Abstract
RNA editing is unique among post-transcriptional processes in plastids, as it exhibits extraordinary phylogenetic dynamics leading to species-specific editing site patterns. The evolutionary loss of a site is considered to entail the loss of the corresponding nuclear-encoded site-specific factor, which prevents the editing of foreign, i.e. heterologous, sites. We investigated the editing of short ‘spliced’ and ‘unspliced’ ndhA gene fragments from spinach in Nicotiana tabacum (tobacco) in vivo using biolistic transformation. Surprisingly, it turned out that the spinach site is edited in the heterologous nuclear background. Furthermore, only exon–exon fusions were edited, whereas intron-containing messages remained unprocessed. A homologue of the spinach site was found to be present and edited in Nicotiana tomentosiformis, representing the paternal parent, but absent from Nicotiana sylvestris, representing the maternal parent of tobacco. Our data show that: (i) the cis-determinants for ndhA editing are split by an intron; (ii) the editing capacity cannot be deduced from editing sites; and (iii) allopolyploidization can increase the editing capacity, which implies that it can influence speciation processes in evolution.
Keywords: allotetraploidy/chloroplast/evolution/RNA editing/RNA splicing
Introduction
Post-transcriptional modification of RNA molecules in plastids and mitochondria represents a key level of nuclear regulatory control of organelle genomes (reviewed in Binder et al., 1996; Barkan and Goldschmidt-Clermont, 2000; Bock, 2000; Monde et al., 2000; Rochaix, 2001). Plastid transcription units are usually multigenic, and primary transcripts are extensively processed to give rise to complex sets of overlapping RNA species. Among the underlying ∼12 activities that are almost exclusively nuclear-encoded, RNA editing is exceptional in several respects. (i) In contrast to other RNA modifications, editing, as found in plant organelles, appears phylogenetically late, probably first in bryophytes (Malek et al., 1996; Freyer et al., 1997). It is not known in prokaryotes and lower eukaryotes. (ii) Editing represents an important means of nuclear regulatory control. It can restore initiation codons, termination codons or amino acid residues that are generally conserved and of structural and/or functional importance for the affected polypeptide or multisubunit assembly (Maier et al., 1996). Quite serious phenotypes appear with impaired editing (Bock et al., 1994; Zito et al., 1997). (iii) In comparison with other plastid expression characteristics, such as RNA splicing for example, RNA editing is evolutionarily highly dynamic. Unlike the phylogenetically well conserved positions and structures of plastid introns, even closely related species can exhibit quite distinct patterns of editing sites (editotypes), which illustrates that such sites can change rapidly during evolution. (iv) Furthermore, no similiarity between cis sequences or predicted secondary structures around editing sites within a given species has been noted, quite in contrast to group II introns (Michel and Ferat, 1995), and factors acting in trans appear to be diverse as well (Hirose and Sugiura, 2001). Obviously, the RNA editing machinery of cell organelles is functionally and phylogenetically highly flexible.
The mechanism, origin and function of RNA editing still remain enigmatic. It has been shown, however, that RNA editing processes in plastids share mechanistic and phylogenetic similarities with mitochondria, which is suggestive of a common evolutionary origin (Maier et al., 1992; Malek et al., 1996; Freyer et al., 1997). In both organelles, it is characterized by C-to-U (in rare cases also by U-to-C) conversions, and circumstantial evidence suggests de- or transamination of cytidine residues as the most plausible mechanism (Maier et al., 1996; Fuchs et al., 2001; Hirose and Sugiura, 2001). No evidence for a plastid location of genes encoding components of the editing machinery has been obtained so far. Trans-factors involved have to be imported from the cytosol and appear to be highly specific for each editing site. The pattern of editing sites is generally species-specific and changes frequently, even between closely related taxa (Zeltz et al., 1993; Freyer et al., 1995, 1997). In the case of a distinct editing site in spinach psbF mRNA, no editing occurs upon introduction into Nicotiana tabacum (tobacco) plastids, suggesting that tobacco is lacking the corresponding editing factor (Bock et al., 1994). Editing of this site, however, is rescued by fusion of tobacco protoplasts with spinach cells (Bock and Koop, 1997). Comparably, the insertion of two maize rpoB editing sites into tobacco chloroplast chromosomes (Reed and Hanson, 1997) has shown that site II, for which a homologue in tobacco exists, is edited efficiently, whereas the heterologous site I, without an equivalent in tobacco, is not. Apparently, editing specificity factors are evolutionarily tightly linked to the presence of the site they are acting on. Loss of a site is supposed to cause the loss of the specificity factor. The remarkable degree of interspecies variation in editing patterns and frequency (Freyer et al., 1997) as well as the general failure to detect heterologous editing in higher plants suggested that editing sites along with their trans-factors co-evolve and form an inseparable evolutionary doublet.
In contrast to the relatively scarce data available on trans-factors, the requirements for cis sequences are better characterized at least for some editing sites. Using a transplastomic approach it has been shown that target sequences for the machinery operating in trans are predominantly, but not exclusively, found immediately upstream of the editing site (Chaudhuri et al., 1995; Bock et al., 1996, 1997; Chaudhuri and Maliga, 1996; Reed et al., 2001). Although these limited sequence stretches are often sufficient to direct proper editing, at least for some sites editing depends on other plastid processes as well. For instance, inhibition of chloroplast translation selectively blocks RNA editing at distinct sites (Karcher and Bock, 1998), whereas others are independent of translational activity (Zeltz et al., 1993; Hess et al., 1994). Aside from translational dependence of individual sites, kinetic aspects of RNA editing have also been investigated. For example, in petB and ycf3 transcripts RNA editing was shown to be a very early processing step, occurring without any preferential order with regard to both splicing and RNA cleavage (Freyer et al., 1993; Hirose et al., 1994; Ruf et al., 1994). Similar observations have been made for sites in plant mitochondria (Sutton et al., 1991; Yang and Mulligan, 1991). An exception has been reported for an editing site in barley, located only 12 bp downstream of the 3′ splice site of the ndhA intron (Lopez et al., 1997; del Campo et al., 2000). As no unspliced ndhA mRNA could be found to be already edited at this site, a strict kinetic relation of the two processes has been inferred. This renders the ndhA message an appealing candidate for studies on the interdependence of different plastid RNA processing events.
We have used a tobacco-based transplastomic approach to investigate the functional and evolutionary relation of splicing and editing on the ndhA transcript from spinach. We confirmed the dependence of ndhA editing on removal of the intron. Surprisingly, we found that tobacco, which lacks the editing site close to the ndhA intron, is capable of efficiently editing the corresponding spinach site, which represents the first example of heterologous editing of a non-endogenous site in plants. Tobacco is an ancient allotetraploid formed between a progenitor of Nicotiana sylvestris (female parent, donor of plastids and mitochondria; Bland et al., 1985; Olmstead and Palmer, 1991) and an ancestor of Nicotiana tomentosiformis (male parent; Godspeed, 1954; Gerstel, 1960, 1963; Kenton et al., 1993; Parokonny and Kenton, 1995; Lim et al., 2000a). In this species, the machinery acting in trans for processing plastid RNAs including the postulated editosome(s) is potentially derived from both parents. This presumably changes number and pattern of the nuclear-encoded editing factors, while the corresponding cis elements originate in only one, the maternal parent. Analysis of the editing capacity of the diploid ancestors of tobacco provided a likely phylogenetic explanation for the observed heterologous editing. The impact of editing upon plant speciation, especially in allopolyploidization and interspecific interbreeding, is discussed.
Results
Integration of the spinach ndhA editing site I into the tobacco plastid genome
The spinach plastid chromosome contains 11 genes coding for subunits of a putative chloroplast NADH dehydrogenase (Schmitz-Linneweber et al., 2001). The ndhA gene is part of a polycistronic transcription unit including altogether seven ndh genes and psaC, which encodes subunit VII of the photosystem I assembly. In the course of precursor processing, a group IIA intron is removed from the ndhA mRNA and the message is edited at two sites (Legen et al., 2001). One of these sites, designated site I, is located only 12 bp downstream of the 3′ intron–exon splice site (Figure 1). This site is also present in various other species, like barley and maize, but absent in tobacco. del Campo et al. (2000) have shown that in barley three of the four editing sites in the ndhA message are edited early in transcript genesis, that is, before splicing and RNA dissection. However, the fourth site, which is the homologue of spinach site I, remained unedited in non-spliced transcripts. This finding is consistent with sequence analysis of cDNA-derived recombinant PCR products spanning the 3′ splice site of the ndhA gene, which all contained the unedited nucleotide residue in maize, and of unspliced versus spliced spinach ndhA cDNAs, of which only the latter were edited (data not shown).
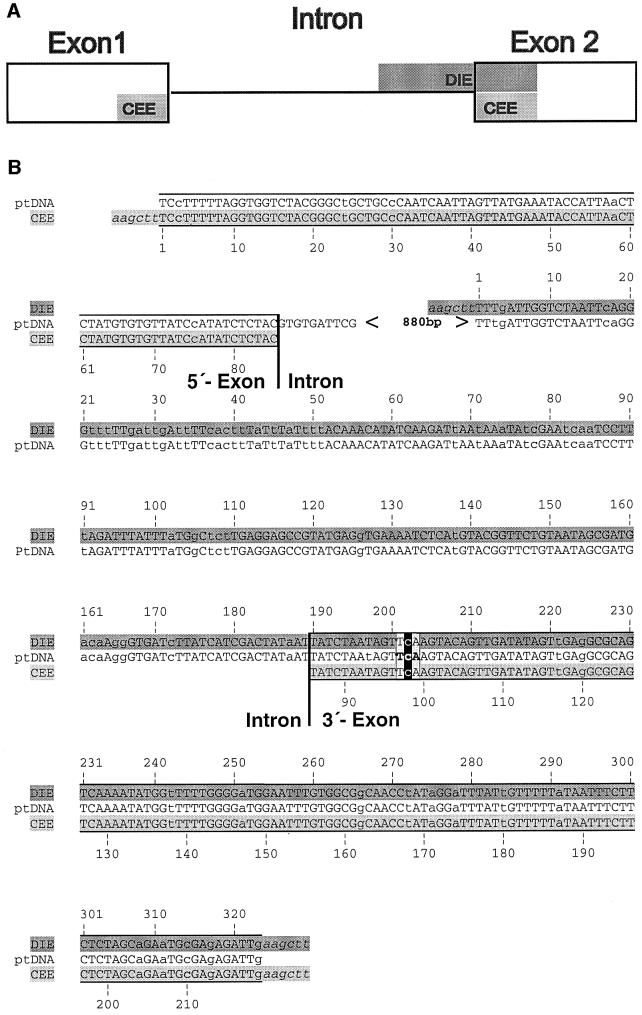
Fig. 1. Alignment of intron-containing and intronless ndhA sequences flanking editing site I. (A) Schematic presentation of the spinach ndhA gene. DNA fragments that are part of constructs pDIE and pCEE are shaded in dark or light grey, respectively. (B) DNA sequences are shown for part of the wild-type spinach ndhA (ptDNA) and ndhA constructs pDIE and pCEE. The pCEE sequence spans the fused 5′ and 3′ exon, whereas the pDIE sequence comprises the 3′ intron–exon border. Exon sequences are boxed. The editing site is marked in black. HindIII linkers used for cloning are in italics. Nucleotides differing from the corresponding tobacco sequence are shown in lower case letters. Shading as in (A).
A spinach plastid DNA fragment spanning the 3′ splice site and another one containing spliced exon sequences were each linked to a selectable marker gene (aadA) conferring resistance to spectinomycin (Goldschmidt-Clermont, 1991; Svab and Maliga, 1993). The intron–exon border fragment was amplified from DNA and the spliced form from cDNA by PCR simultaneously introducing HindIII sites via primer tags. The thymidine at the editing site on the cDNA-derived fragment was turned into a cytidine by a PCR-based mutagenesis scheme. The fragments were inserted into the aadA cassette using a HindIII site in between the aadA coding region and the 3′ regulatory sequence (Figure 2A). Linkage of the gene segments with the selective marker should provide stability of the transgene and also ensure transcription of the ndhA segments. The chimeric aadA–ndhA gene constructs were finally brought into a background of tobacco plastid DNA suitable for plastid transformation, that is, the psbJ–petA intergenic spacer. Insertion of transgenes at this site has no detectable effect on the plant phenotype (Bock et al., 1994).
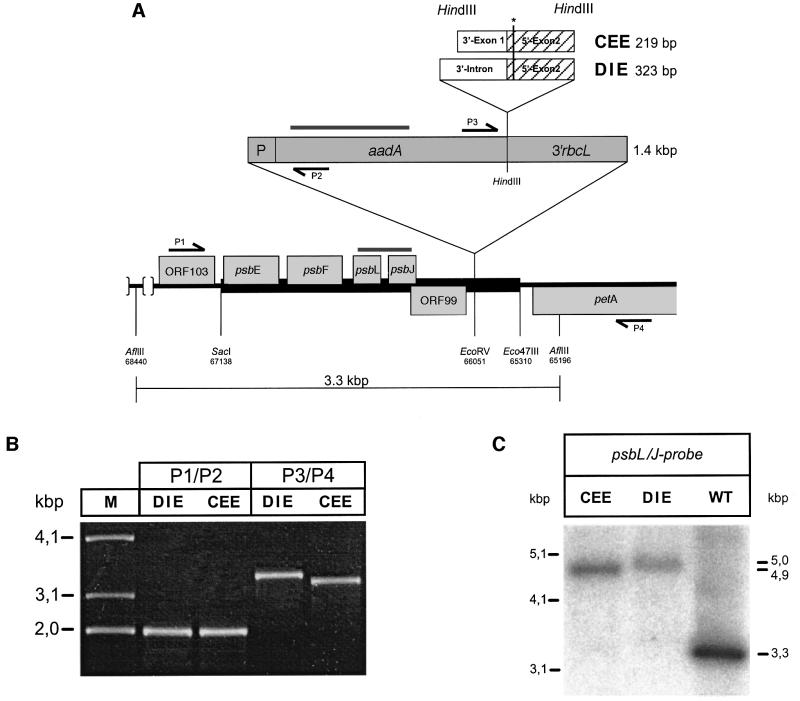
Fig. 2. Introduction of recombinant spinach ndhA constructs into the tobacco plastid chromosome. (A) Plasmid vectors with intron-containing (DIE) and intronless (CEE) spinach ndhA fragments, respectively, designed for plastid transformation of tobacco, were generated by introducing the spectinomycin-conferring aadA cassette into the EcoRV site (nucleotide positions 66053/4 of the tobacco plastid chromosome) of a SacI–Eco47III tobacco plastid DNA fragment (marked as black bar). PCR products CEE and DIE derived from the spinach ndhA gene were cloned into the HindIII site located in between the aadA coding region and its 3′ regulatory region (3′ rbcL). The position of the spinach-editing site is marked by an asterisk. Genes drawn above the line are transcribed from left to right; genes drawn below the line are transcribed in opposite direction. The grey bars indicate regions corresponding to probes used in Southern and northern analysis (Figures 2C and 4). Numbers refer to positions on the tobacco plastid chromosome (DDBJ/EMBL/GenBank accession No. Z00044). psbE, F, L and J encode subunits of photosystem II. They constitute an operon that is transcribed into a polycistronic message. petA encodes cytochrome f. The promoter driving aadA transcription is designated ‘P’. P1–P4 designate primers used for PCR analysis of transformants to check insertion of the aadA–ndhA constructs into the plastid chromosome. The amplification products shown in (B) characterize two spectinomycin-resistant transformants, generated either by bombardment with vector pCEE (line Nt-CEE-5) or pDIE (line Nt-DIE-1). Both 5′ and 3′ flanking sequences were amplified using primer pairs P1–P2 and P3–P4, respectively, demonstrating correct insertion into the plastid chromosome. (C) Five micrograms of total genomic DNA of transplastomic lines Nt-CEE-5 and Nt-DIE-1 as well as of wild-type tobacco were digested with AflIII and separated in a 1% agarose gel. A probe spanning the psbL/J genes (see Figure 2A) was used to detect restriction length polymorphism due to insertion of the respective construct. Signals obtained correspond to calculated fragment lengths (indicated at the right; see also Figure 2A). No signal corresponding to wild-type plastid DNA was detected in the transplastomic plants, suggesting their homoplastomic state. Note that the CEE-containing fragment is 104 bp shorter than the respective DIE fragment.
The two plasmids carrying ndhA exon–exon or intron– exon sequences were designated pCEE and pDIE, respectively (Figure 2A). They were introduced into the tobacco plastid genome biolistically by bombarding sterile leaves with plasmid DNA-coated gold particles. Transformants that were subsequently selected for spectinomycin resistance exhibited a wild-type phenotype. They were screened for correct construct insertion by PCR, Southern and sequence analysis (Figure 2B and C). Southern analyses indicated that the plant material rapidly reached the homoplastomic state for the insertion. Already after the first cycle of regeneration no signal corresponding to the wild-type DNA fragment was detectable (Figure 2C). Among the four and 11 independent lines obtained for constructs DIE and CEE, respectively, two for each construct were selected for further study.
The spinach ndhA site I is edited in tobacco but only in intronless transcripts
Editing of spliced ndhA is virtually 100% in wild-type spinach. To test whether editing occurs in the two chimeric constructs, cDNAs were amplified using primers P3 and ndhAex2rev, and the products were sequenced directly. Surprisingly, it turned out that the spinach CEE transcript was edited in the heterologous tobacco background. Only a small fraction of the sequenced PCR product retained the ‘unedited C’ (Figure 3), indicating that editing is slightly less efficient than in spinach. On the other hand, transcripts of the DIE construct were not edited at all (Figure 3). This suggests that either the partial intron sequence of the DIE construct impairs editing or that sequence elements in exon 1 in the CEE construct are necessary for editing. To exclude the possibility that changes in RNA metabolism resulting from the introduced transgenes cause the observed differences in editing, transcript patterns were analysed in more detail. Taking into account that the editing machinery may only be apt to process a limited number of transcripts, as in the case of the psbL and a rpoB site (Chaudhuri et al., 1995; Reed and Hanson, 1997), depletion of an editing factor may result from an abundant transcript and, hence, editing of an introduced site may only be partial. Therefore, northern hybridization was conducted in order to compare RNA accumulation in the two transplastomic lines CEE and DIE. The transcripts accumulated to identical levels when probed for aadA (Figure 4). RNA filters were also hybridized using psbL/J or ndhA probes to evaluate whether the introduced transgenes influence the expression of the corresponding endogenous transcripts. Although some less abundant transcripts of the psbE operon observed in wild-type do not accumulate in transplastomic plastids, the presence of the only slightly reduced principal psbE/F/L/J transcript (Figure 4, probe psbL/J, lowermost band) clearly shows that the generation of the dominant wild type message is not impaired. When probed for ndhA, both the DIE and CEE plants exhibited almost identical transcript patterns to the wild-type. Expected exceptions are bands originating from transgene insertion (marked by asterisks and arrowheads in Figure 4). In summary, no difference in RNA accumulation of any transcript containing the constructs has been noted between DIE and CEE plants. The data, therefore, suggest strongly that editing of the ndhA site I depends on the structure of the introduced transgene: only the intronless CEE transcript can serve as a target for the editing machinery.
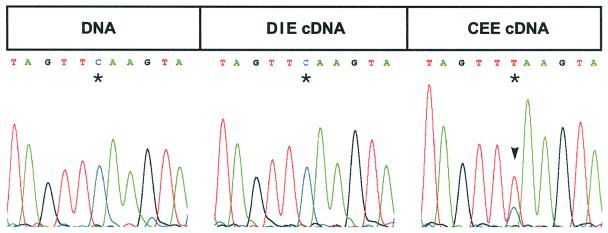
Fig. 3. Only intronless CEE transcripts are subject to editing. To test for editing of the intronless CEE and the intron-containing DIE transcripts, aadA-CEE- and aadA-DIE-specific cDNAs were amplified and sequenced directly. PCR products specific for the inserted spinach ndhA fragments were obtained using primers P3 and ndhAex2rev with total plant cDNA of the transplastomic lines Nt-CEE-5 and Nt-DIE-1 as templates. Spinach ndhA DNA was amplified with primers ndhAex1for and ndhAex2rev. The same primer (ndhAex1for) was used in all sequencing reactions. The relevant short interval of 11 nt from the total chromatograms is shown. The editing site is marked by an asterisk. Base conversion C-to-T (marked by an arrowhead) was observed only in the intronless CEE construct. A smaller C-peak below the T-peak in red indicates that the unedited C is also present in the transcript population.
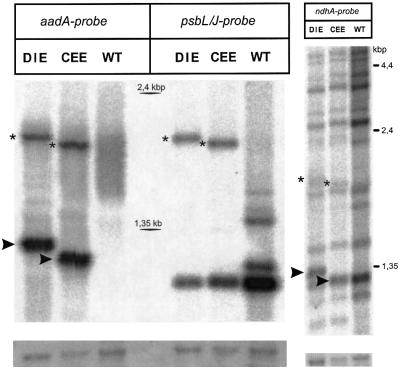
Fig. 4. Both CEE and DEE transcripts accumulate to identical levels. To estimate steady-state RNA levels in transplastomic lines relative to tobacco wild type, 4 µg of total plant RNA from each of wild type, Nt-CEE-5 and Nt-DIE-1 were fractionated in a 1% formaldehyde-containing agarose gel, blotted onto filters and hybridized with radioactively labelled, strand-specific aadA, psbL/J or ndhA probes, respectively. RNA species marked by arrowheads correspond to monocistronic aadA/CEE and aadA/DIE transcripts and are, therefore, detectable with the aadA and also with the ndhA probe. Transcripts of ∼2.1 kb length detected in all three hybridization experiments (marked by asterisks) result from readthrough transcription initiating at the psbE operon promoter (Bock et al., 1994). Their 3′ end is defined by the regulatory sequence of the aadA cassette. All transcripts including the heterologous editing site accumulate to identical levels in DIE and CEE plants. Methylene blue stains of 16S rRNA shown below demonstrate comparable sample loading in all lanes.
Nicotiana tomentosiformis encodes a homologue of ndhA site I that is subject to editing
The unexpected finding that tobacco is capable of editing a spinach site, which the tobacco plastome is lacking, raises the question of the origin of this activity. The comprehensive information on plant mitochondrial and plastid DNA excludes that editing activities are organelle-coded and suggests that the import of nuclear gene products is required. Nicotiana tabacum as an allotetraploid species contains plastids from its maternal parent N.sylvestris and nuclei derived from both N.sylvestris and N.tomentosiformis. To check for the presence or absence of a related editing activity in the diploid ancestors of tobacco, the ndhA DNA and cDNA of N.sylvestris and N.tomentosiformis were amplified and the products sequenced. An alignment of the relevant sequences is shown in Figure 5. As expected, the N.sylvestris ndhA sequence, which is closely related to that of tobacco, possesses no editing site. In contrast, N.tomentosiformis does, and apparently harbours an activity to edit this site as was verified by cDNA sequence analysis (Figure 5). This strongly suggests that N.tabacum inherited the ability to edit the ndhA site from its male parent N.tomentosiformis as a consequence of allotetraploidization, and that this feature was not lost during evolution.
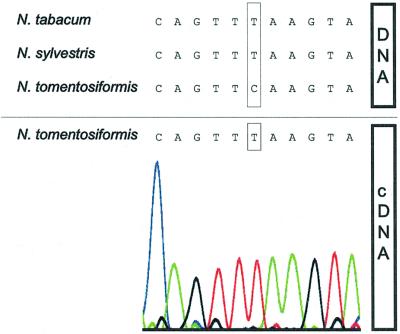
Fig. 5. In contrast to N.sylvestris and N.tabacum, N.tomentosiformis encodes ndhA site I and is capable of editing it. Partial ndhA DNA sequences of the three Nicotiana species investigated are aligned above the solid line. Below the line the ndhA cDNA sequence derived from N.tomentosiformis is shown together with the corresponding chromatogram. The position of ndhA editing site I is boxed.
The inability of tobacco to edit the spinach psbF site and the maize rpoB site I can be ascribed to the lack of the homologous editing sites in the parental genomes
Studies on heterologous editing, notably of spinach psbF and of maize rpoB site I, have shown that tobacco is incapable of processing both sites, which in the first case causes a severe mutant phenotype (Bock et al., 1994; Reed and Hanson, 1997). In the context of the unexpected finding of heterologous ndhA editing, it seemed interesting to check whether the failure to edit these previously investigated sites can be explained by parental genotypes as well. We have, therefore, sequenced the appropriate parts of N.sylvestris and N.tomentosiformis. In both species, psbF contains a phenylalanine-coding triplet at the relevant position, indicating that there is no requirement for the editing machinery to recognize and process this site. For rpoB site I, the corresponding sequences in N.sylvestris and N.tomentosiformis exhibit a thymidine residue at the DNA level as in tobacco. It is, therefore, no surprise that tobacco, like its progenitors, lacks the respective activities and is, therefore, not capable of editing the introduced, heterologous sites. This further corroborates our finding that the editing capacity of tobacco is probably linked to its parental genomes.
Transplastomic CEE plants exhibit an unaltered tobacco editotype
The examination of parental genotypes strongly suggests that present-day N.tabacum inherited its ndhA site I editing capacity from its paternal parent. It is, however, unclear why this activity has been kept for 6 million years, the estimated age of tobacco (Okamura and Goldberg, 1985), without the corresponding target site in the plastid chromosome. One conceivable explanation is that the specificity factor recognizing the ndhA site is also involved in the editing of another site. It is known that transplastomically overexpressed editing sites compete for trans-factors with the endogenous site, which can finally lead to a decrease in editing of the wild-type site, as is the case for the psbL and rpoB site I (Chaudhuri et al., 1995; Reed and Hanson, 1997). We therefore determined the entire editotype of CEE plants in order to test whether any tobacco site is influenced by the highly expressed transgene. None of the sequenced PCR products derived from CEE cDNA for all 31 tobacco editing sites reported (Hirose et al., 1999) exhibited a notable reduction in editing efficiency; that is, all were edited just like in wild type (data not shown). Apparently, expression of the ndhA transgene does not affect processing of any endogenous plastid editing site, although we cannot rigorously rule out the possibility that the factor is depletable, if expression of the transgene is further increased.
Discussion
The post-transcriptional conversion of cytidine residues to uridines in plastid transcripts by RNA editing is mandatory for the correct expression of plastid genetic information (reviewed in Maier et al., 1996; Bock, 2000), as is the correct removal of intervening sequences by RNA splicing (reviewed in Rochaix, 2001). In general, the various plastid RNA processing steps are largely independent of each other (Barkan, 1988; Westhoff and Herrmann, 1988), and in the case of RNA editing, no linkage to other RNA maturation events, e.g. splicing, has been reported previously (Freyer et al., 1993; Ruf et al., 1994). However, in a recent study, cDNA analysis suggested that editing of a distinct site of the barley ndhA message is linked to splicing of an adjacent group II intron (del Campo et al., 2000).
Here, we provide in vivo evidence for a mechanistic and kinetic relation of the two processes in ndhA transcripts. After introduction of spinach 3′ exon sequences, including ndhA editing site I fused to either intron or 5′ exon sequences into tobacco plastid chromosomes, it turned out that only the transcript of the ‘spliced’ exon–exon construct is edited, whereas those of the ‘unspliced’ intron–exon construct remained unedited. Obviously, the intron has to be removed in order to allow editing. An explanation for the strict succession of splicing and editing in the ndhA message may lie in the sequence requirements of the processing machineries involved. In vitro and in vivo data suggest that cis determinants for plastid RNA editing reside immediately upstream of editing sites ranging from only 12 to >40 nt in length (Bock et al., 1996, 1997; Chaudhuri et al., 1996; Hirose and Sugiura, 2001). For ndhA, sequence requirements have not been precisely determined. However, exon–exon constructs apparently provide sufficient sequence interval to allow editing. This raises the question of whether the intron itself interferes with editing or whether part of the cis sequence resides in the first exon.
For the ndhA message of barley it was proposed that part of the intron pairs with the 3′ exon to generate a secondary structure important for splicing but also masking cis determinants for editing (del Campo et al., 2000). In this scenario, it is not necessarily the joining of exons that enables editing, but, as a consequence of splicing, only the demasking of 3′ exon sequences adjacent to the editing site that would then allow the base conversion process. However, the occurrence of such a fold would potentially interfere with tertiary interactions known to occur in group II introns, namely the d–d′ tertiary base pairing, and also with the formation of intron domain VI (Michel and Ferat, 1995), which ultimately would impair splicing. This interaction is not at risk when it is the joining rather than the release of cis sequences that sets the stage for editing. Therefore, the most plausible explanation for the observed strict succession of splicing and editing on the ndhA transcript is that cis sequences separated by the intron have to be brought in contact to each other by splicing in order to allow editing.
For the evolutionary origin of the partite editing cis sequence, two scenarios may be envisaged: either the pres ence of the intron predates the origin of the editing site or the insertion of the intron split pre-existing editing site determinants. In the bryophyte Marchantia polymorpha, ndhA is divided by an intron residing at the very same position as in higher plants, but no editing takes place in this gene or anywhere else in its plastome. This is evidence for an ‘editing-late’ scenario, in which cis sequence elements necessary for ndhA site I editing evolved in both exons, split by a sequence stretch of 1079 bp, the ndhA group II intron.
In general, editing sites in the vicinity of intron–exon boundaries may be subject to diverse selective pressures. (i) After editing, the U residue may be essential for correct translation. (ii) The unedited C residue may be part of a sequence in RNA molecules interacting with splice factors that are known to act on the exon close to IBS–EBS (intron and exon binding site) and d–d′ interactions (Michel and Jacquier, 1987; Jacquier and Jacquesson-Breuleux, 1991; Costa et al., 2000), or may be involved in forming a hitherto uncharacterized interaction necessary for group II intron splicing as proposed by del Campo et al. (2000). (iii) The C residue may also play a role on DNA, for instance as an element of a recognition site for DNA binding proteins. Editing would then not be a mere corrective means of gene expression but rather one increasing the informational and/or structural capacity of certain nucleotide positions, which eventually augments the regulatory flexibility of the plastid genome.
The data presented here also elucidate the crucial role of nuclear genomes in editing. The lack of heterologous editing had suggested that the nuclear-encoded editing factors are only present in the context of their specific target site (Bock et al., 1994; Reed and Hanson, 1997). Here we report a first example that an editing activity can be present despite the absence of the target site: tobacco is capable of editing the spinach ndhA site I, even though its plastid chromosome lacks this site. In addition, we have shown that this site is absent in N.sylvestris, but present and efficiently edited in N.tomentosiformis. This strongly suggests that the nucleus of N.tomentosiformis is the donor of the corresponding tobacco editing activity, as this species represents the male parent of the allotetraploidization event that led to modern tobacco. That the spinach psbF site and maize rpoB site I, which are both absent in tobacco, are not edited when introduced into tobacco chloroplasts (Bock et al., 1994; Reed and Hanson, 1997) is consistent with the finding that they exist neither in N.tomentosiformis nor in N.sylvestris.
The fate of parental genomes after allopolyploidization is quite well documented and found to be complex (Comai, 2000; Wendel, 2000). In many cases, both parental copies persisted after polyploidization. Members of the tobacco β-1,3-glucanase gene family, for example, have been recombined blockwise leading to mixed gene arrangements with respect to parental lines (Sperisen et al., 1991). Other gene pairs have been maintained without apparent major modifications, like those encoding nitrate reductases (Vaucheret et al., 1989), or those coding for subunits of photosystem I (Obokata et al., 1993), or do not notably change in sequence, but only their expression pattern, like endochitinase genes (van Buuren et al., 1992). An important impact on nuclear genes after the evolution of allopolyploids is exerted by organelle genomes (Soltis and Soltis, 1995). Data on natural and synthetic polyploids in Brassica (Song et al., 1993, 1995) revealed that the nuclear genome of tetraploids is more closely related to the parent that contributed the cytoplasm than to the other one. Parental, in particular paternal, nuclear DNA can be lost or silenced quickly within a few generations (e.g. Gastony, 1991; Guo et al., 1996; Liu et al., 1998; Galitski et al., 1999; Lim et al., 2000a). A rare example of prevalence of the male contribution to the allopolyploid genome is the evolution of rDNA in tobacco. Here, gene conversion between parental rDNA clusters favoured almost exclusively the paternal N.tomentosiformis-type units (Borisjuk et al., 1997; Volkov et al., 1999; Lim et al., 2000b).
The different fate of genes after allotetraploidization most likely reflects internal constraints and requirements of the emerging plant. One of these constraints might be the necessity to harmonize nuclear with organellar genomes, a daunting task considering that the number of nuclear genes concerned with organelle functions is estimated to be in the order of at least 20–25% of the nuclear gene complement (Herrmann, 1997; Martin and Herrmann, 1998; Abdallah et al., 2000; The Arabidopsis Genome Initiative, 2000). It is conceivable that some of the paternal genes do not harmonize (well) with the maternal cytoplasm (in uniparental maternal inheritance) or that the mere increase in the number of genes causes dosage problems. A failure to reconcile plastid and nuclear genomes would result in incompatibilities between compartments as known from interspecific genome–plastome hybrids and cybrids (e.g. Stubbe, 1989; Yao et al., 2000; Zubko et al., 2001). Cases in point are classical experiments with the genus Oenothera that have led to the identification of nuclear–plastid disharmony as illustrated by ‘hybrid bleaching’ or ‘hybrid variegation’ (Kutzelnigg and Stubbe, 1974; Stubbe and Herrmann, 1982; Stubbe, 1989). Conceivably, these compartmental incompatibilities can contribute to reproductive barriers between populations and thereby ultimately to speciation. In terms of editing, the progeny of hybridization events between populations with different editotypes may be unable to fully edit their transcriptome leading to severe competitive disadvantages. As a remedy, allotetraploidization might overcome these barriers by providing both parental sets of editing factors. Possibly, this is one of the reasons for the high level of allopolyploidy in the plant kingdom.
In the case of the ndhA-editing site described here, tobacco does not seem to benefit from the paternal editing capacities as it lacks the appropriate target site. The maintenance of this activity is puzzling considering previous data on heterologous editing (Bock et al., 1994; Bock and Koop, 1997; Reed and Hanson, 1997) and the massive genome restructurings encountered in allopolyploid situations. Why did this factor escape the common fate of paternal DNA in allopolyploids, that is, degeneration and deletion, persisting for 6 million years without a target site? It is tempting to speculate that the respective factor plays an additional role in the allotetraploid and maybe also in the paternal plant, either with a different function, or also involved in editing. Analysis of the complete editotype of our transgenic plants largely excludes the possibility that the factor is involved in editing of other plastid sites. A role in the editing of related sequences, i.e. nad1, the mitochondrial homologue to ndhA, however, cannot be excluded (Maier et al., 1992). Further work has to clarify for which task, if any, this factor is employed in tobacco cells.
Materials and methods
Plant material
Tobacco plants (N.tabacum, cv. Petit Havana) were grown under sterile conditions on agar-solidified MS medium (Murashige and Skoog, 1962). Nicotiana sylvestris and N.tomentosiformis were grown on soil for 14 days. Spinach (Spinacia oleracea) was obtained from a local grocery store.
Oligonucleotides
Oligonucleotides used are given in Table I.
Table I. Oligonucleotides.
| Oligonucleotide | Sequence |
|---|---|
| P3 | TATCAGCCCGTCATACTTGAAGC |
| P2 | ACTGCGGAGCCGTACAAATG |
| P1 | CACACAATTTAAGTAGATGCG |
| P4 | CTGTTCTTATTTTACCGGAGG |
| HndhAex1for | CTTTCCAAAGCTTTCCTTTTTAGGTGGTCTACG |
| HndhAex2rev | CTTTCCAAAGCTTCAATCTCTCGCATTCTG |
| HndhAinfor | CTTTCCAAAGCTTTTTGATTGGTCTAATTCAGG |
| aadAT7 | GTAATCGACTCACTATAGGGAACCGGATCAAAGAGT |
| aadAli | GAAGCGGTTATCGCCGAAG |
| ApsbL | TACGACACAATCAAACCCGA |
| AT7psbJ | GATAATACGACTCACTATAGGGTACTAGAGGGATGAACCCAAT |
| mundhAfor | TATCTAACAGTTCAAGTACAGTTGATATAG |
| mundhArev | CTATATCAACTGTACTTGAACTATTAGATA |
| ndhAfor | ACAGGAGATACTCGTTTATT |
| AT7ndhA | GATAATACGACTCACTATAGGGACGAGGTTGTCAATAATAGAT |
Construction of plastid transformation vectors
A 1828 bp SalI–Eco47III tobacco plastid DNA fragment spanning the psbJ–petA intergenic region was cloned into a Bluescript SKII– vector (Stratagene, CA). The resulting recombinant plasmid was linearized with EcoRV and a chimeric aadA gene (Koop et al., 1996) was inserted. A plasmid that carries the aadA cassette in the same orientation as the psbE operon (pBaB) was then linearized with HindIII, which separates the aadA coding sequence from the downstream Chlamydomonas rbcL 3′ regulatory sequence. The HindIII site in pBaB served as target for insertion of intron-containing and intronless spinach ndhA gene fragments generated by PCR. Amplification of the intron-containing ndhA gene was performed using primers HndhAex2rev and HndhAinfor and spinach total DNA as template, yielding a PCR product that is flanked by HindIII restriction sites. The PCR products were digested with HindIII and subsequently cloned into vector pBaB. Plasmids with the insert in the same transcriptional direction as aadA were designated as pDIE.
Construction of the intronless ndhA fragment including the ‘unedited’ C close to the exon–exon border was carried out applying a PCR-based mutagenesis described as follows. Two PCR products were generated using primer pairs HndhAex2rev–mundhAfor and HndhAex1for– mundhArev with spinach cDNA as template. The two mu-primers are reverse complementary to each other and have a C mismatch relative to the T at the editing position of the spinach ndhA-cDNA sequence. This restores the unedited state during amplification. The PCR products were gel-purified using NA 45 DEAE cellulose membrane (Schleicher and Schuell, Dassel, Germany). A mixture of 20 ng of each purified PCR product served as template for a second PCR, this time using primers HndhAex1for and HndhAex2rev. Amplification was performed using a modified procedure of 94°C (for 30 s), 50°C (for 30 s) 72°C (for 5 min) and 30 following cycles with a reduced extension time of 1 min 30 s at 72°C. The resulting DNA fragments were HindIII digested and cloned into vector pBaB in the same way as the intron–exon amplification product, finally yielding vector pCEE.
Plastid transformation and selection for transplastomic lines
Leaves of 14-day-old sterile-grown tobacco seedlings were bombarded with plasmid DNA-coated gold particles using a biolistic gun (PDS-1000/He system, Bio-Rad, CA) and spectinomycin-resistant shoots were selected (Svab and Maliga, 1993). Plastid transformants were identified by PCR using primer pairs P1–P2 and P3–P4, which also tests the correct integration into the plastid genome. DNA isolated from primary regenerates was subjected to PCR and Southern analysis in order to assess transplastome to wild-type plastid chromosome ratios.
Isolation of nucleic acids
Total cellular DNA from tobacco leaves was isolated as described previously (Doyle and Doyle, 1990). Total cellular RNA was extracted with TRIzol reagent (Gibco-BRL, NY) according to the manufacturer’s recommendations.
Northern and Southern analysis
Total RNA from leaves of wild-type tobacco and transplastomic lines was fractionated in 1% agarose gels containing 6.1% formaldehyde. The RNA was transferred onto Hybond-N+ membrane (Amersham Pharmacia, Germany) by capillary blotting and immobilized by UV-crosslinking. Transfer of RNA was checked by staining the membrane with methylene blue. Strand-specific RNA probes were generated based on a PCR approach that introduces a viral T7 RNA polymerase promoter sequence via primers aadAT7, AT7psbJ and AT7ndhA when combined with reverse primers aadAli, ApsbL and ndhAfor, respectively. Without further purification, 200 ng of PCR products then served as a template for in vitro transcription in the presence of [α-32P]UTP. Hybridization was carried out at 65°C in Church buffer [0.25 M Na2HPO4, 7% sodium dodecyl sulfate (SDS)]. Washes were performed at the same temperature in 0.1× standard saline citrate (SSC)/0.1% SDS (1× SSC is 0.15 M NaCl, 0.015 M Na citrate) for 1.5 h with repeated changes of buffer.
Restriction fragments of total tobacco DNA were fractionated in 1% agarose gels and subsequently transferred onto Hybond N+ membrane by capillary blotting. Hybridization of membranes with gene-specific radioactively labelled single-stranded RNA fragments was performed as described above for northern hybridizations.
Synthesis of cDNA
RNA was reverse transcribed using Superscript™ reverse transcriptase (Gibco-BRL, New York) according to Maier et al. (1992).
PCR and DNA sequence analysis
DNA and cDNA templates were amplified according to standard protocols (94°C, 30 s; 53°C, 30 s; 72°C, 1.5 min; 30 cycles) if not otherwise stated. Amplification products were purified from excess primers before sequencing by ethanol precipitation in the presence of 2.5 M NH4Ac. Nucleotide sequences were determined by the dideoxy chain termination method (Sanger et al., 1977) on an ABI 377 system (Applied Biosystems, CA).
Acknowledgments
Acknowledgements
We thank Professor Vera Hemleben for providing N.tomentosiformis seeds and leaf material. We are grateful to Prof. Bartolomé Sabater for stimulating discussions. We also thank Anja Drescher, Holger Hupfer and Julia Legen for helpful comments and Achim Haecker for technical support. This work was supported by the Deutsche Forschungs gemeinschaft (SFB 184 and TR1) and the Fonds der Chemischen Industrie.
References
- Abdallah F., Salamini,F. and Leister,D. (2000) A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci., 5, 141–142. [DOI] [PubMed] [Google Scholar]
- Barkan A. (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J., 7, 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. and Goldschmidt-Clermont,M. (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie, 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Binder S., Marchfelder,A. and Brennicke,A. (1996) Regulation of gene expression in plant mitochondria. Plant Mol. Biol., 32, 303–314. [DOI] [PubMed] [Google Scholar]
- Bland M.M., Matzinger,D.F. and Levings,C.S. (1985) Comparisons of the mitochondrial genome of Nicotiana tabacum with its progenitor species. Theor. Appl. Genet., 69, 535–541. [DOI] [PubMed] [Google Scholar]
- Bock R. (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie, 82, 549–557. [DOI] [PubMed] [Google Scholar]
- Bock R. and Koop,H.U. (1997) Extraplastidic site-specific factors mediate RNA editing in chloroplasts. EMBO J., 16, 3282–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R., Kössel,H. and Maliga,P. (1994) Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J., 13, 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R., Hermann,M. and Kössel,H. (1996) In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J., 15, 5052–5059. [PMC free article] [PubMed] [Google Scholar]
- Bock R., Hermann,M. and Fuchs,M. (1997) Identification of critical nucleotide positions for plastid RNA editing site recognition. RNA, 3, 1194–1200. [PMC free article] [PubMed] [Google Scholar]
- Borisjuk N.V., Davidjuk,Y.M., Kostishin,S.S., Miroshnichenco,G.P., Velasco,R., Hemleben,V. and Volkov,R.A. (1997) Structural analysis of rDNA in the genus Nicotiana. Plant Mol. Biol., 35, 655–660. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S. and Maliga,P. (1996) Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J., 15, 5958–5964. [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Carrer,H. and Maliga,P. (1995) Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J., 14, 2951–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2000) Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol., 43, 387–399. [DOI] [PubMed] [Google Scholar]
- Costa M., Michel,F. and Westhof,E. (2000) A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J., 19, 5007–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo E.M., Sabater,B. and Martin,M. (2000) Transcripts of the ndhH-D operon of barley plastids: possible role of unedited site III in splicing of the ndhA intron. Nucleic Acids Res., 28, 1092–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J. and Doyle,J.L. (1990) Isolation of plant DNA from fresh tissue. Focus, 12, 13–15. [Google Scholar]
- Freyer R., Hoch,B., Neckermann,K., Maier,R.M. and Kössel,H. (1993) RNA editing in maize chloroplasts is a processing step independent of splicing and cleavage to monocistronic mRNAs. Plant J., 4, 621–629. [DOI] [PubMed] [Google Scholar]
- Freyer R., Lopez,C., Maier,R.M., Martin,M., Sabater,B. and Kössel,H. (1995) Editing of the chloroplast ndhB encoded transcript shows divergence between closely related members of the grass family (Poaceae). Plant Mol. Biol., 29, 679–684. [DOI] [PubMed] [Google Scholar]
- Freyer R., Kiefer-Meyer,M.-C. and Kössel,H. (1997) Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl Acad. Sci. USA, 94, 6285–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M., Maier,R.M. and Zeltz,P. (2001) RNA editing in higher plant plastids: oligoribonucleotide SSCP analysis allows the proof of base conversion directly on the RNA level. Curr. Genet., 39, 384–387. [DOI] [PubMed] [Google Scholar]
- Galitski T., Saldanha,A.J., Styles,C.A., Lander,E.S. and Fink,G.R. (1999) Ploidy regulation of gene expression. Science, 285, 251–254. [DOI] [PubMed] [Google Scholar]
- Gastony G.J. (1991) Gene silencing in a polyploid homosporous fern: Paleopolyploidy revisited. Proc. Natl Acad. Sci. USA, 88, 1602–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel D.U. (1960) Segregation in new allopolyploids of Nicotiana. II. Comparison of 6× (N.tabacum × N.tomentosiformis) and 6× (N. tabacum × N.otophora). Genetics, 45, 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel D.U. (1963) Segregation in new allopolyploids of Nicotiana. II. Discordant ratios from individual loci in 6× (N.tabacum × N.tomentosiformis) and 6× (N.tabacum × N.otophora). Genetics, 48, 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godspeed T.H. (1954) The genus Nicotiana. Chronica Botanica Company, Waltham, MA.
- Goldschmidt-Clermont M. (1991) Transgenic expression of amino glycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res., 19, 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Davis,D. and Birchler,J.A. (1996) Dosage effects on gene expression in a maize ploidy series. Genetics, 142, 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann R.G. (1997) Eukaryotism, towards a new interpretation. In Schenk,H.E.A., Herrmann,R.G., Jeon,K.W., Müller,N.E. and Schwemmler,W. (eds), Eukaryotism and Symbiosis. Springer, Berlin, Germany, pp. 73–118.
- Hess W.R., Hoch,B., Zeltz,P., Hübschmann,T., Kössel,H. and Börner,T. (1994) Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell, 6, 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T. and Sugiura,M. (2001) Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J., 20, 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Wakasugi,T., Sugiura,M. and Kössel,H. (1994) RNA editing of tobacco petB mRNAs occurs both in chloroplasts and non-photosynthetic proplastids. Plant Mol. Biol., 26, 509–513. [DOI] [PubMed] [Google Scholar]
- Hirose T., Kusumegi,T., Tsudzuki,T. and Sugiura,M. (1999) RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet., 262, 462–467. [DOI] [PubMed] [Google Scholar]
- Jacquier A. and Jacquesson-Breuleux,N. (1991) Splice site selection and role of the lariat in a group II intron. J. Mol. Biol., 219, 415–428. [DOI] [PubMed] [Google Scholar]
- Karcher D. and Bock,R. (1998) Site-selective inhibition of plastid RNA editing by heat shock and antibiotics: a role for plastid translation in RNA editing. Nucleic Acids Res., 26, 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenton A., Parokonny,A.S., Gleba,Y.Y. and Bennett, M.D. (1993) Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol. Gen. Genet., 240, 159–169. [DOI] [PubMed] [Google Scholar]
- Koop H.U., Steinmüller,K., Wagner,H., Rößler,C., Eibl,C. and Sacher,L. (1996) Integration of foreign sequences into the tobacco plastome via PEG-mediated protoplast transformation. Planta, 199, 193–201. [DOI] [PubMed] [Google Scholar]
- Kutzelnigg H. and Stubbe,W. (1974) Investigations on plastome mutants in Oenothera. 1. General considerations. Sub-Cell. Biochem., 3, 73–89. [Google Scholar]
- Legen J., Schmitz-Linneweber,C., Drescher,A., Hupfer,H., Tillich,M., Herrmann,R.G. and Maier,R.M. (2001) Decoding of the ndhH operon from spinach: an example for the complexity of plastid gene expression in higher plants. Endocytobiosis Cell Res., 14, 11–19. [Google Scholar]
- Lim K.Y., Kovarik,A., Matyasek,R., Bezdek,M., Lichtenstein,C.P. and Leitch,A.R. (2000a) Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma, 109, 161–172. [DOI] [PubMed] [Google Scholar]
- Lim K.Y., Matyasek,R., Lichtenstein,C.P. and Leitch,A.R. (2000b) Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma, 109, 245–258. [DOI] [PubMed] [Google Scholar]
- Liu B., Vega,J.M. and Feldman,M. (1998) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome, 41, 535–542. [DOI] [PubMed] [Google Scholar]
- Lopez C., Freyer,R., Guera,A., Maier,R.M., Martin,M., Sabater,B. and Kössel,H. (1997) Sequence of ndhA gene of barley (Hordeum vulgare L.) plastid (DDBJ/EMBL/GenBank accession nos Y13729 and Y13730). Transcript editing in graminean organs (PGR97–120). Plant Physiol., 115, 313.9380777 [Google Scholar]
- Maier R.M., Neckermann,K., Hoch,B., Akhmedov,N.B. and Kössel,H. (1992) Identification of editing positions in the ndhB transcript from maize chloroplasts reveals sequence similarities between editing sites of chloroplasts and plant mitochondria. Nucleic Acids Res., 20, 6189–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R.M., Zeltz,P., Kössel,H., Bonnard,G., Gualberto,J.M. and Grienenberger,J.M. (1996) RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol., 32, 343–365. [DOI] [PubMed] [Google Scholar]
- Malek O., Lättig,K., Hiesel,R., Brennicke,A. and Knoop,V. (1996) RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J., 15, 1403–1411. [PMC free article] [PubMed] [Google Scholar]
- Martin W. and Herrmann,R.G. (1998) Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol., 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Michel F. and Jacquier,A. (1987) Long range intron–exon and intron–intron pairings involved in self-splicing of class II catalytic introns. Cold Spring Harbor Symp. Quant. Biol., 52, 201–212. [DOI] [PubMed] [Google Scholar]
- Monde R.A., Schuster,G. and Stern,D.B. (2000) Processing and degradation of chloroplast mRNA. Biochimie, 82, 573–582. [DOI] [PubMed] [Google Scholar]
- Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant., 15, 473–497. [Google Scholar]
- Obokata J., Mikami,K., Hayashida,N., Nakamura,M. and Sugiura,M. (1993) Molecular heterogeneity of photosystem I. psaD, psaE, psaF, psaH, and psaL are all present in isoforms in Nicotiana spp. Plant Physiol., 102, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura J.K. and Goldberg,R.G. (1985) Tobacco single-copy DNA is highly homologous to sequences present in the genomes of its diploid progenitors. Mol. Gen. Genet., 198, 290–298. [Google Scholar]
- Olmstead R. and Palmer,J.D. (1991) Chloroplast DNA and systematics in the solanaceae. In Hawkes,J.G., Lester,R.N., Nee,M. and Estrada,N. (eds), Solanaceae III. Taxonomy, Chemistry and Evolution. Kew, Royal Botanic Gardens, Linnean Society of London, London, UK, pp. 161–168.
- Parokonny A.S. and Kenton,A.Y. (1995) Comparative physical mapping and evolution of the Nicotiana tabacum karyotype. In Brandham,P.E. and Bennet,M.D. (eds), Kew Chromosome Conference IV. Royal Botanik Gardens, Kew, London, UK, pp. 301–320.
- Reed M.L. and Hanson,M.R. (1997) A heterologous maize rpoB editing site is recognized by transgenic tobacco chloroplasts. Mol. Cell. Biol., 17, 6948–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed M.L., Peeters,N.M. and Hanson,M.R. (2001) A single alteration 20 nt 5′ to an editing target inhibits chloroplast RNA editing in vivo. Nucleic Acids Res., 29, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.-D. (2001) Posttranscriptional control of chloroplast gene expression. From RNA to photosynthetic complex. Plant Physiol., 125, 142–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Zeltz,P. and Kössel,H. (1994) Complete RNA editing of unspliced and dicistronic transcripts of the intron-containing reading frame IRF170 from maize chloroplasts. Proc. Natl Acad. Sci. USA, 91, 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen,S. and Coulsen,A.R. (1977) DNA sequencing with chain terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Maier,R.M., Alcaraz,J.P., Cottet,A., Herrmann, R.G., and Mache,R. (2001) The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol. Biol., 45, 307–315. [DOI] [PubMed] [Google Scholar]
- Soltis D.E. and Soltis,P.S. (1995) The dynamic nature of polyploid genomes. Proc. Natl Acad. Sci. USA, 92, 8089–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K.M., Tang,K.L. and Osborn,T.C. (1993) Development of synthetic Brassica amphidiploids by reciprocal hybridization and comparison to natural amphidiploids. Theor. Appl. Genet., 86, 811–821. [DOI] [PubMed] [Google Scholar]
- Song K., Lu,P., Tang,K. and Osborn,T.C. (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl Acad. Sci. USA, 92, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperisen C., Ryals,J. and Meins,F. (1991) Comparison of cloned genes provides evidence for intergenomic exchange of DNA in the evolution of a tobacco glucan endo-1,3-β-glucosidase gene family. Proc. Natl Acad. Sci. USA, 88, 1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe W. (1989) Oenothera — an ideal system for studying the interactions of genome and plastome. Plant Mol. Biol. Rep., 7, 245–257. [Google Scholar]
- Stubbe W. and Herrmann,R.G. (1982) Selection and maintainance of plastome mutants and interspecific genome/plastome hybrids from Oenothera. In Edelman,V., Hallick,R.B. and Chua,N.H. (eds), Methods in Chloroplast Molecular Biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, pp. 149–165.
- Sutton C.A., Conklin,P.L., Pruitt,K.D. and Hanson,M.R. (1991) Editing of pre-mRNAs can occur before cis-splicing and trans-splicing in Petunia mitochondria. Mol. Cell. Biol., 11, 4274–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z. and Maliga,P. (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA, 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- van Buuren M., Neuhaus,J.M., Shinshi,H., Ryals,J. and Meins,F.,Jr (1992) The structure and regulation of homeologous tobacco endochitinase genes of Nicotiana sylvestris and N. tomentosiformis origin. Mol. Gen. Genet., 232, 460–469. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vincentz,M., Kronenberger,J., Caboche,M. and Rouze,P. (1989) Molecular cloning and characterisation of the two homologous genes coding for nitrate reductase in tobacco. Mol. Gen. Genet., 216, 10–15. [DOI] [PubMed] [Google Scholar]
- Volkov R.A., Borisjuk,N.V., Panchuk,I.I., Schweizer,D. and Hemleben,V. (1999) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol. Biol. Evol., 16, 311–320. [DOI] [PubMed] [Google Scholar]
- Wendel J.F. (2000) Genome evolution in polyploids. Plant Mol. Biol., 42, 225–249. [PubMed] [Google Scholar]
- Westhoff P. and Herrmann,R.G. (1988) Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur. J. Biochem., 171, 551–564. [DOI] [PubMed] [Google Scholar]
- Yang A.J. and Mulligan,R.M. (1991) RNA editing intermediates of cox2 transcripts in maize mitochondria. Mol. Cell. Biol., 11, 4278–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J.-L. and Cohen,D. (2000) Multiple gene control of plastome-genome incompatibility and plastid DNA inheritance in interspecific hybrids of Zantedeschia. Theor. Appl. Genet., 101, 400–406. [DOI] [PubMed] [Google Scholar]
- Zeltz P., Hess,W.R., Neckermann,K., Börner,T. and Kössel,H. (1993) Editing of the chloroplast rpoB transcript is independent of chloroplast translation and shows different patterns in barley and maize. EMBO J., 12, 4291–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito F., Kuras,R., Choquet,Y., Kössel,H. and Wollman,F.A. (1997) Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol., 33, 79–86. [DOI] [PubMed] [Google Scholar]
- Zubko M.K., Zubko,E.I., Ruban,A.V., Adler,K., Mock,H.P., Misera,S., Gleba,Y.Y. and Grimm,B. (2001) Extensive developmental and metabolic alterations in cybrids Nicotiana tabacum (+ Hyoscyamus niger) are caused by complex nucleo-cytoplasmic incompatibility. Plant J., 25, 627–639. [DOI] [PubMed] [Google Scholar]


