Abstract
The evolutionarily conserved yeast Mec1 and Tel1 protein kinases, as well as the Mec1-interacting protein Ddc2, are involved in the DNA damage checkpoint response. We show that regulation of Tel1 and Ddc2–Mec1 activities is important to modulate both activation and termination of checkpoint-mediated cell cycle arrest. In fact, overproduction of either Tel1 or Ddc2 causes a prolonged cell cycle arrest and cell death in response to DNA damage, impairing the ability of cells to recover from checkpoint activation. This cell cycle arrest is independent of Mec1 in UV-irradiated Tel1-overproducing cells, while it is strictly Mec1 dependent in similarly treated DDC2-overexpressing cells. The Rad53 checkpoint kinase is instead required in both cases for cell cycle arrest, which correlates with its enhanced and persistent phosphorylation, suggesting that unscheduled Rad53 phosphorylation might prevent cells from re-entering the cell cycle after checkpoint activation. In addition, Tel1 overproduction results in transient nuclear division arrest and concomitant Rad53 phosphorylation in the absence of exogenous DNA damage independently of Mec1 and Ddc1.
Keywords: checkpoints/Ddc2/DNA damage/Mec1/Tel1
Introduction
Eukaryotic cells respond to DNA damage and replication blocks by delaying cell cycle progression through a surveillance mechanism known as the DNA damage checkpoint, thus providing the time to restore the correct DNA structure before entering the next cell cycle phase. Failure of DNA repair or checkpoint controls can lead to cell lethality, mutations, genome instability and ultimately cancer (reviewed in Zhou and Elledge, 2000).
In Saccharomyces cerevisiae, the DNA damage checkpoint is a signal transduction cascade involving the evolutionarily conserved protein kinases Mec1, Rad53 and Chk1 (reviewed in Zhou and Elledge, 2000). Rad53 and Chk1 are both phosphorylated in a Mec1-dependent manner (Sanchez et al., 1996; Sun et al., 1996). While Rad53 is required for a proper response to DNA damage in all the cell cycle phases, Chk1 contributes to the activation of the G2/M checkpoint in a Rad53-independent manner (Sanchez et al., 1996, 1999). Mec1 belongs to the phosphatidylinositol-3-kinase motif family, which also includes S.cerevisiae Tel1 (Greenwell et al., 1995; Morrow et al., 1995), Schizosaccharomyces pombe Rad3 (Bentley et al., 1996), Drosophila melanogaster Mei-41 (Hari et al., 1995), human ATM (Savitsky et al., 1995) and ATR (Bentley et al., 1996). Mec1 physically interacts with Ddc2, which undergoes Mec1-dependent phosphorylation both in vivo and in vitro (Paciotti et al., 2000; Rouse and Jackson, 2000; Wakayama et al., 2001). The finding that the DNA damage-induced Mec1-dependent Ddc2 phosphorylation does not require other known checkpoint factors suggests that Mec1, as well as S.pombe Rad3 and human ATM, may be involved in sensing DNA damage (Paciotti et al., 2000; reviewed in O’Connell et al., 2000; Zhou and Elledge, 2000). Additional checkpoint proteins essential for DNA damage-induced cell arrest are Ddc1, Rad17, Mec3, Rad24 and Rad9 (Weinert and Hartwell, 1988; Siede et al., 1993; Weinert et al., 1994; Longhese et al., 1996, 1997; Paulovich et al., 1997). Ddc1, Mec3 and Rad17, like their S.pombe and human homologs, physically interact with each other (Kostrub et al., 1998; Paciotti et al., 1998; Kondo et al., 1999; St Onge et al., 1999; Caspari et al., 2000) and are structurally related to the proliferating cell nuclear antigen (PCNA) clamp (Thelen et al., 1999), which tethers DNA replication proteins to the replicating DNA (Waga and Stillman, 1998). Moreover, Rad24 is homologous to, and interacts with, subunits of the PCNA clamp loader, replication factor C (RF-C) (Griffiths et al., 1995; Lydall and Weinert, 1997; Green et al., 2000). Based on these similarities and genetic and biochemical evidence, Ddc1, Rad17, Mec3 and Rad24 are proposed to work together with Mec1 in sensing damaged DNA molecules and modulating Mec1 activity (reviewed in Longhese et al., 1998; Weinert, 1998; Lowndes and Murguia, 2000). The DNA damage sensing functions are then linked with downstream effectors by Mec1-dependent phosphorylation of Rad9. In fact, Rad9 phosphorylation triggers its interaction with Rad53 and consequent release of active Rad53 kinase (Emili, 1998; Sun et al., 1998; Vialard et al., 1998; Gilbert et al., 2001), thus indicating that Mec1 can regulate both sensing and transducing checkpoint signals.
In addition to their involvement in the checkpoint responses, DDC2, MEC1 and RAD53 are essential for cell viability, and their essential function, but not the checkpoint functions, can be bypassed by increasing the intracellular concentrations of deoxyribonucleotides (dNTPs), either by overexpression of RNR genes encoding ribonucleotide reductase (Desany et al., 1998) or by deletion of the SML1 gene (Zhao et al., 1998), which negatively affects the dNTP pool (Chabes et al., 1999; Zhao et al., 2001).
The Mec1 homolog Tel1 is a protein kinase (Mallory and Petes, 2000) primarily required for telomere length maintenance (Greenwell et al., 1995; Morrow et al., 1995). Several data indicate that it also has a role in the cellular response to DNA damage, which becomes evident in the absence of Mec1. In fact, TEL1 deletion increases the sensitivity of mec1 mutant cells to DNA-damaging agents (Ritchie et al., 1999), and high levels of Tel1 can suppress both cell lethality and hypersensitivity to DNA-damaging agents of mec1Δ cells, suggesting that Tel1 and Mec1 may have partially overlapping functions (Sanchez et al., 1996). Moreover, Tel1 has recently been implicated, together with the Mre11 complex, in a DNA damage checkpoint pathway that responds primarily to double strand breaks and parallels the Mec1 pathway, leading to Rad9 phosphorylation and interaction with Rad53 (D’Amours and Jackson, 2001; Grenon et al., 2001; Usui et al., 2001).
Once the checkpoint is activated by DNA damage, some mechanism must exist to allow cells to resume cell cycle progression when the aberrant DNA structures have been removed. In fact, the inability of cells to recover from checkpoint activation once the damage is repaired would result in cell death in the presence of genotoxic agents. While our knowledge about the mechanisms underlying checkpoint activation is continuously increasing, the processes involved in recovery from DNA damage-induced cell cycle arrest are still completely unknown.
Here we show that regulation of Tel1 and Ddc2–Mec1 activities is important to modulate both the activation of checkpoint-mediated cell cycle arrest and the subsequent recovery. In fact, although by different processes, both TEL1 and DDC2 overexpression cause prolonged cell cycle arrest and cell death after DNA damage, and this correlates with high and persistent levels of Rad53 phosphorylation. In addition, Tel1 overproduction results in Rad53 phosphorylation and nuclear division delay in the absence of exogenous DNA damage, suggesting that Tel1, when present in high levels, might activate the checkpoint even in the absence of external checkpoint signals.
Results
TEL1 overexpression causes prolonged nuclear division delay and phosphorylation of Rad53 in the absence of exogenous DNA damage
The finding that TEL1 overexpression can suppress hypersensitivity of mec1Δ cells to genotoxic agents (Sanchez et al., 1996; see also Figure 3D) indicates that high levels of Tel1 can partially substitute for Mec1 in the response to DNA damage. Furthermore, Tel1 has been implicated together with the Mre11 complex in triggering Rad53 activation and its interaction with Rad9 in response to DNA damage independently of Mec1 (Usui et al., 2001). To gain insights into the function of Tel1 and its involvement in checkpoints, we first analyzed cell cycle progression in the absence of DNA damage of cells carrying the galactose-inducible GAL1–TEL1 fusion at the TEL1 locus. As shown in Figure 1A and B, when cell cultures were arrested in G1 with α-factor in the presence of galactose and released from the block under galactose-induced conditions, GAL1–TEL1 cells progressed through S phase only slightly more slowly than wild type, but then they surprisingly underwent a prolonged arrest with 2C DNA content and undivided nuclei, while nuclear division and cell cycle progression occurred normally in wild-type cells under the same conditions. Similarly, when cell cultures arrested in G2 with nocodazole in the presence of galactose were released from the nocodazole block under galactose-induced conditions, GAL1–TEL1 cells were still arrested with undivided nuclei 240 min after release, while wild-type cells underwent nuclear division within 90 min (Figure 1D). Thus, overexpression of TEL1 causes a G2/M cell cycle arrest in the absence of exogenous DNA damage, suggesting that high levels of Tel1 might lead to activation of a checkpoint pathway. The Tel1-induced cell cycle arrest was transient, since both GAL1–TEL1 and GAL1–TEL1-HA3 cell cultures underwent nuclear division, and resumed budding and DNA replication with wild-type kinetics 12–14 h after a shift to galactose-containing medium, although the Tel1-HA3 level remained unchanged throughout the experiments (data not shown). Moreover, when exponentially growing GAL1–TEL1 cell cultures were shifted to galactose-containing medium for 4 h (when nearly all cells had grown to the dumb-bell shape characteristic of G2/M-arrested cells) and then plated on galactose-containing plates, 70% of cells produced microcolonies with >4 cells or buds within 24 h. Consistent with this observation, Tel1-overproducing cells also showed a slow growth phenotype in the absence of genotoxic agents (Figures 3D, 4C and 5C).
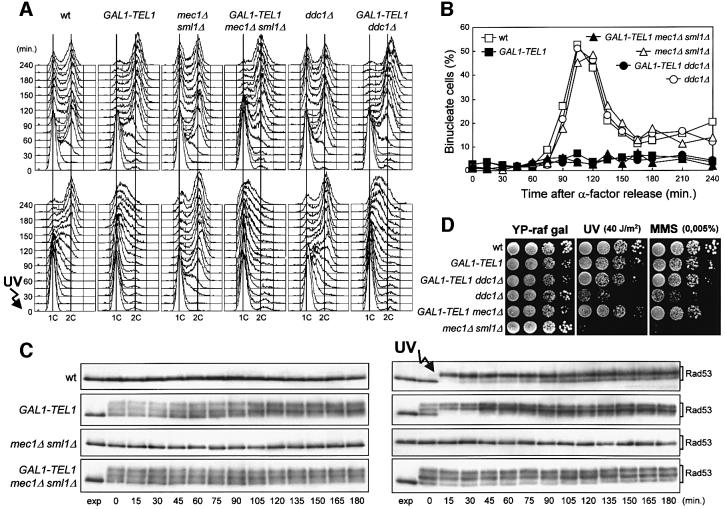
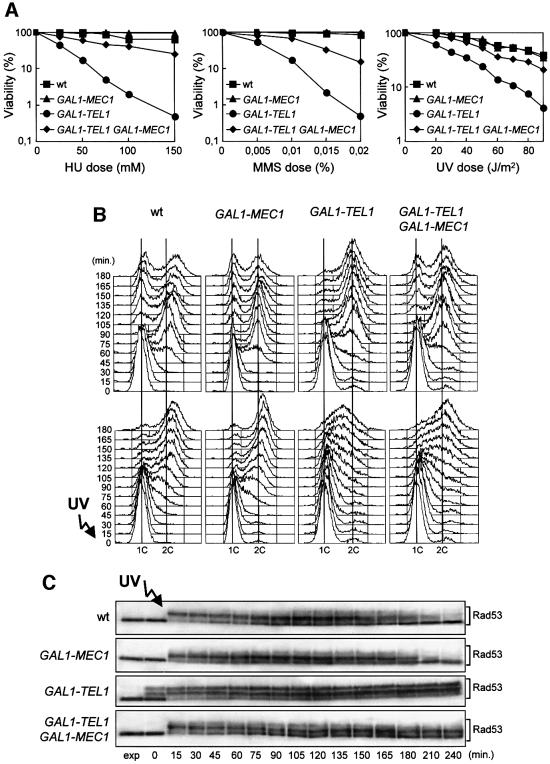
Fig. 3. Effects of TEL1 overexpression in the absence of Mec1 or Ddc1. Strains were as follows: wild type (K699), GAL1–TEL1 (DMP3539/10D), mec1Δ sml1Δ (YLL490), GAL1–TEL1 mec1Δ sml1Δ (DMP3562/2A), ddc1Δ (YLL244) and GAL1–TEL1 ddc1Δ (DMP3575/4A). (A–C) Cell cultures growing logarithmically in YEP-raf were synchronized in G1 with α-factor in the presence of galactose (2 h) and released from the α-factor block at time zero in YEP-raf-gal, or were UV irradiated (40 J/m2) prior to release in YEP-raf-gal. Samples were collected at the times indicated after α-factor release to analyze the DNA content of untreated (top) and UV-treated (bottom) cell cultures by FACS (A), to score the untreated cell cultures for the percentage of binucleate cells by propidium iodide staining (B) and to analyze protein extracts from the untreated (left) and UV-treated (right) cell cultures by western blotting using anti-Rad53 antibodies (C). exp, exponentially growing cells. (D) Serial dilutions of YEP-raf exponentially growing cell cultures were spotted on YEP-raf-gal plates with or without MMS at the concentration indicated. One YEP-raf-gal plate was UV irradiated (UV).
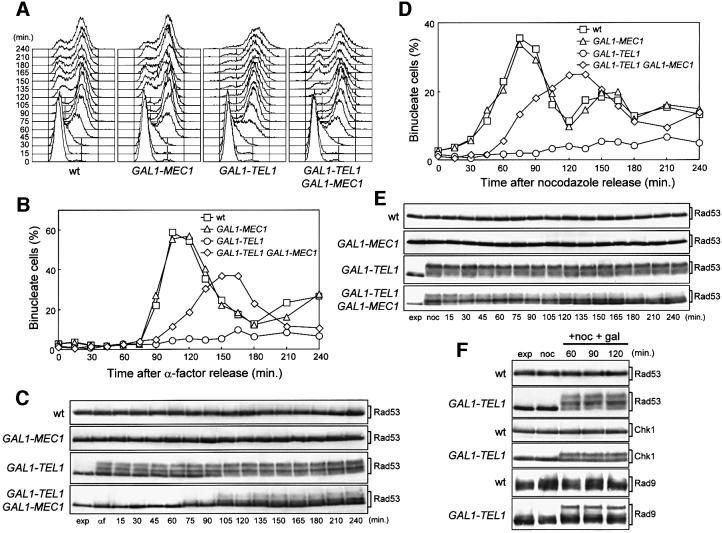
Fig. 1. Effects of TEL1 overexpression during an unperturbed cell cycle. Strains were as follows: wild type (K699), GAL1–MEC1 (YLL516), GAL1–TEL1 (DMP3539/10D) and GAL1–TEL1 GAL1–MEC1 (DMP3539/9D). (A–C) Cell cultures, growing logarithmically in YEP-raf, were synchronized in G1 with α-factor in the presence of galactose (2 h). Release from α-factor block was performed at time zero (C, αf) by transferring cell cultures to fresh YEP-raf-gal medium. Samples were taken at the times indicated after release into the cell cycle to determine the DNA content by fluorescence-activated cell sorter (FACS) analysis (A), the kinetics of nuclear division by direct visualization by propidium iodide staining (B) and the amount and phosphorylation of Rad53 by western blotting of protein extracts with anti-Rad53 antibodies (C). (D and E) Cell cultures, growing logarithmically in YEP-raf, were synchronized in G2 with nocodazole in the presence of galactose (2 h). Release from nocodazole block was performed at time zero (E, noc) by transferring cell cultures to fresh YEP-raf-gal medium. Samples were taken at the times indicated after release from nocodazole to determine the kinetics of nuclear division (D) and Rad53 levels and phosphorylation (E) as described in (B) and (C), respectively. (F) Cell cultures of wild-type (DMP3598/21D) and GAL1–TEL1 (DMP3598/6A) strains were arrested with nocodazole in YEP-raf medium (noc) and resuspended in YEP-raf-gal medium containing 15 µg/ml nocodazole (+noc +gal). Protein extracts were prepared from samples withdrawn at the times indicated and analyzed by western blotting. The slowly migrating protein species specifically reacting with anti-Rad53, anti-MYC (Chk1) and anti-Rad9 antibodies represent phosphorylated forms of the corresponding proteins (Sanchez et al., 1996; Vialard et al., 1998, 1999). exp, exponentially growing cells.
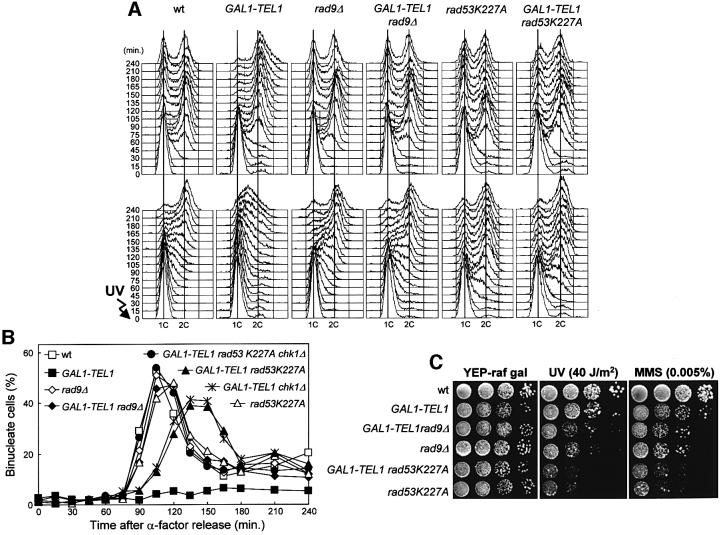
Fig. 4. Cell cycle delay caused by high levels of Tel1 involves Rad53, Rad9 and Chk1. Strains were as follows: wild type (K699), GAL1–TEL1 (DMP3539/10D), rad9Δ (YLL157), GAL1–TEL1 rad9Δ (DMP3575/6B), rad53K227A (DMP3479/2A), GAL1–TEL1 rad53K227A (DMP3479/2B), GAL1–TEL1 chk1Δ (DMP3611/6D) and GAL1–TEL1 rad53K227A chk1Δ (DMP3611/3B). (A and B) Cell cultures growing logarithmically in YEP-raf were synchronized in G1 with α-factor in the presence of galactose (2 h) and released from the α-factor block at time zero in YEP-raf-gal or were UV irradiated (40 J/m2) prior to release in YEP-raf-gal. Samples were collected at the times indicated after α-factor release to analyze the DNA content of untreated (top) and UV-treated (bottom) cell cultures by FACS (A) and to score the untreated cell cultures for the percentage of binucleate cells by propidium iodide staining (B). (C) Serial dilutions of YEP-raf exponentially growing cell cultures were spotted on YEP-raf-gal plates with or without MMS. One YEP-raf-gal plate was UV irradiated (UV).
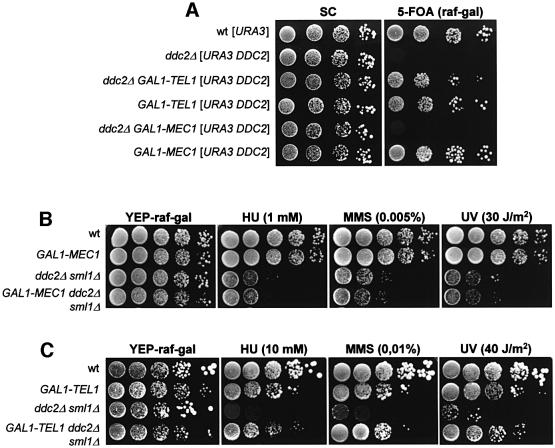
Fig. 5. Genetic interactions between DDC2, MEC1 and TEL1. Strains were as follows: wild type [URA3 YCplac33] (YLL827), ddc2Δ [URA3 DDC2] (YLL275), ddc2Δ GAL1–TEL1 [URA3 DDC2] (DMP3475/3D), GAL1–TEL1 [URA3 DDC2] (YLL943), ddc2Δ [LEU2 GAL1–MEC1] [URA3 DDC2] (YLL930), wild type [LEU2 GAL1–MEC1] [URA3 DDC2] (YLL944), GAL1–MEC1 (YLL516), ddc2Δ sml1Δ (DMP2995/1B), GAL1–MEC1 ddc2Δ sml1Δ (DMP3532/8A), GAL1–TEL1 (DMP3539/10D) and GAL1–TEL1 ddc2Δ sml1Δ (DMP3602/9C). Serial dilutions of YEP-raf exponentially growing cell cultures were spotted on Synthetic Complete (SC)-glucose and 5-Fluoro-orotic acid (5-FoA)-containing SC-raf-gal plates (A) or on YEP-raf-gal plates with or without MMS or HU at the concentrations indicated (B and C). One YEP-raf-gal plate was UV irradiated (UV).
Since activation of the known DNA damage checkpoint pathways leads to phosphorylation of Rad53, which results in changes in Rad53 electrophoretic mobility (Sanchez et al., 1996; Sun et al., 1996), we used western blot analysis to study the Rad53 phosphorylation pattern during the above experiments. Rad53 phosphorylation was clearly detectable in galactose-induced GAL1–TEL1 cells released either from the G1 or the G2 block in the absence of DNA damage, but not in wild-type cells lacking the GAL1–TEL1 fusion under the same conditions (Figure 1C and E). It is worth noting that Rad53 phosphorylation was detectable in GAL1–TEL1 cells both when they were released from the G2 block under galactose-induced conditions (Figure 1E) and when they were kept in G2 by nocodazole treatment after galactose addition (Figure 1F), indicating that high levels of Tel1 induce Rad53 phosphorylation independently of cell cycle progression. Interestingly, the effects of TEL1 overexpression on cell cycle progression were partially counteracted by increased levels of wild-type Mec1 expressed from a GAL1–MEC1 fusion integrated at the URA3 chromosomal locus. In fact, as shown in Figure 1B and D, when galactose-induced α-factor or nocodazole-arrested cells were released into the cell cycle in galactose-containing media, nuclear division occurred faster in GAL1–TEL1 GAL1–MEC1 than in GAL1–TEL1 cells, although with slower kinetics compared with wild-type cells. Moreover, galactose-induced GAL1–TEL1 GAL1–MEC1 cells released from the G1 or G2 blocks contained a reduced amount of Rad53 phosphorylated forms compared with similarly treated GAL1–TEL1 cells (Figure 1C and E), while overexpression of MEC1 did not affect cell cycle progression or Rad53 phosphorylation per se. This suggests that an excess of Tel1 might compete with Mec1 for interactions with downstream effectors in the absence of DNA damage, thus causing unscheduled phosphorylation events and cell cycle arrest.
The above results prompted us to ask whether high levels of Tel1 might induce phosphorylation of checkpoint proteins other than Rad53, such as Ddc1, Ddc2, Chk1 and Rad9, which are all known to undergo Mec1-dependent phosphorylation resulting in electrophoretic mobility shift of the corresponding protein species (Emili, 1998; Paciotti et al., 1998, 2000; Sun et al., 1998; Vialard et al., 1998; Sanchez et al., 1999). Significant amounts of phosphorylated Chk1 and Rad9 could be detected in nocodazole-arrested GAL1–TEL1 cell cultures after addition of galactose (Figure 1F), while Ddc1 and Ddc2 phosphorylations were not influenced by Tel1 overproduction (data not shown), indicating that high levels of Tel1 specifically induce phosphorylation of Rad53, Rad9 and Chk1 in the absence of exogenous DNA damage.
Tel1 overproduction affects the cell response to DNA damage
Since overexpression of TEL1 causes G2/M cell cycle arrest independently of exogenous DNA damage, we investigated its effects in the presence of genotoxic insults. Galactose-induced GAL1–TEL1 cells were more sensitive to hydroxyurea (HU), methyl methanesulfonate (MMS) and UV than isogenic wild-type strains not carrying the GAL1–TEL1 fusion (Figure 2A). This hypersensitivity can be partially suppressed by increasing the level of wild-type Mec1, since GAL1–TEL1 cells concomitantly expressing a GAL1–MEC1 fusion showed lower sensitivity to HU, MMS and UV than GAL1–TEL1 cells (Figure 2A), indicating that high levels of Tel1 might cause the observed phenotypes by competing with physiological amounts of Mec1.
Fig. 2. Response to DNA damage of TEL1-overexpressing cells. Strains were as follows: wild type (K699), GAL1–MEC1 (YLL516), GAL1–TEL1 (DMP3539/10D) and GAL1–TEL1 GAL1–MEC1 (DMP3539/9D). (A) Dose–response killing curves were determined by plating serial dilutions of YEP-raf exponentially growing cell cultures on YEP-raf-gal plates with or without MMS or HU at the concentrations indicated. One set of YEP-raf-gal plates was exposed at the UV doses indicated. Plates were incubated at 25°C and colony-forming units were counted after 3 days. (B and C) Cell cultures growing logarithmically in YEP-raf were synchronized in G1 with α-factor in the presence of galactose (2 h). Cells were released from the α-factor block at time zero in YEP-raf-gal, or were UV irradiated (40 J/m2) prior to release in YEP-raf-gal. Samples of untreated and UV-irradiated cultures were taken at the times indicated after α-factor release to analyze the DNA content by FACS (B) and to determine the level and phosphorylation of Rad53 by western blot analysis of protein extracts from the UV-treated cell cultures with anti-Rad53 antibodies (C). exp, exponentially growing cells.
Moreover, cell cycle progression after UV irradiation was dramatically altered by high levels of Tel1 (Figure 2B). In fact, when galactose-induced cell cultures were arrested in G1, UV irradiated and then released from the G1 block under galactose-induced conditions, GAL1– TEL1 cells dramatically slowed down entry and progression through S phase compared with similarly treated wild-type cells, which completed DNA replication 120 min after UV irradiation (Figure 2B). This correlated with the persistence of high levels of phosphorylated Rad53 in UV-treated GAL1–TEL1 cells even 240 min after release into the cell cycle, while Rad53 phosphorylation began to decrease after 180 min in wild-type cells under the same conditions (Figure 2C).
In agreement with the observed suppression of GAL1– TEL1 cell hypersensitivity to DNA-damaging agents by GAL1–MEC1, overexpression of MEC1 was able to reduce both the DNA damage-induced slowing down of S-phase progression and the concomitant Rad53 phosphorylation in Tel1-overproducing cells. In fact, galactose-induced UV-irradiated GAL1–TEL1 GAL1–MEC1 cells completed DNA replication and showed a reduced amount of phosphorylated Rad53 after release into the cell cycle (Figure 2B and C).
Delay of cell cycle progression and Rad53 phosphorylation caused by high levels of Tel1 do not require Mec1 and Ddc1
The data presented above indicate that an excess of Tel1 kinase causes Rad53, Chk1 and Rad9 phosphorylation, which may be responsible for the G2/M cell cycle arrest in the absence of exogenous insults and for the dramatic slowing down of cell cycle progression in response to DNA damage. Since the G2/M cell cycle arrest caused by Tel1 overproduction can be partially suppressed by increasing Mec1 levels, and an excess of Tel1 can suppress the hypersensitivity to genotoxic agents of mec1Δ cells, Mec1 should not be required for the Tel1-dependent cell cycle arrest. Indeed, neither the G2/M cell cycle arrest nor the Rad53 phosphorylation caused by high levels of Tel1 requires Mec1. In fact, when cells were arrested in G1 in the presence of galactose and then released from the block under galactose-induced conditions, GAL1–TEL1 mec1Δ sml1Δ as well as GAL1–TEL1 cells were still arrested with 2C DNA content and undivided nuclei 240 min after release into the cell cycle, whereas both mec1Δ sml1Δ and wild-type cells progressed through the cell cycle with indistinguishable kinetics (Figure 3A, top and B). Moreover, phosphorylated Rad53 was clearly detectable in GAL1–TEL1 mec1Δ sml1Δ cells throughout the experiment, while it was completely absent in wild-type and mec1Δ sml1Δ cells under the same conditions (Figure 3C, left).
Furthermore, overexpression of TEL1 is epistatic to deletion of MEC1 with respect to hypersensitivity to genotoxic agents (Sanchez et al., 1996; Figure 3D), Rad53 phosphorylation (Figure 3C) and cell cycle progression after DNA damage (Figure 3A, bottom). In fact, when G1-arrested cells were UV irradiated and then released into the cell cycle under galactose-induced conditions, mec1Δ sml1Δ cells completed DNA replication in 45–60 min, while both GAL1–TEL1 and GAL1–TEL1 mec1Δ sml1Δ cell cultures showed a dramatic slowing down of entry and progression through S phase even compared with wild-type cells, which completed DNA replication after 180 min (Figure 3A, bottom). The DNA damage-induced slowing down of cell cycle progression paralleled the presence of phosphorylated Rad53 in UV-irradiated GAL1–TEL1 and GAL1–TEL1 mec1Δ sml1Δ cells throughout the experiment, while Rad53 phosphorylation was completely absent in similarly treated mec1Δ sml1Δ cells (Figure 3C, right).
Since high levels of Tel1 induce a Rad53-dependent cell cycle arrest in the absence of exogenous DNA damage, we asked whether checkpoint proteins thought to be acting at early steps of DNA damage recognition, such as Ddc1, were required for the Tel1-dependent arrest. As shown in Figure 3A and B, when GAL1–TEL1 ddc1Δ cells were arrested in G1 in galactose and then released from the G1 block in galactose-containing medium, they arrested with 2C DNA content and undivided nuclei, while ddc1Δ cells underwent DNA replication and nuclear division with wild-type kinetics under the same conditions. Moreover, when the same cultures were UV irradiated before release from the G1 block under galactose-induced conditions, GAL1–TEL1 ddc1Δ cells showed a dramatic slowing down of cell cycle progression in response to DNA damage, similar to that observed for GAL1–TEL1 cells, while similarly treated ddc1Δ cells progressed through S phase faster than wild type (Figure 3A, bottom). Therefore, Ddc1 is not required for the cell cycle progression delay caused by high levels of Tel1. Accordingly, Tel1 overproduction is also epistatic to ddc1Δ with respect to hypersensitivity to genotoxic agents, since galactose-induced GAL1–TEL1 ddc1Δ cells were as sensitive as GAL1–TEL1 cells and less sensitive than the otherwise isogenic ddc1Δ strains to both UV and MMS (Figure 3D).
Rad53, Rad9 and Chk1 are required for Tel1-dependent cell cycle arrest
Since high levels of Tel1 induce phosphorylation of Rad9, Chk1 and Rad53 in unperturbed conditions, and Rad9 and Rad53 have been implicated in a Tel1-dependent checkpoint (Usui et al., 2001), we asked whether these proteins were required for the cell cycle delay caused by TEL1 overexpression. We therefore analyzed cell cycle progression of GAL1–TEL1 cells carrying the rad9Δ, chk1Δ or the kinase-defective rad53K227A alleles. Rad9 and Rad53 turned out to be necessary for the cell cycle delay caused by TEL1 overexpression both in the presence and absence of DNA damage (Figure 4A and B). In fact, when galactose-induced G1-arrested cells were released from the G1 block in galactose-containing medium, the kinetics of entry and progression through S phase of GAL1– TEL1 rad9Δ and GAL1–TEL1 rad53K227A cells were comparable to those of rad9Δ and rad53K227A cells, respectively (Figure 4A, top). Furthermore, nuclear division occurred with wild-type kinetics in GAL1– TEL1 rad9Δ cells and with some delay in GAL1–TEL1 rad53K227A cells, while GAL1–TEL1 cells under the same conditions were still arrested with undivided nuclei after 240 min (Figure 4B). Similarly, when the same cultures were UV irradiated and then released from the G1 block in galactose-containing medium, GAL1–TEL1 rad9Δ and GAL1–TEL1 rad53K227A cells replicated their DNA with kinetics indistinguishable from those of the rad9Δ and rad53K227A mutants, and much faster than GAL1– TEL1 cells, which dramatically slowed down DNA replication even compared with a similarly treated wild-type strain (Figure 4A, bottom). In agreement with the observation that high levels of Tel1 were unable to suppress the checkpoint defects of rad9Δ and rad53K227A mutants, TEL1 overexpression did not suppress the hypersensitivity to genotoxic agents of rad9Δ and rad53K227A cells (Figure 4C). In fact, UV and MMS sensitivities of galactose-induced GAL1–TEL1 rad9Δ and GAL1–TEL1 rad53K227A cells were indistinguishable from those of the otherwise isogenic rad9Δ and rad53K227A strains (Figure 4C).
Although the rad53K227A mutation completely abolished the dramatic slowing down of entry and progression through S phase of GAL1–TEL1 cells after UV irradiation, GAL1–TEL1 rad53K227A cells still showed a delay of nuclear division in unperturbed conditions compared with the rad53K227A mutants (Figure 4B). This suggests that cell cycle arrest in G2 caused by TEL1 overexpression in the absence of exogenous insults might at least partially depend on proteins acting independently of Rad53. We therefore asked whether it might involve the Chk1 kinase, which is phosphorylated in Tel1-overproducing cells and is specifically required to prevent anaphase entry in cdc13 mutants at restrictive temperature, independently of Rad53 (Sanchez et al., 1999). Indeed, when galactose-induced GAL1–TEL1 chk1Δ cells were released from G1 arrest in galactose-containing media, they underwent nuclear division with kinetics comparable to that of GAL1–TEL1 rad53K227A cells, while GAL1–TEL1 rad53K227A chk1Δ cells divided nuclei with wild-type kinetics and therefore faster than either GAL1–TEL1 rad53K227A or GAL1– TEL1 chk1Δ cells under the same conditions (Figure 4B). This indicates that high levels of Tel1 require both Rad53 and Chk1 to arrest nuclear division completely in the absence of DNA damage.
TEL1, but not MEC1, overexpression suppresses cell lethality and hypersensitivity to genotoxic agents of ddc2Δ cells
Our data indicate that TEL1 overexpression causes Mec1-independent unscheduled phosphorylation of Rad53 and hyperactivation of the checkpoint response, while cell cycle progression and Rad53 phosphorylation in both undamaged and UV-irradiated Mec1-overproducing cells are indistinguishable from wild type (Figures 1 and 2), suggesting that other proteins could be rate limiting for Mec1 activity. A likely candidate for this role could be Ddc2, since it physically interacts with Mec1 and may be required for Mec1 but not for Tel1 functions. As shown in Figure 5, this hypothesis is further strengthened by the observation that while TEL1 overexpression can suppress cell lethality and hypersensitivity to genotoxic agents caused by deletion of DDC2, high levels of Mec1 can not bypass the requirement for Ddc2. In fact, galactose induction of a GAL1–MEC1 fusion did not allow GAL1– MEC1 ddc2Δ cells to lose the URA3 centromeric plasmid carrying the wild-type DDC2 allele and did not change the sensitivity to HU, MMS and UV of ddc2Δ sml1Δ cells (Figure 5A and B). Conversely, galactose-induced GAL1– TEL1 ddc2Δ cells were able to lose the plasmid-borne DDC2 allele (Figure 5A), and GAL1–TEL1 ddc2Δ sml1Δ cells were less sensitive to HU, MMS and UV than otherwise isogenic ddc2Δ sml1Δ cells in galactose-containing medium (Figure 5C). Since both Mec1 and Tel1 are protein kinases (Mallory and Petes, 2000; Paciotti et al., 2000), high levels of Tel1 may be able to overcome the lack of Ddc2 by replacing Mec1 activity, thus implying that Mec1 functions, but not Tel1 functions, are impaired in the absence of Ddc2.
DDC2 overexpression causes an irreversible G2/M arrest after DNA damage
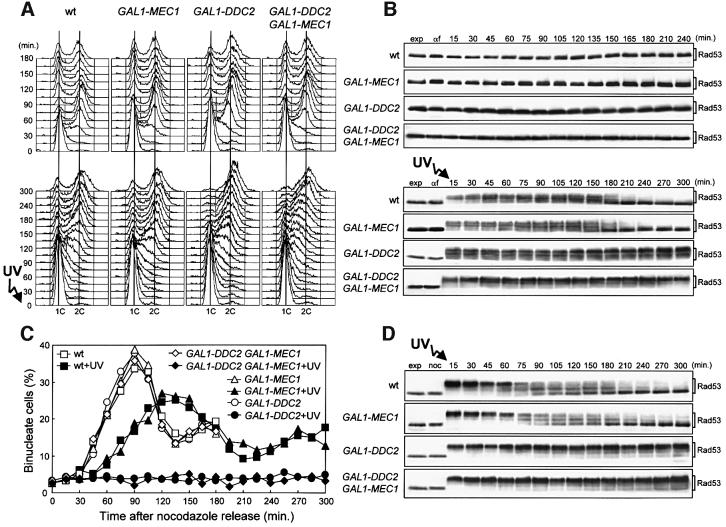
Since Ddc2 may be the rate-limiting factor for Mec1 activity, we analyzed cell cycle progression and Rad53 phosphorylation of cells overexpressing a GAL1–DDC2 fusion integrated in single copy at the LEU2 locus. When galactose-induced cell cultures were blocked in G1 or G2 and then released in galactose-containing medium in the absence of DNA damage, kinetics of entry and progression through S phase (Figure 6A, top) and of nuclear division (Figure 6C) were similar in GAL1–DDC2 and wild-type cells. Moreover, we could not detect Rad53 phosphorylated forms in GAL1–DDC2 cells after release from G1 (Figure 6B, top) or G2 arrests (data not shown) under galactose-induced conditions. Therefore, unlike Tel1 overproduction, high levels of Ddc2 are not able to activate the checkpoint in the absence of DNA damage.
Fig. 6. DDC2 overexpression leads to prolonged G2/M cell cycle arrest after checkpoint activation. Strains were as follows: wild type [URA3 YCplac33] (YLL827), wild type [URA3 GAL1–MEC1] (YLL826), GAL1–DDC2 [URA3 YCplac33] (YLL837) and GAL1–DDC2 [URA3 GAL1–MEC1] (YLL836). (A and B) Cell cultures growing logarithmically in YEP-raf were synchronized with α-factor. Galactose was added 2.5 h before α-factor addition. α-factor-synchronized cells were released from the block at time zero [(B), αf] in YEP-raf-gal or were UV irradiated (40 J/m2) prior to release in YEP-raf-gal. Samples of untreated (top) and UV-treated (bottom) cell cultures were collected at the times indicated after α-factor release to analyze the DNA content by FACS (A) and protein extracts by western blotting, using anti-Rad53 antibodies (B). (C and D) Cell cultures growing logarithmically in YEP-raf were synchronized with nocodazole. Galactose was added 2 h before nocodazole addition. Nocodazole-synchronized cells were released from the block at time zero [(D), noc] in YEP-raf-gal or were UV irradiated (50 J/m2) prior to release in YEP-raf-gal. Samples of untreated and UV-treated cell cultures were collected at the times indicated after nocodazole release to score for the percentage of binucleate cells by propidium iodide staining (C) and to analyze protein extracts from the UV-treated cell cultures by western blotting, using anti-Rad53 antibodies (D). exp, exponentially growing cells. In all the experiments, samples were withdrawn from the UV-treated cultures at times zero and 120 min, and appropriate dilutions were plated on YEPD plates to score for colony-forming units (see text for the percentage of cell survival).
Since the Mec1–Ddc2 complex may be activated by exogenous DNA damage, we then analyzed the response to DNA damage of Ddc2-overproducing cells. Cells with very high levels of Ddc2 due to multiple GAL1–DDC2 fusions integrated at the LEU2 locus had previously been reported to have a slight defect in slowing down DNA replication after DNA damage (Paciotti et al., 2000). Conversely, when galactose-induced cell cultures carrying a single copy of the GAL1–DDC2 fusion integrated at the LEU2 locus were blocked in G1, UV irradiated, and then released from the G1 arrest in galactose-containing medium, the kinetics of entry and progression through S phase were similar to those observed in wild-type cells under the same conditions (Figure 6A, bottom). However, Rad53 phosphorylated forms began to disappear in wild-type and GAL1–MEC1 cultures when DNA replication was completed (180 min after UV irradiation in G1; Figure 6B, bottom) and viability remained high (70% survival). Conversely, UV-treated GAL1–DDC2 cells, which dramatically lost viability (5% survival), still contained high amounts of phosphorylated Rad53 (Figure 6B, bottom) and were mostly arrested with a 2C DNA content (Figure 6A, bottom) and undivided nuclei (data not shown) 300 min after release into the cell cycle. Moreover, when galactose-induced cell cultures were arrested in G2, UV irradiated and then released from the G2 block in galactose-containing medium (Figure 6C and D), GAL1–DDC2 cells lost viability (7% survival) and did not divide nuclei throughout the experiment (300 min), while wild-type cells were mostly viable (82% survival) and underwent nuclear division ∼60 min after UV irradiation. Thus, cells containing high levels of Ddc2 appear to be impaired in undergoing the G2/M transition after DNA damage in G1 or G2. This effect was probably due to the cells’ inability to turn off the checkpoint response, since high amounts of phosphorylated Rad53 were present in GAL1–DDC2 cells until the end of both experiments, while phosphorylated Rad53 was largely absent 210 min after UV irradiation in wild-type cells (Figure 6B, bottom and D).
The above data show that high levels of Ddc2 cause permanent Rad53 phosphorylation and inhibit nuclear division after DNA damage, while the same increase in the amount of Ddc2 is not sufficient to block DNA replication of a damaged template, raising the possibility that, despite Ddc2 high levels, the amount of Mec1 may be limiting under these conditions. This hypothesis is further supported by the finding that co-overexpression of MEC1 and DDC2 also impairs the ability of cells to replicate a damaged template (Figure 6A, bottom). In fact, galactose-induced GAL1–MEC1 GAL1–DDC2 cells released from G1 after UV irradiation entered and progressed through S phase much more slowly than wild-type and GAL1–DDC2 cells under the same conditions. This suggests that while high levels of Ddc2 are sufficient to arrest cells at the G2/M transition, it is necessary to increase the levels of both Mec1 and Ddc2 proteins to arrest DNA replication after DNA damage.
The DNA damage-induced cell cycle arrest caused by high levels of Ddc2 requires Mec1, Ddc1, Rad53 and Rad9
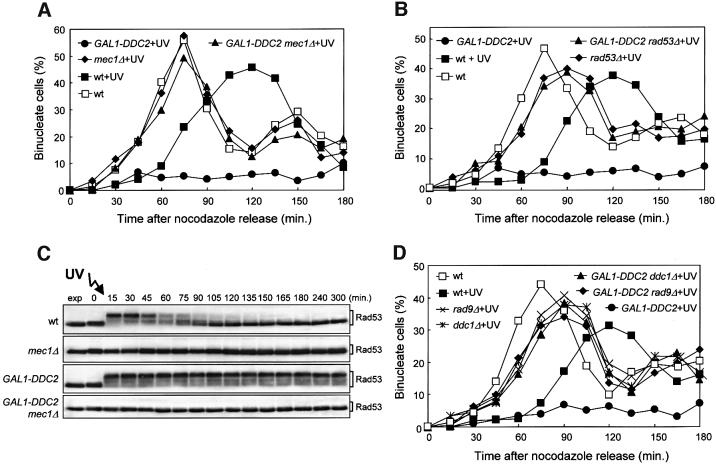
Since high levels of Tel1 can activate the G2/M checkpoint in the absence of genotoxic agents, while overproduc tion of Ddc2 causes a permanent cell cycle arrest in G2 only if DNA has undergone exogenous damage, we asked whether the DNA damage-induced cell cycle arrest mediated by DDC2 overexpression may require proteins different from those involved in the Tel1-dependent arrest. If the DNA damage-induced arrest at the G2/M transi tion in Ddc2-overproducing cells was due to increased Mec1 activity and dependent on the persistence of Rad53 phosphorylated forms, we expected both Mec1 and Rad53 to be absolutely required for this cell cycle arrest. Indeed, when cell cultures were arrested in G2, UV irradiated, and then released from the G2 block in galactose-containing medium, nuclear division occurred in GAL1–DDC2 mec1Δ sml1Δ and GAL1–DDC2 rad53Δ sml1Δ cells with kinetics indistinguishable from those of mec1Δ sml1Δ and rad53Δ sml1Δ mutants (Figure 7A and B). Moreover, Mec1 was also essential for Rad53 phosphorylation in DDC2-overexpressing cells, since we could not detect Rad53 phosphorylated forms in galactose-induced GAL1– DDC2 mec1Δ sml1Δ cells released from the G2 arrest after UV irradiation, while they were present throughout the experiment in similarly treated GAL1–DDC2 cells (Figure 7C).
Fig. 7. Requirement of checkpoint genes for the Ddc2-dependent DNA damage-induced cell cycle arrest. Strains were as follows: wild type (K699), GAL1–DDC2 (YLL279.2), mec1Δ sml1Δ (YLL490), GAL1–DDC2 mec1Δ sml1Δ (DMP3462/2B), rad53Δ sml1Δ (YLL509), GAL1–DDC2 rad53Δ sml1Δ (DMP3356/4A), rad9Δ (YLL157), GAL1–DDC2 rad9Δ (DMP3392/4A), ddc1Δ (YLL244) and GAL1–DDC2 ddc1Δ (DMP3463/10C). Cell cultures growing logarithmically in YEP-raf were synchronized with nocodazole. Galactose was added 2 h before nocodazole addition and nocodazole-synchronized cells were released from the block at time zero in YEP-raf-gal or were UV irradiated (50 J/m2) prior to release in YEP-raf-gal. (A, B and D) Untreated and UV-treated cell cultures were scored at the times indicated for the percentage of binucleate cells by propidium iodide staining. (C) Protein extracts from the UV-treated cell cultures were analyzed by western blotting, using anti-Rad53 antibodies. exp, exponentially growing cells.
As shown in Figure 7D, Ddc1 and Rad9 are also required for the DNA damage-induced G2/M arrest caused by high levels of Ddc2. In fact, nuclear division in galactose-induced GAL1–DDC2 ddc1Δ and GAL1–DDC2 rad9Δ cells released from the G2 arrest after UV irradiation occurred with kinetics comparable to those observed in similarly treated rad9Δ and ddc1Δ cells.
Discussion
The cells must be aware not only of damage, but also of when DNA repair is completed, in order to terminate the checkpoint response and resume cell cycle progression. A key unanswered question is how cells turn off the checkpoint after signals have been removed. It is possible that repair of DNA damage per se might restore normal cell cycle progression and/or active mechanisms might be required to shut off the checkpoint. For example, downstream effectors might become insensitive to the kinase cascade, and/or the checkpoint pathway might be inactivated by dephosphorylating the Rad53 kinase itself. Furthermore, since Mec1 and its interacting factor Ddc2 are required to phosphorylate and activate Rad53, inactivation of the Mec1–Ddc2 complex may be critical to turn off Rad53 and subsequently the checkpoint response. Finally, the Mec1 homolog Tel1, which has been implicated in a Rad53-dependent DNA damage checkpoint pathway that parallels the canonical Mec1 pathway (Usui et al., 2001), might also play a role in termination of the checkpoint response.
A possible way to approach this problem is to study whether and how overproduction of protein kinases and kinase-interacting factors acting upstream of Rad53 can affect cell cycle progression in the absence or in the presence of DNA damage. As discussed below, we found that the levels of Tel1, Mec1 and Ddc2 are important to regulate both activation of the checkpoint and resumption of cell cycle progression after DNA damage-induced cell cycle arrest.
Checkpoint activation in Tel1-overproducing cells
A major difference between the effects of Ddc2 and Tel1 overproduction is that high levels of Tel1 cause phosphorylation of Rad53 and G2/M cell cycle arrest in the absence of exogenous DNA damage, while an excess of Ddc2 has the same effects only when DNA has suffered external insults. The Rad53-dependent cell cycle arrest caused by Tel1 overproduction under otherwise unperturbed conditions may be due to the presence of high levels of active Tel1 kinase resulting in unscheduled phosphorylation events, which in turn cause G2/M transition delay. It is possible that an excess of Tel1 might lead to activation of a Rad53-dependent checkpoint by causing accumulation of DNA damaged molecules and impairment of repair capacity. However, the Mec1 kinase, which has been shown to play a primary role in activation of the checkpoint in response to different kinds of DNA alterations, is not required for the Tel1-dependent cell cycle arrest and Rad53 phosphorylation. Furthermore, Ddc1, which together with Rad24, Rad17 and Mec3 is thought to act at the beginning of the checkpoint pathway by recognizing changes in DNA structure and initiating the Mec1-dependent signal transduction cascade (reviewed in Longhese et al., 1998), is neither phosphorylated nor required for checkpoint activation in Tel1-overproducing cells. This indicates that proteins presumably acting together with Mec1 in the DNA damage-sensing steps of the pathway are not required to support Tel1 functions. We therefore suggest that either overproduction of Tel1 per se might result in increased Tel1 kinase activity that could be sufficient to elicit unscheduled phosphorylation events primarily inhibiting the G2/M transition, or high levels of Tel1 can be activated by endogenous DNA damage that is not sensed in the presence of physiological levels of this kinase. In any case, the Tel1-dependent cell cycle arrest requires both Rad53 and Chk1, which are known to cooperate in the activation of the G2/M DNA damage checkpoint (Sanchez et al., 1999), and Rad9, whose phosphorylation and interaction with Rad53 are required for Rad53 activation (Emili, 1998; Sun et al., 1998; Vialard et al., 1998; Gilbert et al., 2001). Furthermore, Rad9, Rad53 and Chk1 are phosphorylated in Tel1-overproducing cells in the absence of exogenous DNA damage, indicating a tight correlation between phosphorylation of these proteins and their requirement for the Tel1-dependent G2/M arrest. Since it has recently been shown that Rad9 phosphorylation triggers Rad53 activation (Gilbert et al., 2001), it is possible that unregulated levels of Tel1 result in hyperphosphorylation of Rad9, which in turn catalyzes Rad53 phosphorylation, resulting in ectopic checkpoint activation.
It is also worth noting that Tel1 overproduction suppresses the checkpoint defects and the hypersensitivity to DNA-damaging agents of mec1Δ cells. Therefore, increasing the levels of Tel1 uncovers its role in the checkpoint cascade both in the presence and absence of Mec1. In contrast to Mec1, Tel1 functions do not require Ddc2. In fact, TEL1, but not MEC1, overexpression can suppress hypersensitivity to genotoxic agents of ddc2Δ cells, implying that high levels of Tel1 can bypass the requirement for Ddc2. Moreover, the fact that Ddc2 can be co-immunoprecipitated with Mec1, while we were not able to co-immunoprecipitate Tel1 with Ddc2 even in the absence of Mec1 (our unpublished results), indicates that the Ddc2–Mec1 interaction is, in any case, preferential. Interestingly, overexpression of MEC1 can partially suppress the effect of Tel1 overproduction on cell cycle progression and greatly reduces the unscheduled Tel1-dependent Rad53 phosphorylation in the absence of exogenous DNA damage, suggesting that high levels of Tel1 might titrate out factors interacting with Mec1 also in the absence of external insults, thus leading to unregulated and signal-independent checkpoint activation.
DNA damage exacerbates all the phenotypes caused by TEL1 overexpression in the absence of exogenous damage. In fact, Tel1-overproducing cells undergo a dramatic delay in S-phase progression and persistent Rad53 phosphorylation after DNA damage in G1, suggesting that checkpoint activation by an excess of Tel1 in the absence of external insults is not maximal and can be increased by damaging the DNA. This increase is also thought to impair the ability of the cell to recover from the checkpoint-mediated arrest, since Tel1-overproducing cells are hypersensitive to genotoxic agents. Similarly to the results observed in the absence of exogenous DNA damage, Ddc1 and Mec1 are not required for the DNA damage-induced cell cycle arrest in Tel1-overproducing cells, which instead requires Rad53 and Rad9. Moreover, TEL1 overexpression can partially suppress the hypersensitivity to genotoxic agents of mec1 and ddc1 mutants, while it has no effect on that of rad53 and rad9 mutants. Accordingly, recent data have implicated Tel1 together with the Mre11 complex in triggering Rad53 activation and its interaction with Rad9 in response to DNA damage independently of Mec1 and Rad24 (Usui et al., 2001).
High amounts of Ddc2 impair the ability of the cell to recover from DNA damage-induced cell cycle arrest
Neither MEC1 nor DDC2 overexpression alters cell cycle progression or induces Rad53 phosphorylation in the absence of exogenous DNA damage, suggesting that activation by DNA structure signals might be essential for these proteins to function in the checkpoint. In contrast, Tel1 overproduction turns on the checkpoint response independently of external damage and presumed damage sensors, but requires other components of the pathway that function in the signal transduction cascade. Therefore, Tel1 escapes the normal rigorous regulation imposed upon Mec1 and Ddc2, which appear to be normally responsible for the checkpoint response. Moreover, UV-irradiated Ddc2-overproducing cells undergo a prolonged arrest at the G2/M transition, and this correlates with persistent Rad53 phosphorylation and cell death. Since these effects of Ddc2 overproduction are seen only in the presence of Mec1, it is tempting to suggest that high levels of Mec1 per se do not cause any detectable phenotype under the same conditions because Ddc2 may be rate limiting for its activation. Moreover, the effects of Ddc2 overproduction are limited to the DNA damage checkpoint controlling the G2/M transition. In fact, the same excess of Ddc2 causing DNA damage-induced arrest of nuclear division is not sufficient to stop S-phase progression after DNA damage in G1, suggesting that the checkpoint responses in different cell cycle phases might have different requirements for Mec1–Ddc2 activity. This hypothesis is further supported by the finding that overexpression of MEC1 in Ddc2-overproducing cells causes a further slowing down of S-phase progression after DNA damage in G1 compared with wild-type cells. Thus, Mec1 activity becomes rate limiting for hyperactivation of the intra-S checkpoint despite the excess of Ddc2, while this does not appear to be the case for the G2/M transition arrest. Accordingly, we previously showed that overproduction of Mec1 kinase-deficient variants has dominant-negative effects on S-phase entry and progression after DNA damage in G1, but it does not abrogate the G2/M checkpoint (Paciotti et al., 2001). In addition, we described two hypomorphic mec1 mutants that were completely defective in the G1/S and intra-S DNA damage checkpoints, but they were able to activate the G2/M checkpoint (Paciotti et al., 2001). Altogether these data strengthen the hypothesis that the threshold level of Mec1 activity required to slow down replication of a damaged DNA template is higher than that required to block the G2/M transition in the presence of damaged chromosomes.
The finding that both Ddc1 and Rad9 are required for the prolonged DNA damage-induced G2/M arrest caused by DDC2 overexpression indicates that this arrest involves the activation of the canonical DNA damage checkpoint pathway, and that the ability to turn it off is impaired in Ddc2-overproducing cells. The requirement of Ddc1 for the Ddc2-dependent, but not for the Tel1-dependent, cell cycle arrest indicates that Ddc1 is necessary to activate the Ddc2–Mec1 complex in the presence of DNA damage despite the high Ddc2 levels, thus specifically linking DNA damage sensing with Ddc2-mediated hyperactivation of the checkpoint pathway. The hypersensitivity to genotoxic agents of DDC2-overexpressing cells may then be the consequence of their inability to recover from the DNA damage-induced cell cycle arrest.
In summary, although with different features, overproduction of either Tel1 or Ddc2 results in prolonged delay of cell cycle progression, indicating that unbalancing the levels of checkpoint kinases or kinase-interacting factors may hyperactivate the checkpoint. In this view, other proteins important for dephosphorylating Mec1– Ddc2, Tel1 or Rad53 itself may then become rate limiting for recovery from the checkpoint-mediated cell cycle arrest in the presence of unregulated kinase levels. Finally, our finding that the cell cycle arrest caused by Ddc2 and Tel1 overproduction requires Rad53 and correlates with its enhanced and persistent phosphorylation strengthens the hypothesis that unscheduled Rad53 phosphorylation may prevent cells from re-entering the cell cycle after a checkpoint-mediated cell cycle arrest. A search for multicopy suppressors of the cell cycle arrest caused by Tel1 and Ddc2 overproduction may allow the identification of proteins specifically required for termination of the checkpoint response.
Materials and methods
Plasmids, yeast strains and media
The genotypes of all the yeast strains used in this study are listed in Table I. All yeast strains were derivatives of W303 (MATa or MATα, ade2-1, can1-100, trp1-1, leu2-3,112, his3-11,15, ura3).
Table I. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Reference/source |
|---|---|---|
| K699 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 | Longhese et al. (1997) |
| K700 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 | Longhese et al. (1997) |
| YLL157 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad9Δ::URA3 | Longhese et al. (1996) |
| YLL244 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc1Δ::KanMX4 | Longhese et al. (1997) |
| YLL275 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc2Δ::KanMX4 [pML94 URA3 CEN4 DDC2] | Paciotti et al. (2000) |
| YLL279.2 | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1–DDC2::LEU2 | this study |
| YLL490 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1Δ::HIS3 sml1Δ::KanMX4 | Longhese et al. (2000) |
| YLL516 | MATa ade2-1 can1-100 his3-11,15 trp1-1 leu2-3 112 ura3::GAL1–MEC1::URA3 | Paciotti et al. (2001) |
| YLL509 | MATa ade2-1 can1-100 his3-11,15 leu2-3 112 trp1-1 ura3 rad53Δ::HIS3 sml1Δ::KanMX4 | Longhese et al. (2000) |
| YLL826 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 [pML225 URA3 CEN4 GAL1–MEC1] | this study |
| YLL827 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 [YCplac33 URA3 CEN4] | this study |
| YLL836 | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1–DDC2::LEU2 [pML225 URA3 CEN4 GAL1–MEC1] | this study |
| YLL837 | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1–DDC2::LEU2 [YCplac33 URA3 CEN4] | this study |
| YLL848 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 TEL1-HA3::URA3::tel1 | this study |
| YLL930 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc2Δ::KanMX4 [pML94 URA3 CEN4 DDC2] [pML240 LEU2 CEN4 GAL1–MEC1] | this study |
| YLL932 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 GAL1–TEL1-HA3::LEU2::tel1 | this study |
| YLL943 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 GAL1–TEL1::LEU2::tel1 [pML94 URA3 CEN4 DDC2] | this study |
| YLL944 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 [pML94 URA3 CEN4 DDC2] [pML240 LEU2 CEN4 GAL1–MEC1] | this study |
| DMP2995/1B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc2Δ::KanMX4 sml1Δ::KanMX4 | Paciotti et al. (2000) |
| DMP3356/4A | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1–DDC2::LEU2 rad53Δ::HIS3 sml1Δ::KanMX4 | this study |
| DMP3392/4A | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1–DDC2::LEU2 rad9Δ::URA3 | this study |
| DMP3462/2B | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1–DDC2::LEU2 mec1Δ::HIS3 sml1Δ::KanMX4 | this study |
| DMP3463/10C | MATa ade2-1 can1-100 his3-11,15 trp1-1 ura3 leu2-3,112::GAL1–DDC2::LEU2 ddc1Δ::KanMX4 | this study |
| DMP3475/3D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc2Δ::KanMX4 GAL1–TEL1::LEU2::tel1 [pML94 URA3 CEN4 DDC2] | this study |
| DMP3479/2A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad53K227A::KanMX4 | this study |
| DMP3479/2B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad53K227A::KanMX4 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3532/8A | MATa ade2-1 can1-100 his3-11,15 trp1-1 leu2-3,112 ura3::GAL1–MEC1::URA3 ddc2Δ::KanMX4 sml1Δ::KanMX4 | this study |
| DMP3539/9D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1–MEC1::URA3 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3539/10D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3562/2A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1Δ::HIS3 sml1Δ::KanMX4 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3575/4A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ddc1Δ::KanMX4 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3575/6B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad9Δ::URA3 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3598/6A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 DDC1-HA2::LEU2::ddc1 CHK1-MYC18::HIS3 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3598/21D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 DDC1-HA2::LEU2::ddc1 CHK1-MYC18::HIS3 | this study |
| DMP3602/9C | MATa ade2-1 can1-100 his3-11,15 trp1-1 leu2-3,112 ura3 GAL1–TEL1::URA3::tel1 ddc2Δ::KanMX4 sml1Δ::KanMX4 | this study |
| DMP3611/3B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad53K227A::KanMX4 chk1Δ::HIS3 GAL1–TEL1::LEU2::tel1 | this study |
| DMP3611/6D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 chk1Δ::HIS3 GAL1–TEL1::LEU2::tel1 | this study |
Plasmids are indicated in square brackets.
To obtain plasmid pML284.36, containing a TEL1 DNA fragment spanning from the ATG (+1) to position +1476 fused to the GAL1 promoter, the 1476 bp TEL1 fragment was amplified by PCR using yeast genomic DNA as a template, the oligonucleotides PRP222 (5′-CGCGGATCCATATGGAGGATCATGGGATTGTAGAAACT-3′) and PRP223 (5′-GCTCTAGAGTAAGCAAACTTTAAATAGGCGTGCCA-3′) as primers, and Pfu polymerase (Stratagene). The PCR amplification product was then cloned into the BamHI–XbaI sites of plasmid pML95 (Longhese et al., 1997) and controlled by DNA sequence analysis. To construct plasmid pML285, used to generate the TEL1-HA3 allele, a TEL1 fragment spanning from position –1000 to position +900 from the translation initiation codon was amplified by PCR using genomic DNA as template and oligonucleotides PRP213 (5′-GGAATTCAATCATACACGGCAAGCATA-3′) and PRP214 (5′-GGAATTCCTTAAGCGCAGTAGAATGTA-3′) as primers, and then cloned into the EcoRI site of plasmid YIplac211 (Gietz and Sugino, 1988). A NotI restriction site was then introduced by PCR after the TEL1 codon 2 and sequences encoding three tandem hemagglutinin (HA3) epitopes were inserted into it, thus giving rise to plasmid pML285. Plasmid pML322, used to generate the GAL1–TEL1-HA3 allele, was originated by inserting the 1012 bp HindIII TEL1 fragment from pML285 into the HindIII site of plasmid pML284.
Strain YLL850.2, carrying one copy of the GAL1–TEL1 fusion integrated at the TEL1 locus, was obtained by transforming strain K699 with BlpI-digested plasmid pML284.36, and then crossed with strain DMP2760/1A (Longhese et al., 2000) to obtain the meiotic segregants DMP3479/2A, DMP3479/2B and DMP3479/1D. Strains YLL848 and YLL932, carrying one copy of the TEL1-HA3 or GAL1–TEL1-HA3 alleles at the TEL1 locus, were obtained by transforming strain K699 with MfeI-digested plasmid pML285 and BlpI-digested plasmid pML322, respectively. The kinetics of cell cycle progression of these strains were indistinguishable from those of strains K699 and DMP3539/10D, respectively, under both uninduced and galactose-induced conditions. To generate the CHK1 chromosomal deletion, a chk1Δ::HIS3 cassette was constructed by PCR using pFA6a-HIS3 plasmid (Wach et al., 1994) as template, and oligonucleotides PRP190 (5′-TATCATAAGTTGCTGTATATGGGCAGCACGTATTACTATGAGTCTCGTACGCTGCAGGTCGAC-3′) and PRP191 (5′-TGTCTCCATTTTTTTCAGTTG GGAATTAGGATAATATCCCTACAGATAGTATCGATGAATTCGAGCTCG-3′) as primers, followed by transformation of strain K699 with the PCR product, giving rise to strain YLL736, where the 1540 bp of the CHK1 coding region were replaced by the Kluyveromyces lactis HIS3 gene. Strains DMP3611/3B and DMP3611/6D were meiotic segregants from a cross between strains YLL736 and DMP3479/1D. Strain YLL850.2 was also crossed with strain K700, and the derivative DMP3469/2B meiotic segregant was then crossed to strain YLL516 to obtain the DMP3539/9D and DMP3539/10D meiotic segregants. Strain DMP3469/2B was also crossed with strains YLL490 (Longhese et al., 2000), YLL244 (Longhese et al., 1997) and YLL157 (Longhese et al., 1996) to obtain the DMP3562/2A, DMP3575/4A and DMP3575/6B meiotic segregants, respectively. Strain YLL922, carrying the CHK1-MYC18 allele at the CHK1 chromosomal locus, was generated by the PCR one-step tagging method (Knop et al., 1999) using, respectively, plasmid 3746 (K.Nasmyth; IMP, Vienna) as the template and oligonucleotides PRP217 (5′-CTTTAGAATGGAGAAGATTGTTCAAGAAAATTTCAACTATCTGTAGGGATATTATCCTAATTCCCAACTCCGGTTCTGCTGCTAG-3′) and PRP218 (5′-ATAAGTAGA AAGAATTTTTTTTTTTTTTTGATCAGTGCATCTTAACCCTTCTTTTGTCTCCATTTTTTCCTCGAGGCCAGAAGAC-3′) as primers. Since CHK1 alterations do not cause obvious phenotypes, but impair Pds1 phosphorylation (Sanchez et al., 1999), we verified that DNA damage-induced Pds1 phosphorylation was unaffected in CHK1-MYC18 cells. Strains DMP3598/6A and DMP3598/21D were derived from crossing strains YLL922 and DMP3548/3B, which was derived from a cross between strains DMP3198/1A (Paciotti et al., 2000) and DMP3469/2B.
Strain YLL279.2, carrying one copy of the GAL1–DDC2 fusion integrated at the LEU2 locus, was obtained by transforming strain K699 with ClaI-digested plasmid pML105 (Paciotti et al., 2000). Strains YLL826 and YLL827 were obtained by transforming strain K699 with plasmids pML225 (Longhese et al., 2000) and YCplac33 (Gietz and Sugino, 1988), respectively. Strains YLL836 and YLL837 were obtained by transforming strain YLL279.2 with plasmids pML225 and YCplac33, respectively. Strain YLL279.2 was also crossed with strains DMP2855/6B, DMP2946/2C, DMP2946/3A and DMP2854/2B to generate the DMP3356/4A, DMP3392/4A, DMP3463/10C and DMP3462/2B meiotic segregants, respectively. Strains DMP2855/6B, DMP2946/2C, DMP2946/3A and DMP2854/2B were, respectively, meiotic segregants from the crosses YLL509 (Longhese et al., 2000)/K700, YLL157/K700, YLL244/K700 and YLL490/K700. Strain DMP3475/3D was a meiotic segregant from a cross between strains YLL275 (Paciotti et al., 2000) and DMP3469/1A, which was derived from a cross between strains DMP3539/10D and K700. Strain YLL930 was obtained by transformation of strain YLL275 with plasmid pML240 (Longhese et al., 2000). Strain YLL943 was obtained by transformation of strain 3539/10D with plasmid pML94 (Paciotti et al., 2000). Strain YLL944 was obtained by transformation of strain K699 with plasmids pML240 (Longhese et al., 2000) and pML94 (Paciotti et al., 2000). Strain DMP3532/8A was a meiotic segregant from a cross of strains YLL516 and DMP2995/6C, which was derived from a cross between strains DMP2995/1B (Paciotti et al., 2000) and K700. Strain DMP3602/9C was derived from the cross between strains YLL2995/6C and DMP3539/10D.
The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR. Standard yeast genetic techniques and media were according to Rose et al. (1990). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, 50 mg/l adenine) supplemented with 2% glucose (YEPD) or 2% raffinose (YEP-raf) or 2% raffinose and 1% galactose (YEP-raf-gal). Transformants carrying the KanMX4 cassette were selected on YEPD plates containing 400 µg/ml G418 (US Biological).
Evaluation of endogenous and galactose-induced Ddc2, Tel1 and Mec1 protein levels
To compare Ddc2, Tel1 and Mec1 protein levels expressed from the GAL1 promoter under galactose-induced conditions to the endogenous levels of the same proteins, isogenic W303 derivative strains expressing tagged DDC2-HA3 (Paciotti et al., 2000), MEC1-HA9 (Paciotti et al., 2000) and TEL1-HA3 alleles (Table I) either from their own promoters or from the GAL1 promoter were grown in raffinose-containing medium and then shifted to raf-gal-medium for 4 h. The same amounts of protein extracts prepared from the above cultures were loaded onto an SDS–polyacrylamide gel and subjected to western blot analysis with anti-HA antibodies. Films were then scanned and bands quantified with NIH Image. By this analysis, the levels of Mec1, Tel1 and Ddc2 produced under galactose-induced conditions by the strains carrying the GAL1 constructs were ∼20-, 20- and 25-fold higher, respectively, than the corresponding endogenous levels.
Other techniques
Synchronization experiments, total protein extract preparation and western blot analysis were performed as described in Paciotti et al. (2000).
Acknowledgments
Acknowledgements
We wish to thank Francisca Lottersberger for construction of the CHK1-MYC tagged allele, N.Lowndes for antibodies against Rad9, C.Santocanale and J.Diffley for antibodies against Rad53, S.Piatti for critical reading of the manuscript, and all the members of our laboratory for useful discussions and criticisms. This work was supported by grants from Telethon–Italy (E.1247) and Associazione Italiana per la Ricerca sul Cancro to M.P.L., from Associazione Italiana per la Ricerca sul Cancro and Cofinanziamento 1999 MURST-Università di Milano-Bicocca to G.L. and from CNR Target Project on Biotechnology Grant CT.97.01180.PF49(F).
References
- Bentley N.J., Holtzman,D.A., Flaggs,G., Keegan,K.S., Demaggio,A., Ford,J.C., Hoekstra,M. and Carr,A.M. (1996) The Schizosaccharo myces pombe rad3 checkpoint gene. EMBO J., 15, 6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Caspari T., Dahlen,M., Kanter-Smoler,G., Lindsay,H.D., Hofman,K., Papadimitriou,K., Sunnerhagen,P. and Carr,A.M. (2000) Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol., 20, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A., Domkin,V. and Thelander,L. (1999) Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem., 274, 36679–36683. [DOI] [PubMed] [Google Scholar]
- D’Amours D. and Jackson,S.P. (2001) The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev., 15, 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B.A., Alcasabas,A.A., Bachant,J.B. and Elledge,S.J. (1998) Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev., 12, 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A. (1998) MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell, 2, 183–189. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base-pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gilbert C.S., Green,C.M. and Lowndes,N.F. (2001) Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell, 8, 129–136. [DOI] [PubMed] [Google Scholar]
- Green C.M., Erdjument-Bromage,H., Tempst,P. and Lowndes,N.F. (2000) A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol., 10, 39–42. [DOI] [PubMed] [Google Scholar]
- Greenwell P.W., Kronmal,S.L., Porter,S.E., Gassenhuber,J., Obermaier,B. and Petes,T.D. (1995) TEL1, a gene involved in controlling telomere length in S.cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell, 82, 823–829. [DOI] [PubMed] [Google Scholar]
- Grenon M., Gilbert,C.S. and Lowndes,N.F. (2001) Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nature Cell Biol., 3, 844–847. [DOI] [PubMed] [Google Scholar]
- Griffiths D.J., Barbet,N.C., McCready,S., Lehmann,A.R. and Carr,A.M. (1995) Fission yeast RAD17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J., 14, 5812–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari K.L., Santerre,A., Sekelsky,J.J., McKim,K.S., Boyd,J.B. and Hawley,R.S. (1995) The mei-41 gene of D.melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell, 82, 815–821. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Kondo T., Matsumoto,K. and Sugimoto,K. (1999) Role of a complex containing Rad17, Mec3 and Ddc1 in the yeast DNA damage checkpoint pathway. Mol. Cell. Biol., 19, 1136–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrub C.F., Knudsen,K., Subramani,S. and Enoch,T. (1998) Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J., 17, 2055–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M.P., Fraschini,R., Plevani,P. and Lucchini,G. (1996) Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage and they define a functional link between Mec3 and DNA primase. Mol. Cell. Biol., 16, 3235–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M.P., Paciotti,V., Fraschini,R., Zaccarini,R., Plevani,P. and Lucchini,G. (1997) The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J., 16, 5216–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M.P., Foiani,M., Muzi Falconi,M., Lucchini,G. and Plevani,P. (1998) DNA damage checkpoint in budding yeast. EMBO J., 17, 5525–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M.P., Paciotti,V., Neecke,H. and Lucchini,G. (2000) Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics, 155, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes N.F. and Murguia,J.R. (2000) Sensing and responding to DNA damage. Curr. Opin. Genes Dev., 10, 17–25. [DOI] [PubMed] [Google Scholar]
- Lydall D. and Weinert,T. (1997) G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol. Gen. Genet., 256, 638–651. [DOI] [PubMed] [Google Scholar]
- Mallory J.C. and Petes,T.D. (2000) Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl Acad. Sci. USA, 97, 13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow D.M., Tagle,D.A., Shiloh,Y., Collins,F.S. and Hieter,P. (1995) TEL1, an S.cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell, 82, 831–840. [DOI] [PubMed] [Google Scholar]
- O’Connell M.J., Walworth,N.C. and Carr,A.M. (2000) The G2-phase DNA-damage checkpoint. Trends Cell Biol., 10, 296–303. [DOI] [PubMed] [Google Scholar]
- Paciotti V., Lucchini,G., Plevani,P. and Longhese,M.P. (1998) Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J., 17, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V., Clerici,M., Lucchini,G. and Longhese,M.P. (2000) The checkpoint protein Ddc2, functionally related to S.pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev., 14, 2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Paciotti V., Clerici,M., Scotti,M., Lucchini,G. and Longhese,M.P. (2001) Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol., 21, 3913–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A.G., Margulies,R.U., Garvik,B.M. and Hartwell,L.H. (1997) RAD9, RAD17 and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics, 145, 45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K.B., Mallory,J.C. and Petes,T.D. (1999) Interactions of TLC1 (which encodes the RNA sub-unit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rouse J. and Jackson,S.P. (2000) LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signaling pathway in Saccharomyces cerevisiae. EMBO J., 19, 5801–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Desany,B.A., Jones,W.J., Liu,Q., Wang,B. and Elledge,S.J. (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science, 271, 357–360. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Bachant,J., Wang,H., Hu,F.H., Liu,D., Tezlaff,M. and Elledge,S.J. (1999) Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science, 286, 1166–1171. [DOI] [PubMed] [Google Scholar]
- Savitsky K. et al. (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science, 286, 1749–1753. [DOI] [PubMed] [Google Scholar]
- Siede W., Friedberg,A.S. and Friedberg,E.C. (1993) RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 7985–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge R.P., Udell,C.M., Casselman,R. and Davey,S. (1999) The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol. Biol. Cell, 10, 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Fay,D.S., Marini,F., Foiani,M. and Stern,D.F. (1996) Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev., 10, 395–406. [DOI] [PubMed] [Google Scholar]
- Sun Z., Hsiao,J., Fay,D.S. and Stern,D.F. (1998) Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science, 281, 272–274. [DOI] [PubMed] [Google Scholar]
- Thelen M.P., Venclovas,C. and Fidelis,K. (1999) A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell, 96, 769–770. [DOI] [PubMed] [Google Scholar]
- Usui T., Ogawa,H. and Petrini,J. (2001) A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell, 7, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Vialard J.E., Gilbert,C.S., Green,C.M. and Lowndes,N.F. (1998) The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J., 17, 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Waga S. and Stillman,B. (1998) The DNA replication forks in eukaryotic cells. Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- Wakayama T., Kondo,T., Ando,S., Matsumoto,K. and Sugimoto,K. (2001) Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. (1998) DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell, 94, 555–558. [DOI] [PubMed] [Google Scholar]
- Weinert T.A. and Hartwell,L.H. (1988) The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science, 241, 317–322. [DOI] [PubMed] [Google Scholar]
- Weinert T.A., Kiser,G.L. and Hartwell,L.H. (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev., 8, 652–665. [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller,E.G.D. and Rothstein,R. (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell, 2, 329–340. [DOI] [PubMed] [Google Scholar]
- Zhao X., Chabes,A., Domkin,V., Thelander,L. and Rothstein,R. (2001) The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J., 20, 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge,S.J. (2000) The DNA damage response: putting checkpoints in perspective. Nature, 408, 433–439. [DOI] [PubMed] [Google Scholar]