the cell cycle. Cells containing either wild-type CDC6 or CDC6-d2 under the control of the GAL1,10 promoter were grown to mid-log phase in raffinose-containing medium and then transferred to fresh medium containing galactose. At the time intervals indicated in Figure 1B, samples were taken and the DNA content was measured by flow cytometry. Figure 1A shows that overexpression of wild-type Cdc6 does not prevent cell proliferation. As shown in Figure 1B, this overexpression does have an effect on cell cycle kinetics as can be seen by the increase in the fraction of cells with a 2C DNA content, consistent with previous work (Bueno and Russell, 1992). The expression of Cdc6-d2, however, prevents proliferation (Figure 1A) and cells accumulate with an apparent 2C DNA content (Figure 1B). In other experiments, when Cdc6-d2 was expressed after release from a nocodazole arrest, cells were unable to complete mitosis, indicating that Cdc6-d2 can block cell cycle progression after S phase is complete (data not shown). Thus, unlike Cdc6-d1, which inhibited S phase but not mitosis, Cdc6-d2 prevents passage through mitosis but does not prevent DNA replication.
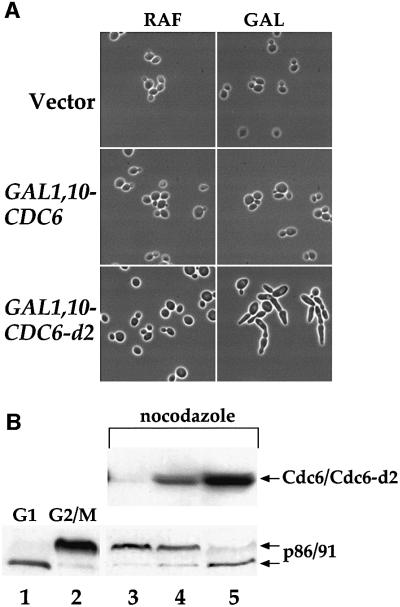
Microscopic examination revealed that cells expressing GAL1,10–CDC6-d2 developed abnormal cell morphology characterized by the presence of highly elongated buds (Figure 2A). This morphology is similar to that of cdc4 temperature-sensitive mutant cells at the restrictive temperature (Hartwell et al., 1973, 1974; Hereford and Hartwell, 1974; Hartwell, 1976). It is also similar to the phenotype of cells overexpressing the Cdc28-Clb kinase inhibitor (CKI) Sic1 (Schwob et al., 1994; Dahmann et al., 1995; Noton and Diffley, 2000) and of cells bearing mutations that compromise Cdc28-Clb activity (Schwob et al., 1994), presumably because Cdc28-Clb kinase activity inhibits polarized bud growth induced by Cln-Cdc28 kinase (Lew and Reed, 1993, 1995). This phenotype suggested that Cdc6-d2 was causing a G2/M arrest by inhibiting Cdc28-Clb kinase. Cdc6 can interact with and inhibit Cdc28-Clb kinase in vitro (Elsasser et al., 1996; Calzada et al., 2000) and in vivo (Desdouets et al., 1998; Calzada et al., 2000); thus, it was possible that the dominant-negative effect of overexpression of Cdc6-d2 was the result of Cdc28-Clb kinase inhibition. Consistent with this, removal of the N-terminal 47 amino acids of Cdc6-d2, which contain the Cdc28-Clb-interacting domain, eliminates the growth arrest phenotype (data not shown).
Fig. 2. (A) YGP28 (vector), YGP24 (GAL1,10–CDC6) or YGP25 (GAL1,10–CDC6-d2) cells were grown in YPRaf to mid-log phase and then transferred into either fresh YPRaf or YPGal and allowed to grow for 6 h. Three microlitres from each culture were examined by phase contrast microscopy. (B) YGP28 (lane 3), YGP24 (lane 4) and YGP25 (lane 5) cells were grown in YPRaf to mid-log phase and then blocked in G2/M with nocodozole. Once 90% of the cells had a large budded phenotype they were transferred into YPGal also containing nocodazole. After a further 2.5 h, samples were taken and processed for immunoblotting to examine the phosphorylation status of the DNA polymerase α subunit p86/91. Control samples to visualize the usual pattern of p86/91 phosphorylation in G1 (lane 1) and G2/M (lane 2) cells are also shown.
The second largest subunit (B subunit) of DNA polymerase α primase is phosphorylated late in the cell cycle by Cdc28-Clb kinase (Foiani et al., 1995). This phosphorylation causes a shift in mobility in SDS– polyacrylamide gels, and inhibition of Cdc28-Clb kinase in nocodazole-arrested cells is sufficient to cause dephosphorylation of the B subunit (Desdouets et al., 1998). Overexpression of wild-type Cdc6 in nocodazole-arrested cells can drive dephosphorylation of B subunit when SCFCDC4 is inactive (Desdouets et al., 1998). However, Figure 2B, lane 4, shows that overexpression of wild-type Cdc6 in nocodazole-arrested cells does not lead to dephosphorylation of B subunit when SCFCDC4 is active. By contrast, overexpression of the mutant Cdc6-d2 leads to efficient dephosphorylation of B subunit even when SCFCDC4 is active (Figure 2B, lane 5).
These results are consistent with the idea that overexpression of Cdc6-d2 blocks cells in G2/M by inactivating Cdc28-Clb kinase. CDK inhibition could be due to direct action of Cdc6 on Cdc28. Alternatively, since both Cdc6 (Piatti et al., 1995; Elsasser et al., 1996; Drury et al., 1997) and the CKI Sic1 (Schwob et al., 1994; Knapp et al., 1996; Verma et al., 1997b) are targeted for degradation by SCFCDC4, the effects might be indirect. That is, overexpression of the mutant Cdc6 might, in some way, interfere with SCFCDC4, causing inappropriate accumulation of Sic1 and consequent cell cycle arrest. Such a mechanism has been shown to explain how overexpression of budding yeast Cdc6 drives re-replication in fission yeast (Wolf et al., 1999a). However, as shown in Figure 1A, overexpression of Cdc6-d2 is still lethal in cells lacking Sic1, thus, the dominant-negative phenotype is not due to stabilization of Sic1.
Cdc6-d2 proteolysis does not occur in G2/M but is normal in G1 and S phases
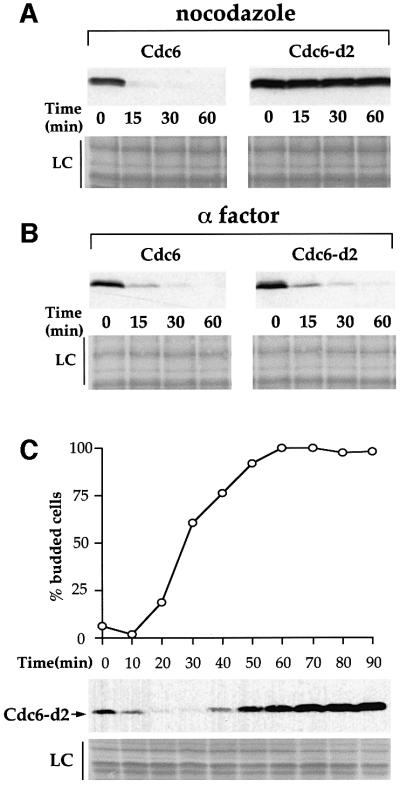
The results described above suggest that Cdc6-d2 overexpression prevents entry into mitosis by inactivating Cdc28-Clb kinase. The wild-type Cdc6 protein appears to be less efficient at inhibiting Cdc28-Clb kinase since its overproduction does not block cell proliferation. One possible explanation for this difference might be that the mutant protein is more stable than wild-type Cdc6. This would allow the protein to accumulate to higher levels and, thus, inhibit Cdc28-Clb kinase more effectively. This possibility is consistent with the fact that Cdc6-d2 overexpression in a CDC4+ background results in a phenotype very similar to the overexpression of wild-type Cdc6 in a cdc4 mutant background (Drury et al., 1997; Sanchez et al., 1999). It is also consistent with the fact that Cdc6-d2 accumulates to higher levels than wild-type Cdc6 when expressed in nocodazole-arrested cells (Figure 2B). To test this hypothesis directly, we examined the rate at which wild-type and mutant proteins disappeared in cells arrested in mitosis. In Figure 3A, cells were first arrested with nocodazole in raffinose-containing medium and Cdc6 or Cdc6-d2 was expressed by switching cells to galactose-containing medium for 30 min. Transcription was then repressed by transferring cells to glucose-containing medium and further translation was blocked with cycloheximide. The disappearance of Cdc6 was examined by immunoblot. This experiment shows that, as described previously, the wild-type protein disappears rapidly in nocodazole-arrested cells; however, the mutant Cdc6-d2 protein is almost completely stable in these cells. This experiment indicates that Cdc6-d2 is resistant to mode 3 proteolysis.
Fig. 3. YGP24 (GAL1,10–CDC6) and YGP25 (GAL1,10–CDC6-d2) cells were grown in YPRaf until mid-log phase, the cultures were then split into two and blocked with either nocodazole (A) or α factor (B). After blocking GAL1,10–CDC6 or GAL1,10–CDC6-d2 was induced by the addition of galactose. Thirty minutes later, transcription from the GAL1,10 promoter and translation were repressed (T = 0). Further samples were taken at the times indicated and processed for immuno blotting. LC is the loading control showing a region of Ponceau-S-stained membrane corresponding to 55–66 kDa. (C) YP25 cells were grown in YPRaf and blocked with α factor to synchronize them. Once the cells were blocked galactose was added to induce expression of GAL1,10–CDC6-d2. After 30 min the cells were washed and released from the block into YPRaf/Gal. Samples were taken at the time of release and every subsequent 10 min to be processed for immuno blotting and to assess the budding index.
To determine whether this effect on Cdc6 proteolysis is a specific cell cycle effect or some more general defect, we examined the proteolysis of wild-type and mutant Cdc6 proteins at other points in the cell cycle. In α-factor-arrested cells Cdc6 proteolysis occurs by a pathway (mode 1) that is distinct from mode 3; it is independent of SCFCDC4 and does not require any of the CDK consensus sites in Cdc6. Figure 3B shows that Cdc6 and Cdc6-d2 are both degraded at very similar rates in α-factor-arrested cells. Thus, Cdc6-d2 is not defective as a substrate for mode 1 proteolysis while it is completely defective as a substrate for mode 3 proteolysis.
Mode 2 proteolysis during late G1 phase and S phase is more rapid than mode 3 proteolysis but, like mode 3, requires SCFCDC4. We were, therefore, interested in determining whether Cdc6-d2 was generally defective in all SCFCDC4 proteolysis or if it is specifically defective in mode 3 proteolysis. To test this, cells were arrested in G1 with α factor, transferred to galactose-containing medium in α factor and released from the α-factor arrest into fresh galactose-containing medium. In this experiment, therefore, Cdc6 is constitutively expressed during and after release from the G1 arrest. We have shown previously that the very rapid mode 2 proteolysis drastically reduces the steady-state levels of Cdc6 from approximately the time of bud emergence until the end of S phase in such an experiment (Drury et al., 1997, 2000). Figure 3C shows that Cdc6-d2 mutant protein is also unable to accumulate during this time interval. The reduction in Cdc6-d2 levels during S phase is very similar to that seen with the wild-type protein (Drury et al., 2000; and data not shown) indicating that Cdc6-d2 can still be targeted for proteolysis during S phase. At later time points in this experiment, Cdc6-d2 accumulates to quite high levels, consistent with a defect in degradation later in the cell cycle. Thus, Cdc6-d2 is specifically defective as a substrate for mode 3 proteolysis and is not affected for mode 1 or 2 proteolysis.
The mutation in Cdc6-d2 identifies a CDK consensus site required for mode 3 proteolysis
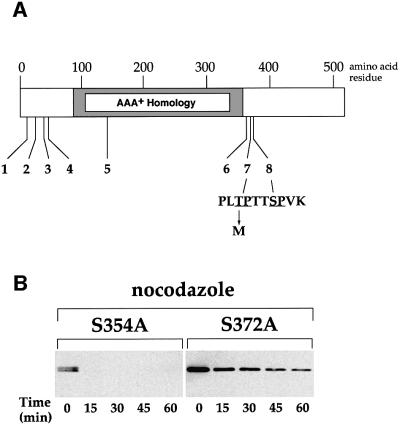
DNA sequence analysis of the CDC6-d2 allele identified a single point mutation causing a change from C to T at bp 1103, which resulted in a replacement of threonine at amino acid 368 with methionine (Figure 4A). Cdc6 contains eight SP or TP motifs that could potentially serve as CDK phosphorylation sites; the mutation in Cdc6-d2 destroys the seventh of these sites. This site is not a ‘complete’ consensus site since it does not contain a basic residue at the +3 position (Langan et al., 1989; Shenoy et al., 1989). Cdc6 is a member of a superfamily of ATPases (the AAA+ family) characterized by an extensive region of homology (Perkins and Diffley, 1998; Neuwald et al., 1999). The mutation in Cdc6-d2 occurs ∼30 amino acids C-terminal to this region of similarity. In a 50 amino acid residue region surrounding this mutation, there are six proline and three glycine residues, suggesting that this region may be unstructured. There are two other SP motifs in this region and we were interested in determining whether mutation of these sites also affected the stability of Cdc6. To examine this we used site-directed mutagenesis to construct two alleles of CDC6, changing serine 354 to alanine in one case (S354A) and serine 372 to alanine in the other (S372A). Figure 4C shows that Cdc6-S354A was highly unstable in nocodazole-arrested cells, similar to wild-type Cdc6. However, Cdc6-S372A was significantly more stable under these conditions, though less stable than Cdc6-d2 (compare Figure 3A with 4B). Thus CDK sites seven and eight both contribute to proteolysis during mitosis.
Fig. 4. (A) A schematic diagram to show the relative positions of the eight S/TP motifs in Cdc6. The amino acid sequence around sites 7 and 8 is shown and the arrow denotes a T to M substitution at amino acid position 368 caused by the mutation in CDC6-d2. Amino acid residue numbers are at the top of the diagram; the region of similarity in various AAA+ family members is indicated as a grey box (Perkins and Diffley, 1998; Neuwald et al., 1999). (B) GAL1,10 promoter shut-off experiments with cells blocked in G2/M by nocodazole and expressing either GAL1,10–CDC6-S354A (phosphorylation site 6) or GAL1,10–CDC6-S372A (phosphorylation site 8). Transcription and translation were repressed and a sample was taken (T = 0) to be processed for immunoblotting. Subsequent samples were taken at the times indicated.
Cdc6 contains two separate Cdc4p-interaction domains
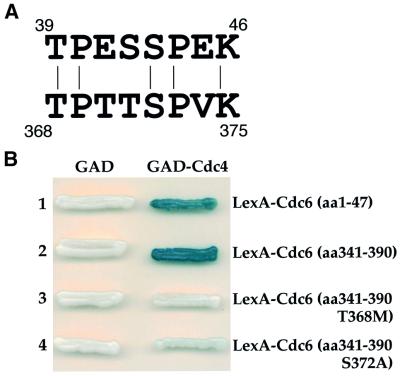
Our previous analysis had suggested that the N-terminal 47 amino acids of Cdc6 played an important role in SCFCDC4-dependent proteolysis, thus it was somewhat surprising to find that sequences in the middle of the protein were also involved in proteolysis. We showed previously that the N-terminal 47 amino acids of Cdc6 interacted with Cdc4 in a two-hybrid assay and that the strongest interaction required sequences between amino acids 17 and 47 (Drury et al., 1997). We noticed a short region of sequence similarity between this region and the region encompassing the mutation in Cdc6-d2 (Figure 5A). This sequence contains an incomplete CDK consensus site (without the basic residue at +3) followed by a complete consensus site. We were interested in whether this sequence comprises a hitherto unidentified Cdc4-interaction domain. To address this, we fused a 50 amino acid polypeptide encompassing this sequence to the LexA DNA binding domain and examined its ability to interact with Cdc4 in a two-hybrid experiment. Figure 5B shows that, like the N-terminal 47 amino acids of Cdc6, amino acids 341–390 interacted strongly with Cdc4, indicating that this region can act as a Cdc4-interaction domain in vivo.
Fig. 5. (A) Similarity of a Cdc4-interaction domain in the N-terminus of Cdc6 with sequence around the mutation in Cdc6-d2. The numbers denote the amino acid in Cdc6 at the start and finish of the two sequences. (B) Two-hybrid analysis with these two regions of Cdc6.
Interaction of SCFCDC4 with substrates such as Sic1 requires phosphorylation at CDK sites (Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997a). We considered the possibility that mutation of CDK sites 7 and 8, which causes stabilization of Cdc6 in nocodazole-arrested cells, might affect interaction with Cdc4. To examine this, polypeptides corresponding to amino acid residues 341–390 but containing single amino acid substitutions in the CDK consensus sites were tested for their ability to interact with Cdc4 in the two-hybrid system. Figure 5B shows that mutation of either phosphorylation site dramatically reduced Cdc4 interaction. The T368M mutant (from Cdc6-d2), which in the context of the full-length protein is completely stable in nocodazole-arrested cells (Figure 3A), shows no detectable interaction with Cdc4, while the S372A mutant, which is more stable but not completely stable (Figure 4B), still shows weak interaction. Thus, there is a strong correlation between stability in G2/M and ability to interact with Cdc4.
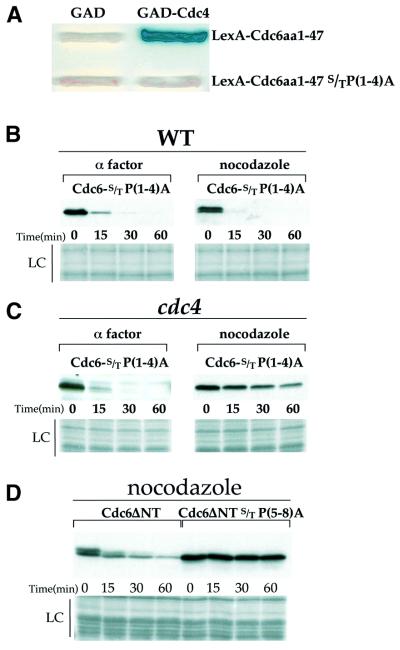
The results described above strongly suggest that Cdc6 contains an additional Cdc4-interaction domain between amino acids 341 and 390, which is critical for mode 3 proteolysis. The existence of a functionally important Cdc4-interaction domain in addition to the N-terminal site is supported by several other experiments. First, Figure 6A shows that a polypeptide containing the N-terminal 47 amino acids of Cdc6 interacts with Cdc4 in the two-hybrid system and, consistent with the importance of CDK phosphorylation, mutation of the four SP/TP motifs in this polypeptide abrogates this interaction. However, as shown in Figure 6B, when these four mutations are engineered into the full-length Cdc6 protein, the resulting protein is as rapidly degraded in nocodazole-arrested cells as it is in α-factor-arrested cells. As shown in Figure 6C, degradation of this protein in nocodazole-arrested cells is still completely dependent upon Cdc4, arguing strongly that there must be an additional Cdc4-interaction site in the protein. Secondly, we had previously shown that deletion of the N-terminal 47 amino acids of Cdc6 significantly stabilized the protein in nocodazole-arrested cells. We noted, however, that this truncated protein was still degraded, although the degradation was significantly slower (Drury et al., 1997). Furthermore, the degradation of this truncated protein was faster in nocodazole-arrested cells than it was in α-factor-arrested cells. In these original experiments, we were not adding cycloheximide at the time of transcriptional repression to prevent further translation of stable messages. Using cycloheximide, in Figure 6D we show that this truncated protein is, indeed, degraded in nocodazole-arrested cells although the rate of degradation is considerably slower than the rate for full-length protein (compare Figures 3A and 6D). Moreover, mutation of the remaining four CDK consensus sites in this truncated protein renders it completely stable in nocodazole-arrested cells. Although we do not know why the truncated protein is degraded more slowly than the full-length protein, we note that this truncation also removes a putative nuclear localization signal that resides in the N-terminal 47 amino acids (Jong et al., 1996). Regardless, these experiments, taken together, demonstrate the existence of a second, functionally important Cdc4-interaction domain between amino acids 341 and 390, which contains two critical CDK phosphorylation sites.
Fig. 6. (A) The requirement for the S/TP motifs in the N-terminal fragment of Cdc6 was tested in a two-hybrid assay with Cdc4p. The top row shows the two-hybrid interaction between the N-terminus of Cdc6 and Cdc4p described previously. In the second row the N-terminus but with the S/TP motifs mutated to A[S/TP(1–4)A] does not interact with GAD-Cdc4. (B) Promoter shut-off experiments with Cdc6-S/TP(1–4)A in cells blocked in G1 or G2/M show that these motifs are not required for instability in G1 or G2/M. However, this instability is dependent on CDC4 (C) in G2/M since Cdc6-S/TP(1–4)A is stable in cdc4ts cells blocked in nocodazole. (D) Cdc6ΔNT is more stable than the full-length protein. It is, however, degraded at a slower rate as can be seen above in a promoter shut-off in G2/M blocked cells. Mutation of the remaining S/TP(5–8)A in this truncated protein stabilizes it.
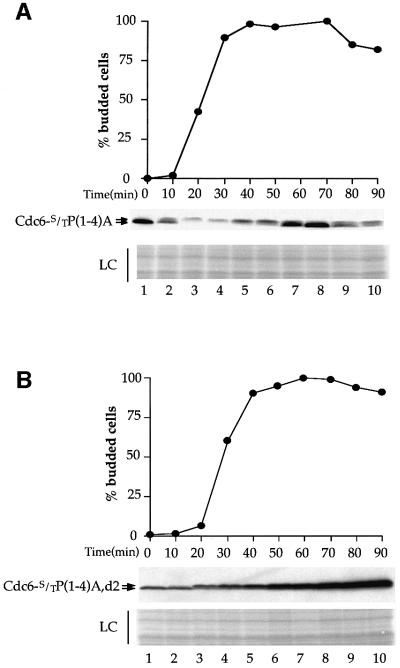
Our results thus far show that Cdc6 contains two Cdc4-interaction domains; one is located in the N-terminal 47 amino acids (Cdc4 site 1) and one is located between amino acids 341 and 390 (Cdc4 site 2). We have shown that mutation which abrogates Cdc4 site 2 interaction (i.e. Cdc6-d2) prevents SCFCDC4-dependent mode 3 degradation in nocodazole-arrested cells but not SCFCDC4-dependent mode 2 degradation during S phase. Conversely, mutations that abrogate Cdc4 site 1 interaction [i.e. Cdc6S/TP(1–4)A] do not affect SCFCDC4-dependent mode 3 degradation in nocodazole-arrested cells. Thus, we were interested in determining whether the degradation of this mutant during S phase by mode 2 was affected. Figure 7A shows that the levels of this mutant protein, when constitutively expressed, are significantly reduced during S phase although these levels are not reduced to the same extent as the wild-type protein, suggesting some role for site 1 in mode 2 proteolysis. However, this experiment, together with the experiment in Figure 3C, shows that neither of the two Cdc4-interaction sites is sufficient to account fully for mode 2. Thus, we considered the possibility that either site can target Cdc6 for mode 2 proteolysis. To test this, we examined the proteolysis of a Cdc6 mutant in which both Cdc4-interaction sites are inactivated by mutation of the important CDK consensus sites. To do this, all four CDK sites in the N-terminus, together with CDK site 7, were mutated. Figure 7B shows that, unlike mutants in either site 1 or site 2 alone, this protein is stable during S phase. Thus, these two Cdc4-interaction sites act redundantly during S phase.
Fig. 7. (A) YLD66 cells grown in YPRaf were synchronized with α factor. Once the cells were blocked, galactose was added to induce the Cdc6-S/TP(1–4)A protein. After a further 30 min the cells were released from the block into YPRaf/Gal. Samples were taken at the time of release (T = 0) and every 10 min after for a duration of 90 min to assess the budding index and for immunoblotting to detect levels of Cdc6-S/TP(1–4)A. (B) A similar experiment was performed with strain YLD69 [GAL1,10–CDC6-S/TP(1–4)A,d2].
Discussion
The proteolysis of the budding yeast Cdc6 protein is under surprisingly complex regulation. Previously, we distinguished two distinct modes of SCFCDC4-dependent Cdc6 proteolysis during late G1/early S phase and during G2/M primarily on the basis of degradation rate. Here, we have extended this distinction by showing that Cdc6 is targeted for SCFCDC4-dependent degradation by two different SCFCDC4-interaction domains, which are regulated differently during the cell cycle. One of these interaction domains, in the N-terminal 47 amino acids, is only utilized during late G1 and S phases while the other, located in the middle of the primary amino acid sequence is utilized from late G1 through mitosis (see Figure 7A). As a consequence, Cdc6 proteolysis during late G1/S phase is not dependent upon either domain individually but requires one or the other domain while proteolysis in G2/M is entirely dependent upon the second domain.
The mechanism by which these two SCFCDC4 elements are differentially regulated is unclear. It is possible that SCFCDC4 itself changes during the cell cycle, either in its subunit composition or by some post-translational modification, so that it is unable to recognize site 1 in G2/M. Although we cannot rule out this possibility, there is currently no evidence to support this idea. Alternatively, some additional regulation may act through Cdc6 itself to determine the availability of these interaction sites. The rapid mode 2 proteolysis normally begins near the onset of S phase and our previous work showed that, although it required CDK activity, it did not require the activity of the S-phase-promoting cyclins, Clb5 and Clb6. Moreover, mode 2 Cdc6 proteolysis was not delayed in strains lacking Clb5 and Clb6. This suggested that the G1 cyclins may play a direct role in mode 2 degradation. In nocodazole-arrested cells, when Cdc6 is degraded via mode 3, the Clns are present at very low levels because mitotic cyclins like Clb2 repress their expression (Amon et al., 1993). Thus, the inactivity of site 1 in nocodazole-arrested cells may indicate that this site is a specific substrate of the Cln kinase and is not phosphorylated by Clb-associated kinases. We think this is unlikely for two reasons. First, the sequences of the two sets of CDK sites are very similar and second, overexpression of Cln2 in nocodazole-arrested cells does not destabilize Cdc6-d2 (L.S.Drury and J.F.X.Diffley, data not shown).
We suggest another possibility. In addition to several CDK consensus sites and a Cdc4-interacting domain, the N-terminal 47 amino acids also contain a region that interacts very tightly with Clb, but not with Cln-associated Cdc28 (Elsasser et al., 1996). The tight association of Cdc28-Clb with this N-terminal region may act to prevent either phosphorylation of the N-terminus or association of the phosphorylated N-terminus with Cdc4. Further work will be required to separate the Clb-Cdc28 binding domain from the Cdc4-interacting domain and test this hypothesis.
The complexity of Cdc6 proteolysis suggests that elimination of the protein from cells is likely to be important. Mode 2 degradation acts to eliminate Cdc6 rapidly from cells before DNA replication begins and, thus, appears to play a role in preventing the relicensing of origins that have already fired. The experiments in this paper, together with the results of Bueno and Russell (1992) suggest an additional reason for removing Cdc6 rapidly from cells: when not degraded, Cdc6 can interfere with progression into mitosis. In the experiments described here, the high Cdc6 levels generated in the CDC6-d2 mutant appear to prevent entry into mitosis. It is possible that even when expressed at lower levels, stable Cdc6 might cause some mitotic defect. Cdc4 is required to degrade the CKI Sic1 in G1 phase to allow cells to enter S phase. Interestingly, Cdc4 is still essential in cells lacking Sic1 (Schwob et al., 1994) and such double mutants (cdc4ts,sic1Δ) are defective in progressing through mitosis (Goh and Surana, 1999). Perhaps the inability to degrade Cdc6 contributes to this defect.
The ability of stable Cdc6 to prevent mitosis correlates with an apparent reduction in CDK activity in vivo as shown by dephosphorylation of DNA polymerase α. Given that this loss of CDK activity in vivo appears to be independent of Sic1 and that Cdc6 can interact with and inhibit Cdc28-Clb kinases in vivo and in vitro, it is most likely that stable Cdc6 inhibits mitosis by directly inhibiting Cdc28 kinase. The reason for this CKI activity is unclear. The domain responsible for kinase inhibition can be removed from Cdc6 without affecting its ability to assemble pre-RCs and support normal DNA replication (Drury et al., 1997). It is, therefore, likely that CDK inhibition by Cdc6 is, in some way, a redundant activity. It is possible that this activity of Cdc6 acts with other mechanisms to ensure full inhibition of Cdc28 activity at the end of mitosis. It is also possible that the tight interaction of Cdc28 with Cdc6 may inhibit Cdc6 function; however, elimination of this interaction does not lead to over-replication because several redundant mechanisms prevent reassembly of pre-RCs later in the cell cycle (Diffley, 2001).
In both budding and fission yeasts, Cdc6 (cdc18) is targeted for degradation by CDK-dependent, SCFCDC4-mediated polyubiquitylation. Whether the fission yeast protein has a similar organization of SCFCDC4-interaction sites remains to be determined. However, like budding yeast Cdc6, fission yeast cdc18 normally disappears during S phase and remains unstable in cells arrested in mitosis (Kelly et al., 1993; Kominami and Toda, 1997; Baum et al., 1998; Jallepalli et al., 1998; Lopez-Girona et al., 1998; Wolf et al., 1999b). In vertebrates, Cdc6 proteolysis may be more complex. Chromatin-bound Cdc6 appears to remain stable during S phase (Williams et al., 1997; Coverley et al., 2000) though soluble Cdc6 is degraded at this time (Coverley et al., 2000). Additionally, human Cdc6 appears to be targeted for degradation at the end of mitosis by the APC/C (Mendez and Stillman, 2000; Petersen et al., 2000). Thus, although details may differ, proteolysis at some point during the cell cycle may be a common feature of Cdc6 regulation.
In conclusion, we have shown that Cdc6 proteolysis via SCFCDC4 occurs through two different Cdc4-interaction sites, which are regulated differently during the cell cycle. This represents a novel way in which the rate of a protein’s degradation can be modulated by a single ubiquitylation pathway.
Materials and methods
Strains and media
Strains (listed in Table I) were all derived from W303-1a. Cells were routinely grown in YP containing glucose (YPD), raffinose (YPRaf) or galactose (YPGal) at 2% (Rose et al., 1989). Cell cycle blocks with nocodazole or α factor were as described previously (Diffley et al., 1994).
Table I. Yeast strains.
| Strain | Genotype |
|---|---|
| YGP24 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, GAL1,10–CDC6, (HIS3) |
| YGP25 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, GAL1,10–CDC6-d2, (HIS3) |
| YGP28 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, pRS303 (HIS3) |
| YGP54 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, GAL1,10–CDC6S372A, (HIS3) |
| YLD65 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, GAL1,10- CDC6-S/TP (1–4)A, (HIS3) |
| YLD66 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, cdc4-1, GAL1,10–CDC6-S/TP (1–4)A, (HIS3) |
| YLD67 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, GAL1,10–CDC6 ΔNT (LEU2) |
| YLD68 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, GAL1,10–CDC6ΔNT S/TP (5–8)A, (LEU2) |
| YLD69 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3 112 can1-100, GAL1,10–CDC6-S/TP (1–4)A,d2, (HIS3) |
| L40 | MATa his3Δ200 trp1-901 leu2-3 112 ade2 lys2-801am URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3 |
Plasmid constructs
The plasmids containing GAL1,10–CDC6 have been described previously (Drury et al., 1997). To make the plasmid containing the GAL1,10– CDC6-S/TP(1–4)A allele (pLD25), site-directed mutagenesis was performed using a QuikChange™ site-directed mutagenesis kit (Stratagene). The starting template was GAL1,10–CDC6 in pRS303 and sites were mutated sequentially with the oligonucleotide primers T7A, T23A, T39A and S43A (see Table II). The final product was sequenced to ensure that only the directed mutations were present. The same strategy was used to make the S354A and the S372A alleles using oligonucleotide primers 95856 and 95854, respectively (Table II). To make the GAL1,10–CDC6-S/TP(1–4)A,d2 allele the smaller PstI fragment was subcloned from pGP26 (pRS303-GAL1,10–CDC6-d2) into the larger PstI fragment from pLD25 and the orientation was confirmed by sequencing.
Table II. Oligonucleotide primers.
| Primer | Sequence 5′–3′ |
|---|---|
| 49362 | CGCGGATCCGTTGCGTATATCATTGCATC |
| 67395 | AAAAAGGATCCATGCAGTTTGGCTCACAGTCT |
| 49628 | AAAAAGGATCCGTATGTCAGCTATACCAAT |
| 67393 | GTTGCAGTTCACAGATGTTGCACCAGAATCCGCGCCAGAA |
| 124310 | AAAAAGAATTCAGTATCGAAATCTATGAGTT |
| 124311 | TTTTTGTCGACCTATATGTAGTTCAAGCCTATTT |
| 95854 | CCTTTGACGCCAACTACTGCCCCGGTAAAGAAATCG |
| 95856 | GAAAAGCGGTTTCTGCTGGCGCCAACAACAAGAGGATCATTG |
| T7A | GTCAGCTATACCAATAGCCCCAACTAAGCGTATC |
| T23A | CTATTTGACGATGCTCCAGCGGCGCCTCCACGACCTTTG |
| T39A | GTTGCAGTTCACAGATGTTGCACCAGAATCCGCGCCAGAA |
| S43A | GATGTTACACCAGAATCCGCGCCAGAAAAACTGCAGTTTGG |
The plasmid pGP19 is pRS305 containing the GAL1,10–CDC6ΔNT subcloned from pLD4 (Drury et al., 1997). pTS4 was made by using the oligonucleotide primers 67395 and 49362 to PCR the CDC6ΔNT gene from pLD14 [GAL1,10–CDC6S/TP(1–8)A] so that it contains S/TP(5–8)A (T.Seki, personal communication).
To make the two-hybrid constructs, the oligonucleotide primers 124310 and 124311 were used to PCR a 150 bp fragment from CDC6, CDC6-d2 or CDC6S372A and cloned into pBTM116 using the EcoRI and SalI restriction enzyme recognition sites. Similarly, to analyse the N-terminal Cdc4-interaction domain, the oligonucleotide primers 49628 and 67393 were used to PCR a fragment encoding the first 47 amino acids from CDC6 from pLD25 and these were subcloned into pBTM116. The pACT vector encoding GAD-Cdc4 was described previously (Drury et al., 1997).
Cell cycle experiments
Cell cycle synchronization was performed as described previously (Diffley et al., 1994). α-factor block and release experiments entailed washing the blocked cells twice with 2 vol of fresh medium and returning them to fresh medium at the same cell density.
GAL1,10 promoter induction, repression and protein stability experiments
For induction of the GAL1,10 promoter, cells were grown in YP medium containing raffinose as a carbon source. Galactose was added to a final concentration of 2%. The time of induction was generally 30 min. Transcriptional repression was achieved by the addition to the cell suspension of glucose to a final concentration of 2%. At the same time cycloheximide was added to a final concentration of 1 mg/ml to prevent further translation of any mRNA.
Cell extracts, immunoblotting and loading controls
Protein extracts from trichloroacetic acid-fixed yeast cells were made as described (Desdouets et al., 1998). The samples were boiled in sample buffer for 3 min and 5–10 µl was loaded on 10% a SDS gel and submitted to PAGE. For immunoblot analysis the proteins were transferred on to Hybond ECL membrane (Amersham) and blocked with 5% dried milk in Tris-buffered saline containing 0.1% Tween-20. Detection of wild-type Cdc6 and the various mutant Cdc6 proteins was performed using the monoclonal 9H/85 at 5 µg/ml and detection of the DNA polymerase α subunit p86/91 was with the 6D2 antibody (Desdouets et al., 1998). The proteins were detected using ECL reagents (Amersham) as per the manufacturer’s instructions. To ensure equal sample loading and even transfer from the polyacrylamide gel to the membrane the blots were stained with Ponceau S (Sigma) and scanned before antibody incubation.
Two-hybrid analysis
Various pBTM116 and pACT constructs were transformed into the yeast strain L40 (Hollenberg et al., 1995). The transformants were tested for protein–protein interaction by streaking on selective agar plates buffered with phosphate buffer to pH 7.0 and containing 5-bromo-chloroindol-3-yl β-d-galactopyranoside (X-Gal) at a concentration of 40 µg/ml.
FACS analysis
Samples for flow cytometric analysis were collected and processed as described previously (Santocanale and Diffley, 1996). Analysis was performed using a Becton Dickinson FACScan.
Acknowledgments
Acknowledgements
We thank members of the laboratory for helpful discussions. This work was supported by the Imperial Cancer Research Fund.
References
- Amon A., Tyers,M., Futcher,B. and Nasmyth,K. (1993) Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell, 74, 993–1007. [DOI] [PubMed] [Google Scholar]
- Baum B., Nishitani,H., Yanow,S. and Nurse,P. (1998) Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J., 17, 5689–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno A. and Russell,P. (1992) Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J., 11, 2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada A., Sanchez,M., Sanchez,E. and Bueno,A. (2000) The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem., 275, 9734–9741. [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski,R.S., Dante,M., Shero,J.H. and Hieter,P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene, 110, 119–122. [DOI] [PubMed] [Google Scholar]
- Cocker J.H., Piatti,S., Santocanale,C., Nasmyth,K. and Diffley,J.F.X. (1996) An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature, 379, 180–182. [DOI] [PubMed] [Google Scholar]
- Coverley D., Pelizon,C., Trewick,S. and Laskey,R.A. (2000) Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci., 113, 1929–1938. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Diffley,J.F.X. and Nasmyth,K.A. (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of origins to a pre-replicative state. Curr. Biol., 5, 1257–1269. [DOI] [PubMed] [Google Scholar]
- Desdouets C., Santocanale,C., Drury,L.S., Perkins,G., Foiani,M., Plevani,P. and Diffley,J.F.X. (1998) Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase α. EMBO J., 17, 4139–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detweiler C.S. and Li,J.J. (1997) Cdc6p establishes and maintains a state of replication competence during G1 phase. J. Cell Sci., 110, 753–763. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X. (1996) Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev., 10, 2819–2830. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X. (2001) DNA replication: building the perfect switch. Curr. Biol., 11, R367–R370. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D. and Blow,J.J. (1999) The regulation of replication origin activation. Curr. Opin. Genet. Dev., 9, 62–68. [DOI] [PubMed] [Google Scholar]
- Drury L.S., Perkins,G. and Diffley,J.F.X. (1997) The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J., 16, 5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L.S., Perkins,G. and Diffley,J.F.X. (2000) The cyclin dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol., 10, 231–240. [DOI] [PubMed] [Google Scholar]
- Dutta A. and Bell,S.P. (1997) Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell. Dev. Biol., 13, 293–332. [DOI] [PubMed] [Google Scholar]
- Elsasser S., Lou,F., Wang,B., Campbell,J.L. and Jong,A. (1996) Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol. Biol. Cell, 7, 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S., Chi,Y., Yang,P. and Campbell,J.L. (1999) Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol. Biol. Cell, 10, 3263–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman R.M., Correll,C.C., Kaplan,K.B. and Deshaies,R.J. (1997) A complex of Cdc4p, Skp1p and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell, 91, 221–230. [DOI] [PubMed] [Google Scholar]
- Foiani M., Liberi,G., Lucchini,G. and Plevani,P. (1995) Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol. Cell. Biol., 15, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P.Y. and Surana,U. (1999) Cdc4, a protein required for the onset of S phase, serves an essential function during G(2)/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 5512–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. (1976) Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J. Mol. Biol., 104, 803–817. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Mortimer,R.K., Culotti,J. and Culotti,M. (1973) Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics, 74, 267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Culotti,J., Pringle,J.R. and Reid,B.J. (1974) Genetic control of the cell division cycle in yeast. Science, 183, 46–51. [DOI] [PubMed] [Google Scholar]
- Hereford L.M. and Hartwell,L.H. (1974) Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J. Mol. Biol., 84, 445–461. [DOI] [PubMed] [Google Scholar]
- Hollenberg S.M., Sternglanz,R., Cheng,P.F. and Weintraub,H. (1995) Identification of a new family of tissue-specific basic helix–loop–helix proteins with a 2-hybrid system. Mol. Cell. Biol., 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli P.V., Tien,D. and Kelly,T.J. (1998) sud1(+) targets cyclin-dependent kinase-phosphorylated Cdc18 and Rum1 proteins for degradation and stops unwanted diploidization in fission yeast. Proc. Natl Acad. Sci. USA, 95, 8159–8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong A., Young,M., Chen,G.C., Zhang,S.Q. and Chan,C. (1996) Intracellular location of the Saccharomyces cerevisiae CDC6 gene product. DNA Cell Biol., 15, 883–895. [DOI] [PubMed] [Google Scholar]
- Kelly T.J., Martin,G.S., Forsburg,S.L., Stephen,R.J., Russo,A. and Nurse,P. (1993) The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell, 74, 371–382. [DOI] [PubMed] [Google Scholar]
- Knapp D., Bhoite,L., Stillman,D.J. and Nasmyth,K. (1996) The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol. Cell. Biol., 16, 5701–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K. and Toda,T. (1997) Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev., 11, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Langan T.A., Gautier,J., Lohka,M., Hollingsworth,R., Moreno,S., Nurse,P., Maller,J. and Sclafani,R.A. (1989) Mammalian growth-associated H1 histone kinase: a homolog of the cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol. Cell. Biol., 9, 3860–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J. and Reed,S.I. (1993) Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol., 120, 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J. and Reed,S.I. (1995) Cell cycle control of morphogenesis in budding yeast. Curr. Opin. Genet. Dev., 5, 17–23. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A., Mondesert,O., Leatherwood,J. and Russell,P. (1998) Negative regulation of cdc18 DNA replication protein by cdc2. Mol. Biol. Cell, 9, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny C.J., Partridge,J.F., Mikesell,G.E., Creemer,D.P. and Breeden,L.L. (1997) A novel Mcm1-dependent element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev., 11, 1277–1288. [DOI] [PubMed] [Google Scholar]
- Mendez J. and Stillman,B. (2000) Chromatin association of human origin recognition complex, cdc6 and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol., 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A., Aravind,L., Spouge,J. and Koonin,E. (1999) AAA+: A class of chaperone-like ATPases associated with the assembly, operation and diassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- Noton E.A. and Diffley,J.F.X. (2000) CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol. Cell, 5, 85–95. [DOI] [PubMed] [Google Scholar]
- Pasero P. and Schwob,E. (2000) Think global, act local—how to regulate S phase from individual replication origins. Curr. Opin. Genet. Dev., 10, 178–186. [DOI] [PubMed] [Google Scholar]
- Perkins G. and Diffley,J.F.X. (1998) Nucleotide dependent prereplicative complex assembly by Cdc6p, a homologue of eukaryotic and prokaryotic clamp-loaders. Mol. Cell, 2, 23–32. [DOI] [PubMed] [Google Scholar]
- Petersen B.O. et al. (2000) Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev., 14, 2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Lengauer,C. and Nasmyth,K. (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J., 14, 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1989) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanchez M., Calzada,A. and Bueno,A. (1999) The Cdc6 protein is ubiquitinated in vivo for proteolysis in Saccharomyces cerevisiae. J. Biol. Chem., 274, 9092–9097. [DOI] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F.X. (1996) ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J., 15, 6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Schwob E., Bohm,T., Mendenhall,M.D. and Nasmyth,K. (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell, 79, 233–244. [DOI] [PubMed] [Google Scholar]
- Shenoy S., Choi,J.-K., Bagrodia,S., Copeland,T.D., Maller,J.M. and Shalloway,D. (1989) Purified maturation promoting factor phosphorylates pp60c-src at sites phosphorylated during fibroblast mitosis. Cell, 57, 763–774. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Craig,K.L., Tyers,M., Elledge,S.J. and Harper,J.W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell, 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA-replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Verma R., Annan,R.S., Huddleston,M.J., Carr,S.A., Reynard,G. and Deshaies,R.J. (1997a) Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science, 278, 455–460. [DOI] [PubMed] [Google Scholar]
- Verma R., Feldman,R.M. and Deshaies,R.J. (1997b) SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34 and cyclin/CDK activities. Mol. Biol. Cell, 8, 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.S., Shohet,R.V. and Stillman,B. (1997) A human protein related to yeast Cdc6p. Proc. Natl Acad. Sci. USA, 94, 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D.A., McKeon,F. and Jackson,P.K. (1999a) Budding yeast Cdc6p induces re-replication in fission yeast by inhibition of SCF(Pop)-mediated proteolysis. Mol. Gen. Genet., 262, 473–480. [DOI] [PubMed] [Google Scholar]
- Wolf D.A., McKeon,F. and Jackson,P.K. (1999b) F-box/WD-repeat proteins pop1p and Sud1p/Pop2p form complexes that bind and direct the proteolysis of cdc18p. Curr. Biol., 9, 373–376. [DOI] [PubMed] [Google Scholar]
- Zhou C. and Jong,A. (1990) CDC6 mRNA fluctuates periodically in the yeast cell cycle. J. Biol. Chem., 265, 19904–19909. [PubMed] [Google Scholar]
- Zwerschke W., Rottjakob,H.-W. and Küntzel,H. (1994) The Saccharo myces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J. Biol. Chem., 269, 23351–23356. [PubMed] [Google Scholar]